Comprehensive Bioinformatics Analysis Combined with Wet-Lab Experiments to Find Target Proteins of Chinese Medicine Monomer
Abstract
:1. Introduction
2. Results
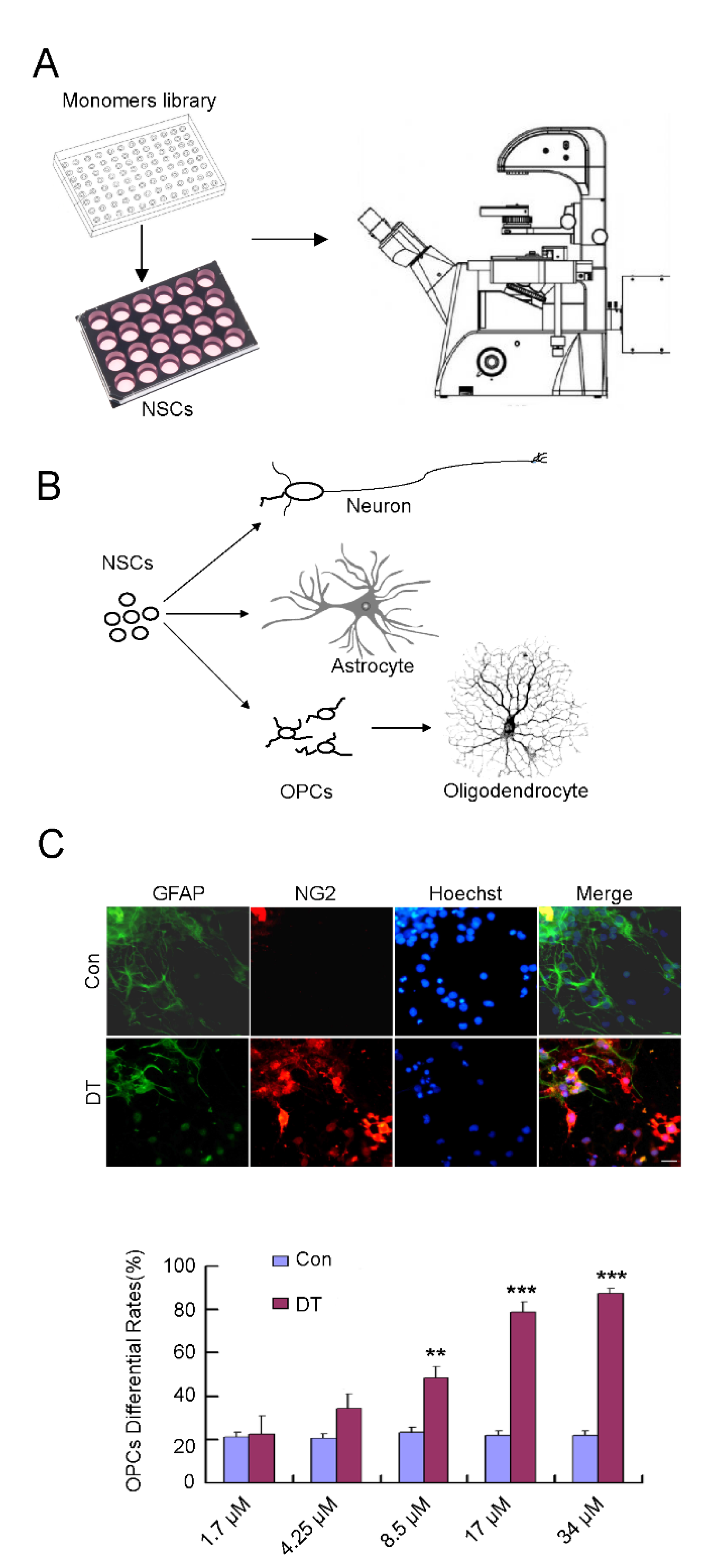
2.1. DT Promotes the Generation of OPCs from NSCs
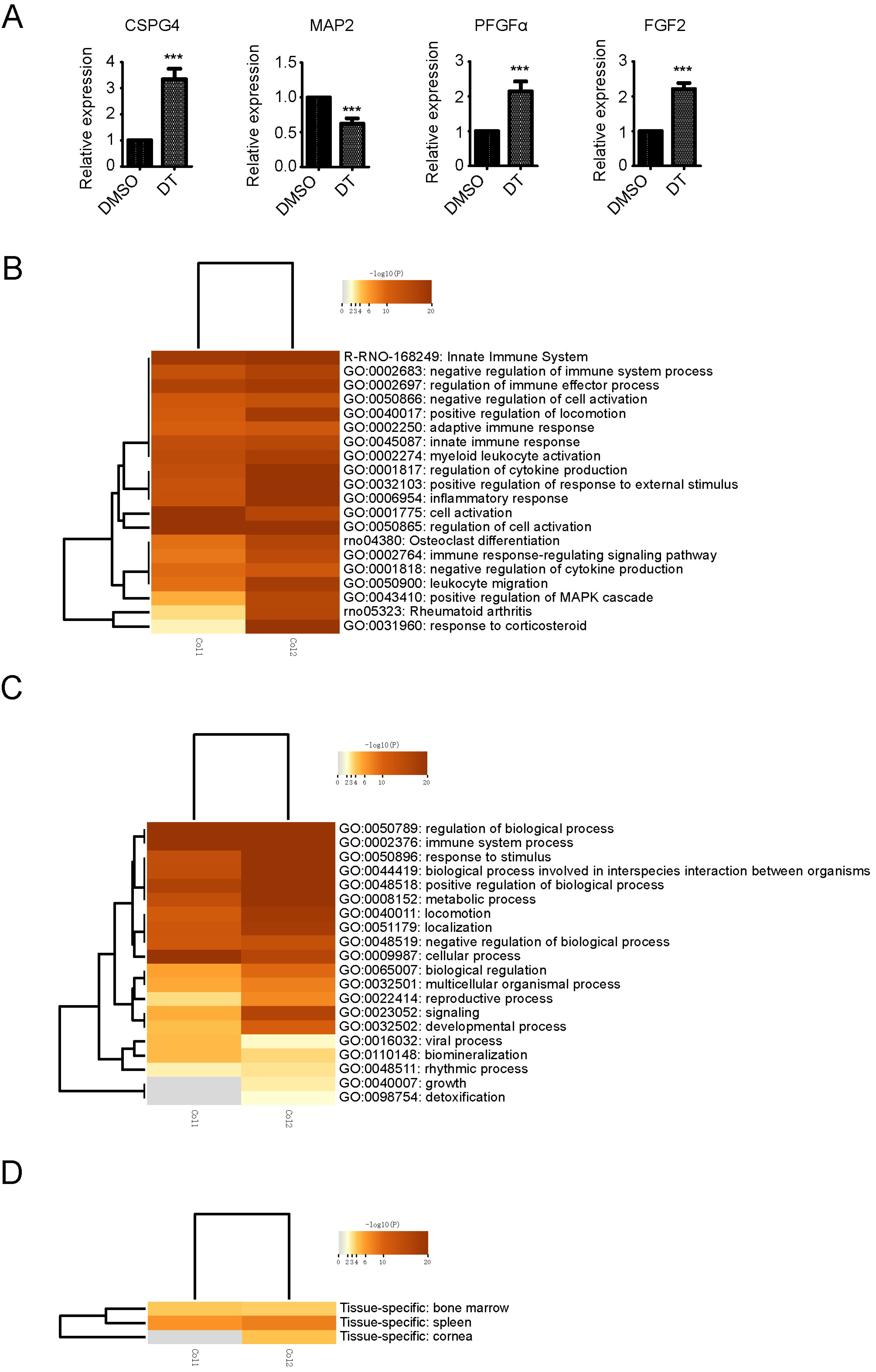
2.2. Transcriptome Analysis in DT-Induced NSCs Differentiation to OPCs
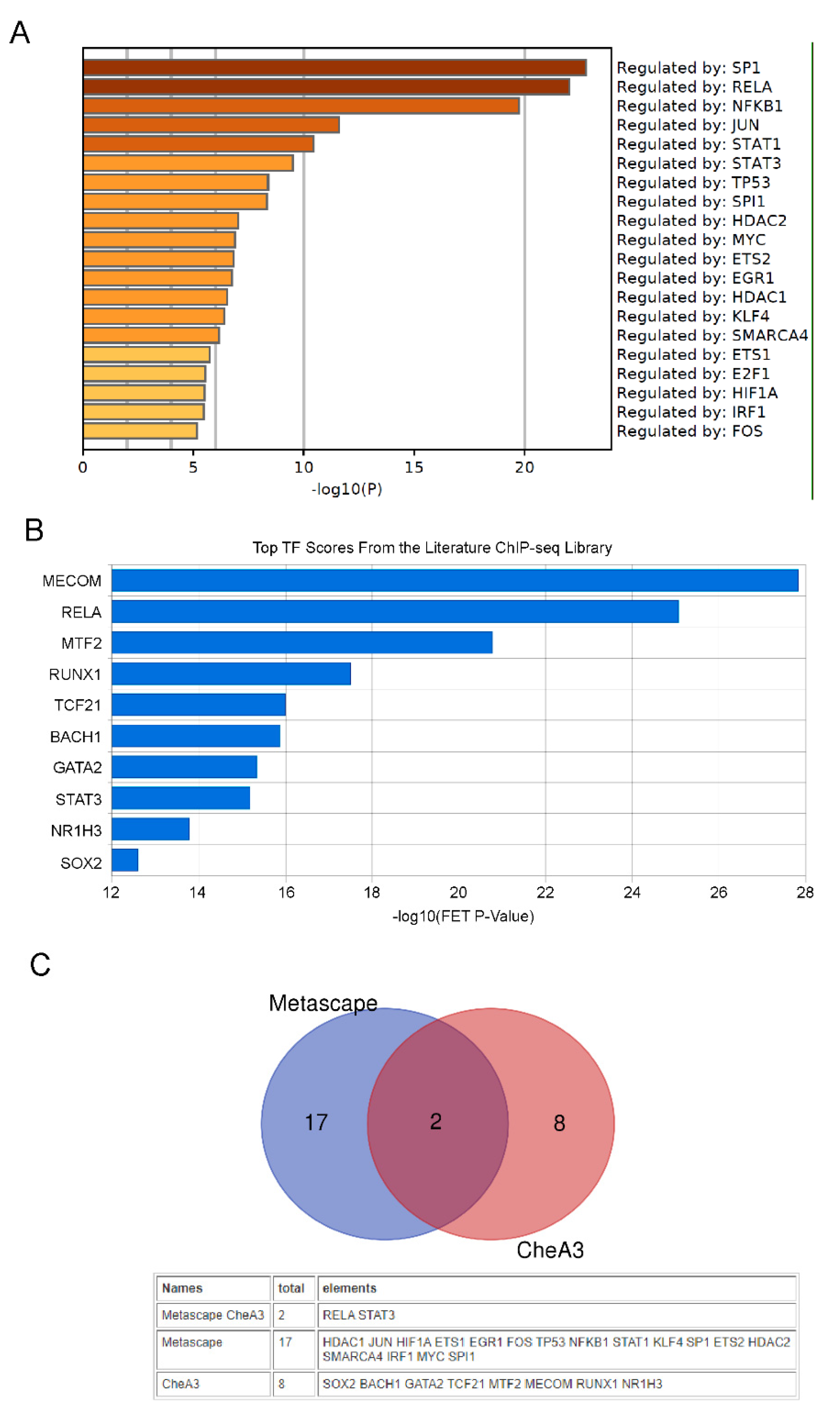
2.3. Transcription Factor Analysis
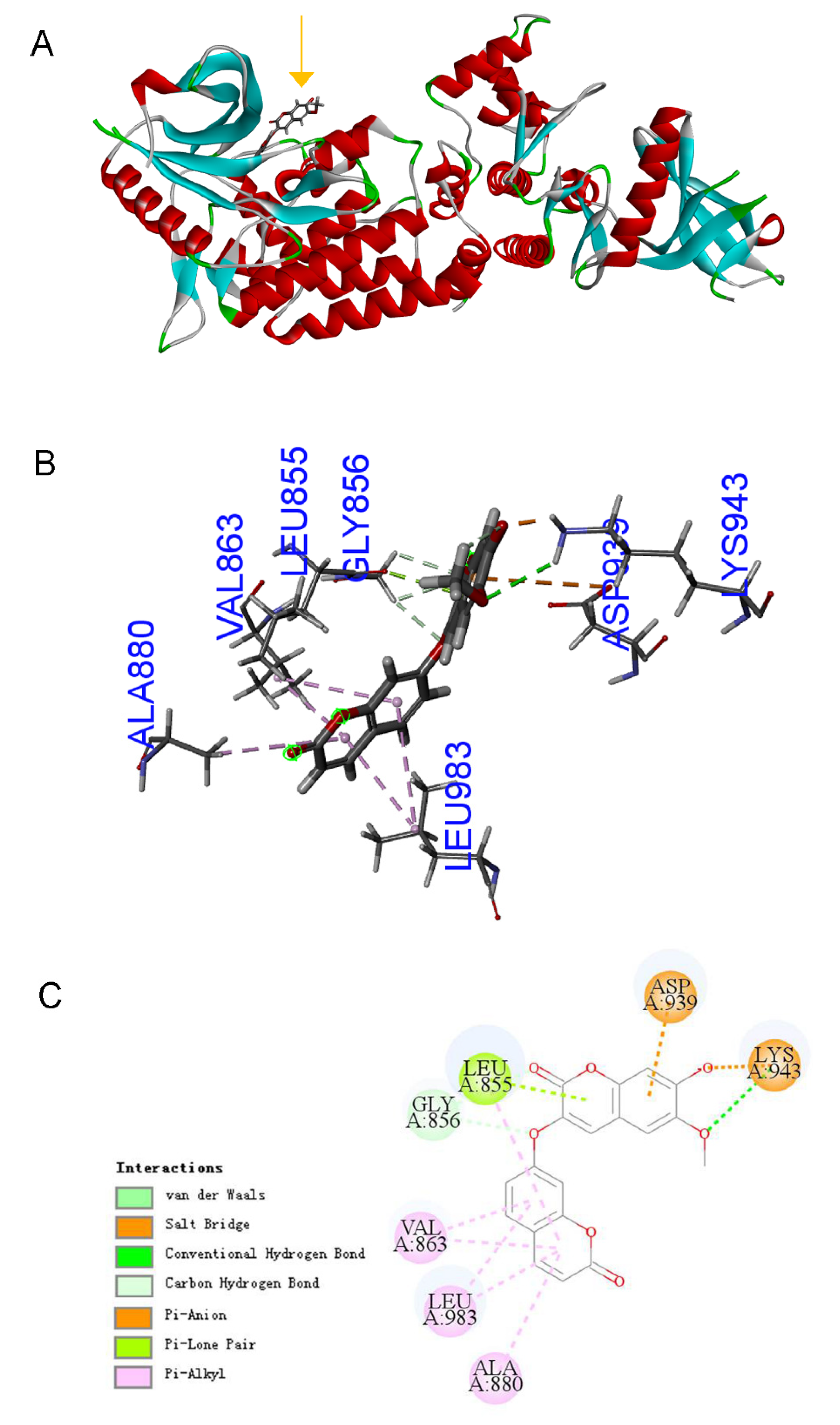
2.4. Bioinformatics Predicts That DT Is a JAK2 Agonist
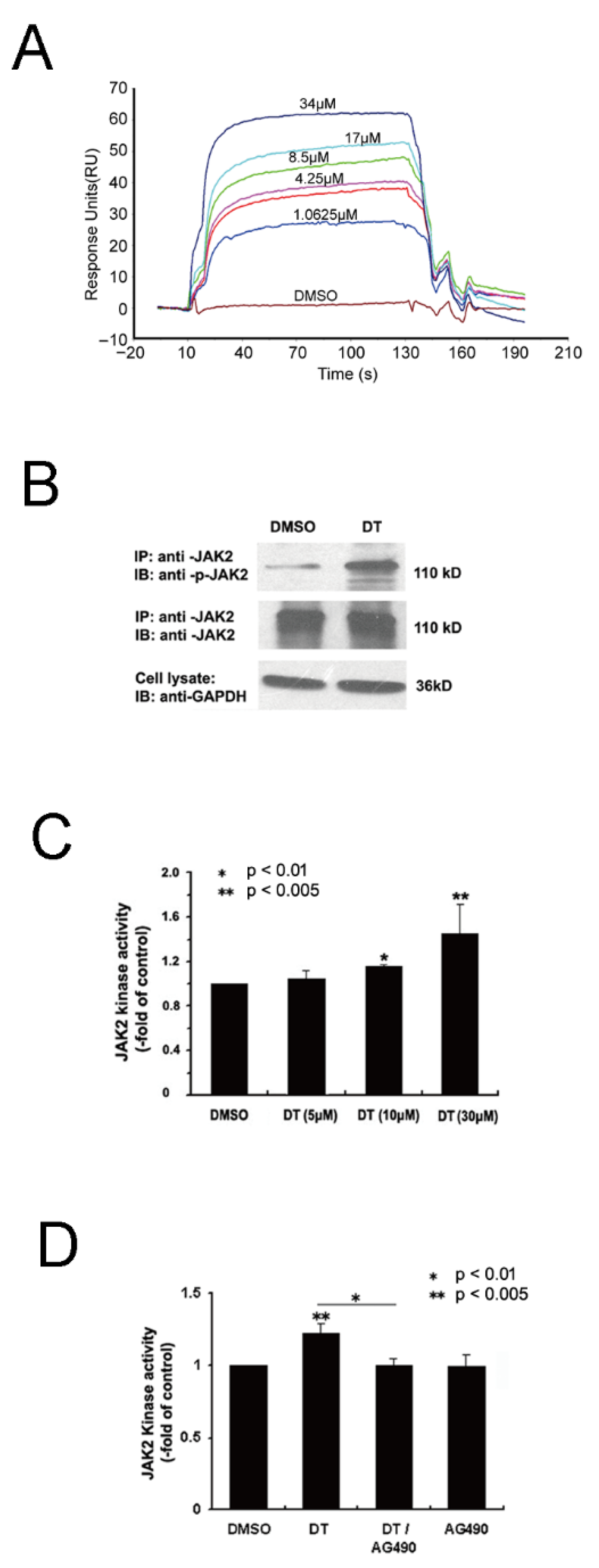
2.5. Wet-Lab Experiments Prove That DT Is a JAK2 Agonist
3. Discussion
4. Materials and Methods
4.1. NSC Culture
4.2. Traditional Chinese Medicine Monomers Library
4.3. Compound Screens
4.4. Immunofluorescence
4.5. High-Throughput RNA Sequencing
4.6. Bioinformatics Analysis and Target Prediction
4.7. Inverse Virtual Screening
4.8. Immunoprecipitation and Immunoblotting
4.9. Determination of Binding Affinity of DT to JAK2 Using Splasmon Resonance Techniques
4.10. JAK2 In Vitro Kinase Activity Assay
4.11. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
References
- Reich, D.S.; Lucchinetti, C.F.; Calabresi, P.A. Multiple Sclerosis. N. Engl. J. Med. 2018, 378, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Disease, G.B.D.; Injury, I.; Prevalence, C. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar]
- Chang, A.; Tourtellotte, W.W.; Rudick, R.; Trapp, B.D. Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N. Engl. J. Med. 2002, 346, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Nicaise, A.M.; Wagstaff, L.J.; Willis, C.M.; Paisie, C.; Chandok, H.; Robson, P.; Fossati, V.; Williams, A.; Crocker, S.J. Cellular senescence in progenitor cells contributes to diminished remyelination potential in progressive multiple sclerosis. Proc. Natl. Acad. Sci. USA 2019, 116, 9030–9039. [Google Scholar] [CrossRef]
- Spitzer, S.O.; Sitnikov, S.; Kamen, Y.; Evans, K.A.; Kronenberg-Versteeg, D.; Dietmann, S.; de Faria, O., Jr.; Agathou, S.; Karadottir, R.T. Oligodendrocyte Progenitor Cells Become Regionally Diverse and Heterogeneous with Age. Neuron 2019, 101, 459–471 e5. [Google Scholar] [CrossRef]
- Bouab, M.; Paliouras, G.N.; Aumont, A.; Forest-Berard, K.; Fernandes, K.J. Aging of the subventricular zone neural stem cell niche: Evidence for quiescence-associated changes between early and mid-adulthood. Neuroscience 2011, 173, 135–149. [Google Scholar] [CrossRef]
- Beyer, F.; Samper Agrelo, I.; Kury, P. Do Neural Stem Cells Have a Choice? Heterogenic Outcome of Cell Fate Acquisition in Different Injury Models. Int. J. Mol. Sci. 2019, 20, 455. [Google Scholar] [CrossRef]
- Biswas, S.; Chung, S.H.; Jiang, P.; Dehghan, S.; Deng, W. Development of glial restricted human neural stem cells for oligodendrocyte differentiation in vitro and in vivo. Sci. Rep. 2019, 9, 9013. [Google Scholar] [CrossRef]
- Wang, J.; Pol, S.U.; Haberman, A.K.; Wang, C.; O′Bara, M.A.; Sim, F.J. Transcription factor induction of human oligodendrocyte progenitor fate and differentiation. Proc. Natl. Acad. Sci. USA 2014, 111, E2885–E2894. [Google Scholar] [CrossRef]
- Copray, S.; Balasubramaniyan, V.; Levenga, J.; de Bruijn, J.; Liem, R.; Boddeke, E. Olig2 overexpression induces the in vitro differentiation of neural stem cells into mature oligodendrocytes. Stem Cells 2006, 24, 1001–1010. [Google Scholar] [CrossRef]
- Marei, H.E.; Shouman, Z.; Althani, A.; Afifi, N.; Lashen, S.; Hasan, A.; Caceci, T.; Rizzi, R.; Cenciarelli, C.; Casalbore, P. Differentiation of human olfactory bulb-derived neural stem cells toward oligodendrocyte. J. Cell. Physiol. 2018, 233, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Spencer, D.; Yu, D.; Morshed, R.A.; Li, G.; Pituch, K.C.; Gao, D.X.; Bertolino, N.; Procissi, D.; Lesniak, M.S.; Balyasnikova, I.V. Pharmacologic modulation of nasal epithelium augments neural stem cell targeting of glioblastoma. Theranostics 2019, 9, 2071–2083. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Tang, X.; Bai, Y.F.; Wang, S.; An, J.; Wu, Y.; Xu, Z.D.; Zhang, Y.A.; Chen, Z. Dopaminergic precursors differentiated from human blood-derived induced neural stem cells improve symptoms of a mouse Parkinson′s disease model. Theranostics 2018, 8, 4679–4694. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Keenan, A.B.; Torre, D.; Lachmann, A.; Leong, A.K.; Wojciechowicz, M.L.; Utti, V.; Jagodnik, K.M.; Kropiwnicki, E.; Wang, Z.; Ma′ayan, A. ChEA3: Transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019, 47, W212–W224. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.M. Nonequivalent response to bromodomain-targeting BET inhibitors in oligodendrocyte cell fate decision. Chem. Biol. 2014, 21, 804–806. [Google Scholar] [CrossRef] [PubMed]
- Ishii, A.; Fyffe-Maricich, S.L.; Furusho, M.; Miller, R.H.; Bansal, R. ERK1/ERK2 MAPK signaling is required to increase myelin thickness independent of oligodendrocyte differentiation and initiation of myelination. J. Neurosci. Off. J. Soc. Neurosci. 2012, 32, 8855–8864. [Google Scholar] [CrossRef]
- Zhang, P.; Chebath, J.; Lonai, P.; Revel, M. Enhancement of oligodendrocyte differentiation from murine embryonic stem cells by an activator of gp130 signaling. Stem Cells 2004, 22, 344–354. [Google Scholar] [CrossRef]
- Yu, S.; Guo, H.; Gao, X.; Li, M.; Bian, H. Daphnoretin: An invasion inhibitor and apoptosis accelerator for colon cancer cells by regulating the Akt signal pathway. Biomed. Pharmacother. 2019, 111, 1013–1021. [Google Scholar] [CrossRef]
- Chen, C.A.; Liu, C.K.; Hsu, M.L.; Chi, C.W.; Ko, C.C.; Chen, J.S.; Lai, C.T.; Chang, H.H.; Lee, T.Y.; Lai, Y.L.; et al. Daphnoretin modulates differentiation and maturation of human dendritic cells through down-regulation of c-Jun N-terminal kinase. Int. Immunopharmacol. 2017, 51, 25–30. [Google Scholar] [CrossRef]
- Jayavelu, A.K.; Schnoder, T.M.; Perner, F.; Herzog, C.; Meiler, A.; Krishnamoorthy, G.; Huber, N.; Mohr, J.; Edelmann-Stephan, B.; Austin, R.; et al. Splicing factor YBX1 mediates persistence of JAK2-mutated neoplasms. Nature 2020, 588, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Lindemans, C.A.; Calafiore, M.; Mertelsmann, A.M.; O’Connor, M.H.; Dudakov, J.A.; Jenq, R.R.; Velardi, E.; Young, L.F.; Smith, O.M.; Lawrence, G.; et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 2015, 528, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Han, Y.; Liu, Y.; Zhao, Z.; Ma, F.; Cui, A.; Zhang, F.; Liu, Z.; Xue, Y.; Bai, J.; et al. CREBZF as a Key Regulator of STAT3 Pathway in the Control of Liver Regeneration in Mice. Hepatology 2020, 71, 1421–1436. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Dunne, C.; Hewson, J.; Wohl, C.; Wheatley, M.; Peterson, A.C.; Reynolds, B.A. Multipotent CNS stem cells are present in the adult mammalian spinal cord and ventricular neuroaxis. J. Neurosci. Off. J. Soc. Neurosci. 1996, 16, 7599–7609. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Zhu, Y.; Yue, C.; Yang, Q.; Zhang, Z. Comprehensive Bioinformatics Analysis Combined with Wet-Lab Experiments to Find Target Proteins of Chinese Medicine Monomer. Molecules 2022, 27, 6105. https://doi.org/10.3390/molecules27186105
Xu X, Zhu Y, Yue C, Yang Q, Zhang Z. Comprehensive Bioinformatics Analysis Combined with Wet-Lab Experiments to Find Target Proteins of Chinese Medicine Monomer. Molecules. 2022; 27(18):6105. https://doi.org/10.3390/molecules27186105
Chicago/Turabian StyleXu, Xiaohui, Yunyi Zhu, Changling Yue, Qianwen Yang, and Zhaohuan Zhang. 2022. "Comprehensive Bioinformatics Analysis Combined with Wet-Lab Experiments to Find Target Proteins of Chinese Medicine Monomer" Molecules 27, no. 18: 6105. https://doi.org/10.3390/molecules27186105
APA StyleXu, X., Zhu, Y., Yue, C., Yang, Q., & Zhang, Z. (2022). Comprehensive Bioinformatics Analysis Combined with Wet-Lab Experiments to Find Target Proteins of Chinese Medicine Monomer. Molecules, 27(18), 6105. https://doi.org/10.3390/molecules27186105







