Identification of Novel Natural Inhibitors to Human 3-Phosphoglycerate Dehydrogenase (PHGDH) for Cancer Treatment
Abstract
1. Introduction
2. Results
2.1. In-Silico-Based Screening of Phenolic Compounds Library against Coenzyme and Substrate Binding Sites of Human 3-Phosphoglycerate Dehydrogenase (PHGDH)
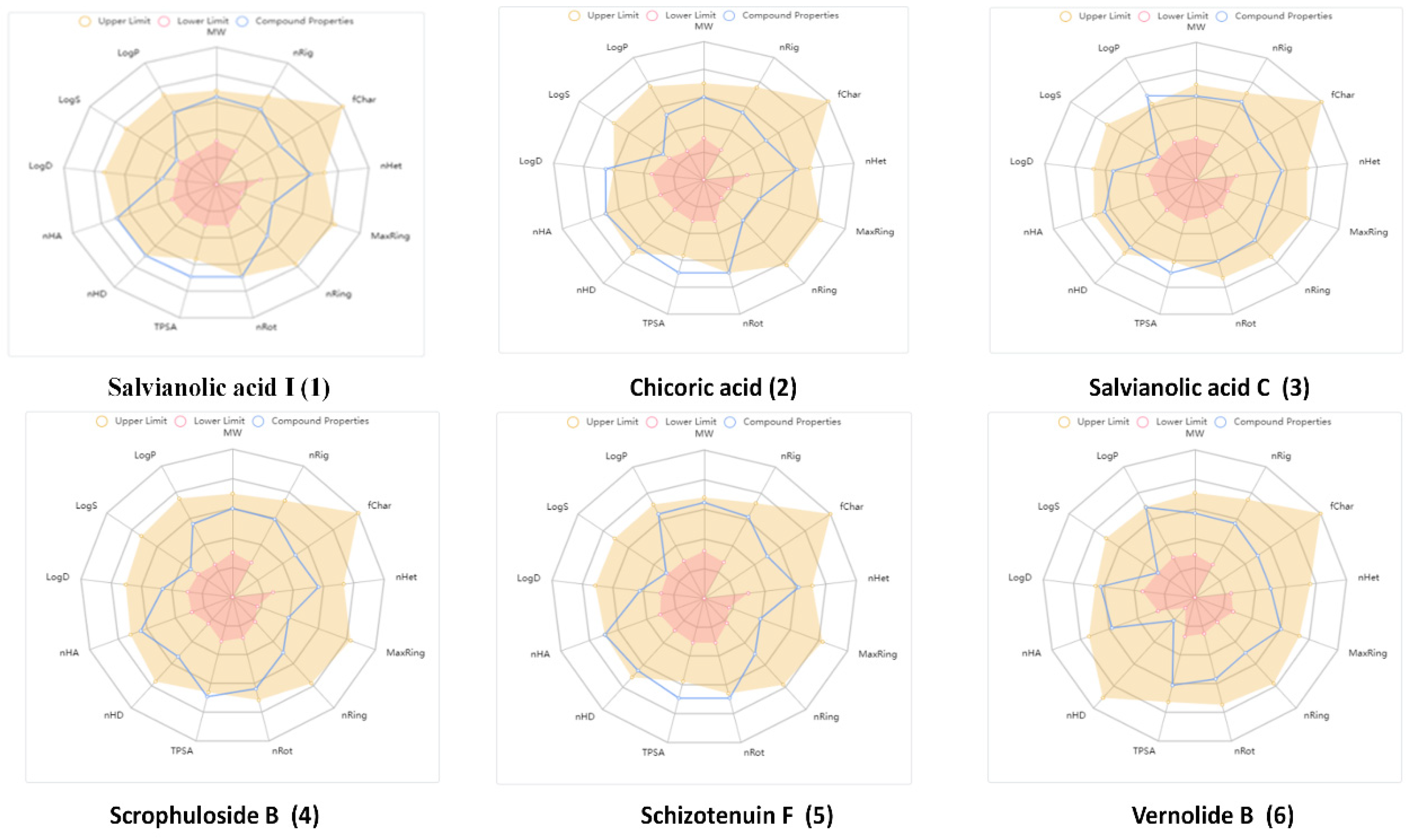
2.1.1. Computational Analysis of Physicochemical Properties of Hit Compounds
2.1.2. ADMET Profiling of Hit Compounds
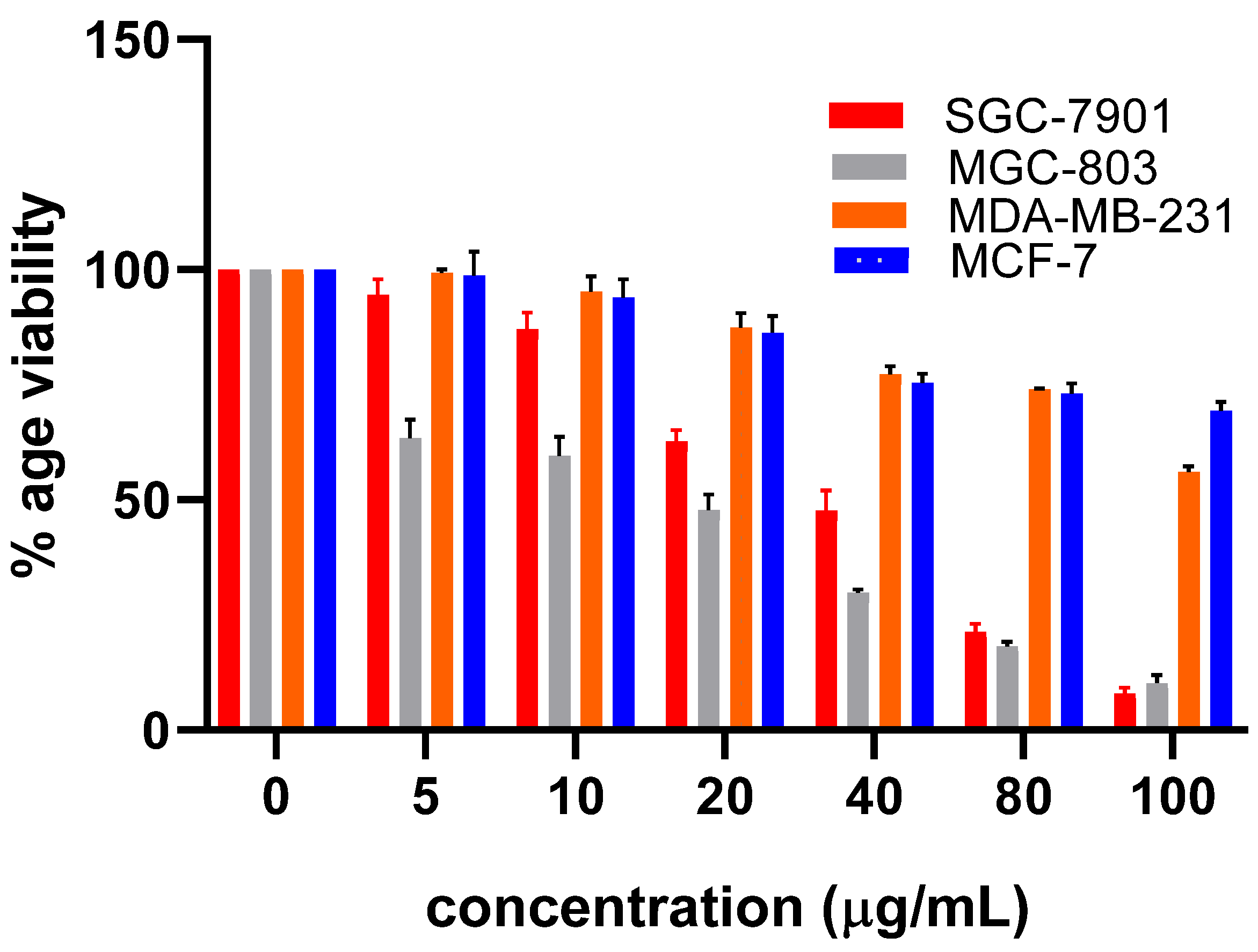
2.2. Evaluation of Cytotoxic Potential of Hit Compounds against PHGDH Overexpressing Gastric Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Structure Retrieval and Preparation of PHGDH
4.2. Phytochemical Library Preparation
4.3. Docking Analysis
4.4. Cell Culture
4.5. MTT Cytotoxicity Assay
4.6. ADMET and Drug Likeness Analysis of Compounds
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Sarfraz, I.; Rasul, A.; Hussain, G.; Hussain, S.M.; Ahmad, M.; Nageen, B.; Jabeen, F.; Selamoglu, Z.; Ali, M. Malic enzyme 2 as a potential therapeutic drug target for cancer. IUBMB Life 2018, 70, 1076–1083. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lei, K.; Han, F.J.E.R.M.P.S. Tumor microenvironment: Recent advances in various cancer treatments. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3855–3864. [Google Scholar] [PubMed]
- El-Sayes, N.; Vito, A.; Mossman, K. Tumor Heterogeneity: A Great Barrier in the Age of Cancer Immunotherapy. Cancers 2021, 13, 806. [Google Scholar] [CrossRef]
- Vazquez, A.; Kamphorst, J.J.; Markert, E.K.; Schug, Z.T.; Tardito, S.; Gottlieb, E. Cancer metabolism at a glance. J. Cell Sci. 2016, 129, 3367–3373. [Google Scholar] [CrossRef]
- Torresano, L.; Nuevo-Tapioles, C.; Santacatterina, F.; Cuezva, J.M. Metabolic reprogramming and disease progression in cancer patients. Biochim. Biophys. Acta. Mol. Basis Dis. 2020, 1866, 165721. [Google Scholar] [CrossRef]
- Pan, S.; Fan, M.; Liu, Z.; Li, X.; Wang, H. Serine, glycine and onecarbon metabolism in cancer (Review). Int. J. Oncol. 2021, 58, 158–170. [Google Scholar] [CrossRef]
- Mattaini, K.R.; Sullivan, M.R.; Vander Heiden, M.G. The importance of serine metabolism in cancer. J. Cell Biol. 2016, 214, 249–257. [Google Scholar] [CrossRef]
- Mullarky, E.; Lucki, N.C.; Beheshti Zavareh, R.; Anglin, J.L.; Gomes, A.P.; Nicolay, B.N.; Wong, J.C.; Christen, S.; Takahashi, H.; Singh, P.K.; et al. Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc. Natl. Acad. Sci. USA 2016, 113, 1778–1783. [Google Scholar] [CrossRef]
- Pacold, M.E.; Brimacombe, K.R.; Chan, S.H.; Rohde, J.M.; Lewis, C.A.; Swier, L.J.; Possemato, R.; Chen, W.W.; Sullivan, L.B.; Fiske, B.P.; et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat. Chem. Biol. 2016, 12, 452–458. [Google Scholar] [CrossRef]
- Rathore, R.; Schutt, C.R.; Van Tine, B.A. PHGDH as a mechanism for resistance in metabolically-driven cancers. Cancer Drug Resist. 2020, 3, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.Y.; Feng, K.R.; Wang, F.; Zhang, J.W.; Cheng, J.F.; Lin, G.Q.; Gao, D.; Tian, P. A retrospective overview of PHGDH and its inhibitors for regulating cancer metabolism. Eur. J. Med. Chem. 2021, 217, 113379. [Google Scholar] [CrossRef]
- Ma, X.; Li, B.; Liu, J.; Fu, Y.; Luo, Y. Phosphoglycerate dehydrogenase promotes pancreatic cancer development by interacting with eIF4A1 and eIF4E. J. Exp. Clin. Cancer Res. CR 2019, 38, 66. [Google Scholar] [CrossRef]
- Weinstabl, H.; Treu, M.; Rinnenthal, J.; Zahn, S.K.; Ettmayer, P.; Bader, G.; Dahmann, G.; Kessler, D.; Rumpel, K.; Mischerikow, N.; et al. Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) Inhibitor BI-4924 Disrupts Serine Biosynthesis. J. Med. Chem. 2019, 62, 7976–7997. [Google Scholar] [CrossRef] [PubMed]
- Arlt, B.; Mastrobuoni, G.; Wuenschel, J.; Astrahantseff, K.; Eggert, A.; Kempa, S.; Deubzer, H.E. Inhibiting PHGDH with NCT-503 reroutes glucose-derived carbons into the TCA cycle, independently of its on-target effect. J. Enzym. Inhib. Med. Chem. 2021, 36, 1282–1289. [Google Scholar] [CrossRef]
- Montrose, D.C.; Saha, S.; Foronda, M.; McNally, E.M.; Chen, J.; Zhou, X.K.; Ha, T.; Krumsiek, J.; Buyukozkan, M.; Verma, A.; et al. Exogenous and Endogenous Sources of Serine Contribute to Colon Cancer Metabolism, Growth, and Resistance to 5-Fluorouracil. Cancer Res. 2021, 81, 2275–2288. [Google Scholar] [CrossRef]
- Tajan, M.; Hennequart, M.; Cheung, E.C.; Zani, F.; Hock, A.K.; Legrave, N.; Maddocks, O.D.K.; Ridgway, R.A.; Athineos, D.; Suarez-Bonnet, A.; et al. Serine synthesis pathway inhibition cooperates with dietary serine and glycine limitation for cancer therapy. Nat. Commun. 2021, 12, 366. [Google Scholar] [CrossRef] [PubMed]
- Chien, L.H.; Wu, C.T.; Deng, J.S.; Jiang, W.P.; Huang, W.C.; Huang, G.J. Salvianolic Acid C Protects against Cisplatin-Induced Acute Kidney Injury through Attenuation of Inflammation, Oxidative Stress and Apoptotic Effects and Activation of the CaMKK-AMPK-Sirt1-Associated Signaling Pathway in Mouse Models. Antioxidants 2021, 10, 1620. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Scagel, C.F. Chicoric acid: Chemistry, distribution, and production. Front. Chem. 2013, 1, 40. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhang, X.; Zhai, H.; Zhang, D.; Ma, S. Chicoric acid (CA) induces autophagy in gastric cancer through promoting endoplasmic reticulum (ER) stress regulated by AMPK. Biomed. Pharmacother. 2019, 118, 109144. [Google Scholar] [CrossRef]
- Sevimli-Gur, C.; Yesil-Celiktas, O. Cytotoxicity screening of supercritical fluid extracted seaweeds and phenylpropanoids. Mol. Biol. Rep. 2019, 46, 3691–3699. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.X.; Ju, H.Q.; Liu, Z.X.; Chen, D.L.; Wang, Y.; Zhao, Q.; Wu, Q.N.; Zeng, Z.L.; Qiu, H.B.; Hu, P.S.; et al. ME1 Regulates NADPH Homeostasis to Promote Gastric Cancer Growth and Metastasis. Cancer Res. 2018, 78, 1972–1985. [Google Scholar] [CrossRef]
- Xian, Y.; Zhang, S.; Wang, X.; Qin, J.; Wang, W.; Wu, H. Phosphoglycerate dehydrogenase is a novel predictor for poor prognosis in gastric cancer. Onco. Targets. Ther. 2016, 9, 5553–5560. [Google Scholar] [PubMed]
- Wang, Q.; Liberti, M.V.; Liu, P.; Deng, X.; Liu, Y.; Locasale, J.W.; Lai, L. Rational Design of Selective Allosteric Inhibitors of PHGDH and Serine Synthesis with Anti-tumor Activity. Cell Chem. Biol. 2017, 24, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, R.; Christensen, M.H. MolDock: A new technique for high-accuracy molecular docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef]
- Zheng, M.; Guo, J.; Xu, J.; Yang, K.; Tang, R.; Gu, X.; Li, H.; Chen, L. Ixocarpalactone A from dietary tomatillo inhibits pancreatic cancer growth by targeting PHGDH. Food Funct. 2019, 10, 3386–3395. [Google Scholar] [CrossRef]
- Rasul, A.; Yu, B.; Zhong, L.; Khan, M.; Yang, H.; Ma, T. Cytotoxic effect of evodiamine in SGC-7901 human gastric adenocarcinoma cells via simultaneous induction of apoptosis and autophagy. Oncol. Rep. 2012, 27, 1481–1487. [Google Scholar]
- Dong, J.; Wang, N.N.; Yao, Z.J.; Zhang, L.; Cheng, Y.; Ouyang, D.; Lu, A.P.; Cao, D.S. ADMETlab: A platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminform. 2018, 10, 29. [Google Scholar] [CrossRef]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef]
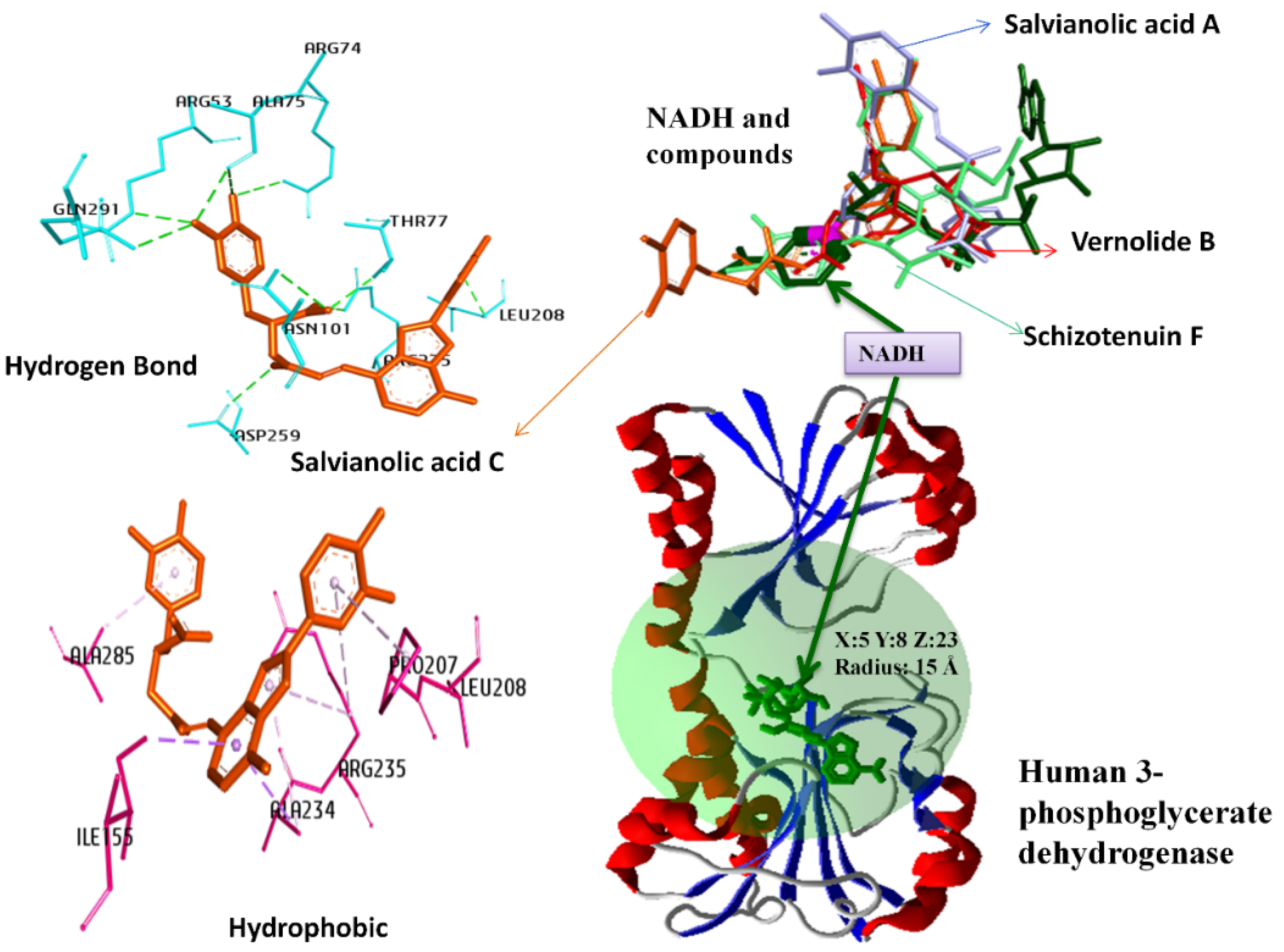
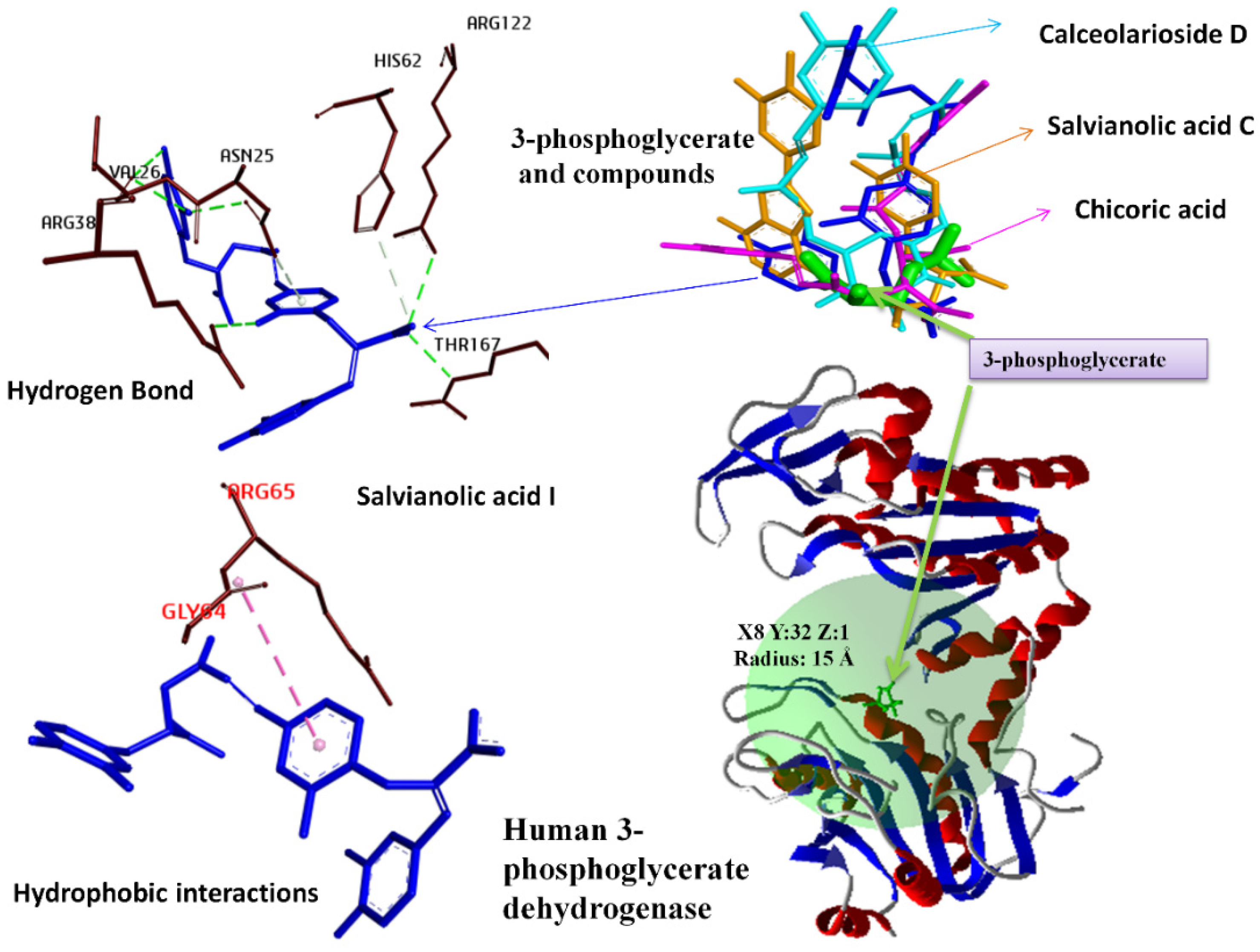
| Nicotinamide Adenine Dinucleotide (NAD+) Binding Cavity | 3-Phosphoglyceric Acid Binding Cavity | ||||||
|---|---|---|---|---|---|---|---|
| Compounds | PubChem CID | MolDock Score | HBond | Compounds | PubChem CID | MolDock Score | HBond |
| BI-4924 | 138756831 | −154.91 | −12.89 | BI-4924 | 138756831 | −122.20 | −13.66 |
| NCT 503 | 118796328 | −144.58 | −1.008 | NCT 503 | 118796328 | −117.15 | −3.14 |
| CBR-5884 | 4674993 | −138.48 | −4.401 | CBR-5884 | 4674993 | −113.39 | −5.55 |
| Salvianolic Acid C | 13991590 | −189.11 | −19.18 | Salvianolic acid I | 10459878 | −160.87 | −14.28 |
| Schizotenuin F | 10347565 | −182.47 | −12.19 | Chicoric acid | 5281764 | −159.05 | −19.22 |
| Vernolide B | 73076547 | −174.98 | −4.77 | Scrophuloside B | 13991590 | −157.89 | −8.24 |
| Salvianolic acid A | 5281793 | −173.90 | −12.43 | Salvianolic Acid C | 14015431 | −150.78 | −19.58 |
| Salvianolic Acid N | 49769102 | −168.28 | −16.93 | Tungtungmadic Acid | 70697815 | −146.01 | −22.12 |
| Salvianolic acid I | 10459878 | −168.17 | −16.88 | 3,5-Di-O-caffeoyl-muco-quinic acid | 6475855 | −144.32 | −19.87 |
| Apigenin-7-O-gentiobioside | 10841200 | −167.10 | −25.07 | Salvianolicacid D | 73060756 | −141.63 | −11.90 |
| 3,5-Di-O-caffeoyl-muco-quinic acid | 6475855 | −164.60 | −13.56 | Schizotenuin F | Unknown 1 | −141.59 | −18.25 |
| Chicoric acid | 5281764 | −164.37 | −19.62 | Salvianolic acid A | 5281793 | −140.21 | −13.02 |
| Tungtungmadic Acid | 70697815 | −164.34 | −14.49 | Salvianolic acid K | 10482829 | −138.74 | −17.69 |
| Salvianolic Acid C (L1) | Schizotenuin F (L2) | Vernolide B (L3) | Salvianolic Acid A (L4) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | Interacting Group | D | T | C | Interacting Group | D | T | C | Interacting Group | D | T | C | Interacting Group | D |
| Hydrogen Bond | Conventional Hydrogen Bond | ARG53:NE—L1:O5 | 2.87 | Hydrogen Bond | Conventional Hydrogen Bond | GLY78:N—L2:O11 | 3.10 | Hydrogen Bond | Co. HB | THR77:OG1—L3:O8 | 3.10 | Hydrogen Bond | Conventional Hydrogen Bond | ASP80:OD1—L4:O6 | 3.08 |
| ARG74:NH1—L1:O7 | 3.32 | LEU208:N—L2:O8 | 2.99 | GLY78:N—L3:O7 | 2.98 | GLY153:N—L4:O4 | 2.91 | ||||||||
| THR77:OG1—L1:O4 | 2.72 | ARG235:NH1—L2:O4 | 2.76 | L3:O6—CYS233:O | 2.83 | ARG154:NH1—L4:O2 | 3.08 | ||||||||
| ASN101:ND2—L1:O4 | 3.27 | ALA285:N—L2:O3 | 3.46 | Ca. HB | ALA234:CA—L3:O6 | 3.30 | GLY156:N—L4:O4 | 3.10 | |||||||
| LEU208:N—L1:O6 | 2.81 | L2:O4—ALA234:O | 2.78 | L3:C14—PRO98:O | 3.54 | ARG235:NE—L4:O6 | 2.60 | ||||||||
| ARG235:NH1—L1:O8 | 2.90 | L2:O4—HIS282:NE2 | 3.07 | L3:C16—ALA234:O | 3.13 | L4:O6—ASP80:OD2 | 3.27 | ||||||||
| GLN291:NE2—L1:O5 | 3.16 | L2:O12—THR206:O | 2.97 | Hydrophobic | Alkyl | ALA105—L3:C22 | 3.68 | L4:O9—THR206:O | 2.60 | ||||||
| L1:O5—ALA75:O | 2.91 | Hydrophobic | Alkyl | ALA105—L2 | 4.08 | ARG154—L3 | 4.61 | L4:O10—CYS233:O | 3.37 | ||||||
| L1:O7—ALA75:O | 3.22 | ILE155—L2 | 4.99 | PRO207—L3 | 5.32 | Ca. HB | ARG154:CD—L4:O2 | 3.06 | |||||||
| L1:O10—ASP259:OD2 | 3.39 | PRO207—L2 | 5.00 | L3—ILE155 | 4.52 | Pi-DHB | ARG235:NH1—L4 | 4.02 | |||||||
| Hydrophobic | Pi-Alkyl | L1—ALA234 | 4.55 | ARG235—L2 | 5.46 | L3:C9—ARG154 | 3.92 | Hydrophobic | Pi-Alkyl | L4—ILE155 | 5.21 | ||||
| L1—ARG235 | 5.08 | L3:C9—ILE155 | 4.82 | L4—ALA234 | 4.57 | ||||||||||
| L1—ALA285 | 4.09 | L3:C10—PRO207 | 4.48 | L4—ARG235 | 5.05 | ||||||||||
| L1—PRO207 | 4.74 | L3:C19—PRO207 | 5.03 | ||||||||||||
| L1—ARG235 | 5.35 | L3:C19—ARG235 | 2.94 | ||||||||||||
| Pi-S | ILE155:CD1—L1 | 3.70 | L3:C23—LEU208 | 4.91 | |||||||||||
| ALA234:CB—L1 | 3.81 | L3:C23—ARG235 | 4.11 | ||||||||||||
| Salvianolic Acid I (L5) | Chicoric Acid (L6) | Scrophuloside B (L1) | Salvianolic Acid C (L7) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | Interacting Group | D | T | C | Interacting Group | D | T | C | Interacting Group | D | T | C | Interacting Group | D |
| Hydrogen Bond | Conventional Hydrogen Bond | ARG122:HH12—L5O11 | 2.08 | Hydrogen Bond | Conventional Hydrogen Bond | ASN25:HD22—L6:O8 | 1.81 | Hydrogen Bond | Conventional Hydrogen Bond | ARG65:HE—L1:O4 | 2.80 | Hydrogen Bond | Conventional Hydrogen Bond | ASN25:HD21—L7:O1 | 2.089 |
| ARG170:HE—L5O11 | 1.66 | ARG38:HH12—L6:O8 | 2.72 | ARG65:HH21—L1:O4 | 1.88 | ARG38:HH22—L7:O4 | 2.311 | ||||||||
| ARG170:HH21—L5O11 | 2.76 | ARG65:HN—L6:O9 | 1.73 | ARG122:HH12—L1:O8 | 1.69 | ARG65:HN—L7:O9 | 2.258 | ||||||||
| L5H1—VAL26:O | 1.70 | ARG65:HE—L6:O5 | 2.28 | ARG170:HE—L1:O2 | 2.24 | ARG122:HH22—L7:O2 | 2.139 | ||||||||
| L5H2—VAL26:O | 1.74 | ARG65:HH21—L6:O3 | 1.44 | ARG170:HH21—L1:O4 | 2.20 | ARG170:HH21—L7:O5 | 2.768 | ||||||||
| Ca. HB | HIS62:HE1—L5O12 | 2.95 | ARG122:HH12—L6:O5 | 2.55 | L1:H6—ASP23:OD2 | 2.97 | L7:H1—GLY166:O | 2.072 | |||||||
| THR167:HA—L5O12 | 2.56 | ARG122:HH12—L6:O6 | 1.63 | L1:H6—GLY166:O | 2.09 | L7:H2—GLY166:O | 1.651 | ||||||||
| H | A-Pi S | GLY64:C,O; ARG65:N—L5 | 5.21 | ARG122:HH22—L6:O5 | 1.58 | Ca. HB | HIS62:HE1—L1:O10 | 2.35 | L7:H5—ASN25:O | 2.562 | |||||
| ARG170:HE—L6:O4 | 1.57 | GLY64:HA2—L1:O7 | 2.23 | L7:H6—TYR75:OH | 2.991 | ||||||||||
| ARG170:HH21—L6:O3 | 1.76 | HIS172:HE1—L1:O8 | 2.77 | L7:H7—MET28:O | 2.472 | ||||||||||
| ARG170:HH21—L6:O4 | 2.68 | HB-E | Pi-C | ARG38:NH1—L1 | 3.32 | Ca. HB | HIS62:HE1—L7:O3 | 3.065 | |||||||
| L6:H2—ASP23:OD2 | 2.08 | Pi-D HB | ARG38:NH2—L1 | 3.46 | GLY64:HA1—L7:O9 | 3.087 | |||||||||
| L6:H3—TYR75:OH | 2.29 | E | Pi-C | ARG65:NH2—L1 | 3.99 | L7:H11—HIS62:NE2 | 2.184 | ||||||||
| L6:H4—GLY372:O | 3.06 | HB | Pi-DHB | ASN25:HD21—L1 | 2.64 | H | Pi-A | L7—LEU63 | 5.042 | ||||||
| Ca. HB | HIS169:HE1—L6:O4 | 2.90 | Hydrophobic | Pi-A | L1—ARG65 | 4.97 | |||||||||
| L6:H8—HIS62:NE2 | 2.09 | Pi-A | L1—PRO27 | 4.74 | |||||||||||
| H | A-Pi S | GLY395:C,O; GLY396:N—L6 | 4.02 | ||||||||||||
| Pi-A | L6—ARG65 | 3.90 | |||||||||||||
| Physicochemical Property | Optimal | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|---|
| Molecular Weight (MW) | 100~600 | 538.11 | 474.08 | 492.11 | 474.15 | 552.13 | 434.19 |
| nHA | 0~12 | 12 | 12 | 10 | 10 | 12 | 8 |
| nHD | 0~7 | 7 | 6 | 6 | 4 | 6 | 1 |
| nRot | 0~11 | 11 | 11 | 8 | 9 | 12 | 7 |
| nRing | 0~6 | 3 | 2 | 4 | 3 | 3 | 3 |
| MaxRing | 0~18 | 6 | 6 | 9 | 6 | 6 | 13 |
| nHet | 1~15 | 12 | 12 | 10 | 10 | 12 | 8 |
| fChar | −4~4 | 0 | 0 | 0 | 0 | 0 | 0 |
| nRig | 0~30 | 23 | 18 | 25 | 21 | 23 | 19 |
| TPSA | 0~140 | 211.28 | 208.12 | 177.89 | 151.98 | 200.28 | 104.43 |
| logS | −4~0.5 | −3.679 | −3.503 | −3.867 | −3.406 | −3.875 | −4.02 |
| logP | 0~3 | 2.074 | 1.672 | 3.641 | 1.846 | 2.491 | 2.973 |
| logD | 1~3 | 1.389 | 3.427 | 2.265 | 1.808 | 1.643 | 2.774 |
| Medicinal chemistry | Pfizer Rule | Accepted | Accepted | Accepted | Accepted | Accepted | Accepted |
| Category | Property | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|---|
| Absorption | Caco-2 > −5.15 | −6.271/poor | −6.450/poor | −5.629/poor | −5.966/poor | −5.954/poor | −4.772/excellent |
| Pgp-Inhibitor | --- | --- | --- | --- | --- | -- | |
| Pgp-Substrate | --- | --- | --- | -- | --- | --- | |
| HIA | + | + | + | ++ | + | --- | |
| Distribution | PPB | 96.077% | 98.828% | 99.640% | 98.413% | 96.746% | 86.919% |
| BBB | --- | --- | --- | -- | --- | - | |
| Metabolism | CYP1A2-Inhibitor | - | --- | + | - | + | -- |
| CYP1A2-Substrate | --- | --- | --- | --- | --- | +++ | |
| CYP3A4-Inhibitor | --- | --- | -- | - | -- | -- | |
| CYP3A4-Substrate | --- | --- | --- | -- | --- | + | |
| CYP2C19-Inhibitor | + | - | + | + | + | + | |
| CYP2C19-Substrate | -- | +++ | ++ | ++ | ++ | -- | |
| CYP2C9-Inhibitor | --- | --- | --- | -- | --- | --- | |
| CYP2D6-Inhibitor | -- | -- | -- | + | -- | -- | |
| CYP2C9-Substrate | --- | --- | -- | -- | -- | + | |
| CYP2D6-Substrate | --- | -- | --- | -- | --- | + | |
| Excretion | Clearance | 3.954/Low | 4.971/Low | 10.679/ Moderate | 5.328/ Moderate | 8.282/ Moderate | 11.696/ Moderate |
| Toxicity | hERG | --- | --- | --- | - | --- | --- |
| H-HT | ++ | -- | -- | --- | - | ++ | |
| Ames | --- | --- | -- | -- | --- | +++ | |
| DILI | ++ | -- | +++ | -- | +++ | +++ | |
| FDAMDD | --- | --- | -- | --- | -- | --- | |
| For the classification models, the prediction probability values are represented with different symbols: 0–0.1(---), 0.1–0.3(--), 0.3–0.5(-), 0.5–0.7(+), 0.7–0.9(++), and 0.9–1.0(+++). Usually, the token ‘+++’ or ‘++’ represents the molecule that is more likely to be toxic or defective, while ‘--’ or ‘-’ represents nontoxic or appropriate. | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadiqa, A.; Rasul, A.; Hassan, M.; Sultana, S.; Jabeen, F. Identification of Novel Natural Inhibitors to Human 3-Phosphoglycerate Dehydrogenase (PHGDH) for Cancer Treatment. Molecules 2022, 27, 6108. https://doi.org/10.3390/molecules27186108
Sadiqa A, Rasul A, Hassan M, Sultana S, Jabeen F. Identification of Novel Natural Inhibitors to Human 3-Phosphoglycerate Dehydrogenase (PHGDH) for Cancer Treatment. Molecules. 2022; 27(18):6108. https://doi.org/10.3390/molecules27186108
Chicago/Turabian StyleSadiqa, Ayesha, Azhar Rasul, Mudassir Hassan, Salma Sultana, and Farhat Jabeen. 2022. "Identification of Novel Natural Inhibitors to Human 3-Phosphoglycerate Dehydrogenase (PHGDH) for Cancer Treatment" Molecules 27, no. 18: 6108. https://doi.org/10.3390/molecules27186108
APA StyleSadiqa, A., Rasul, A., Hassan, M., Sultana, S., & Jabeen, F. (2022). Identification of Novel Natural Inhibitors to Human 3-Phosphoglycerate Dehydrogenase (PHGDH) for Cancer Treatment. Molecules, 27(18), 6108. https://doi.org/10.3390/molecules27186108







