Integrated System Pharmacology Approaches to Elucidate Multi-Target Mechanism of Solanum surattense against Hepatocellular Carcinoma
Abstract
1. Introduction of Hepatocellular Carcinoma
2. Results
2.1. In Vitro Toxicological Analysis Hepatocellular Carcinoma and Normal Cells
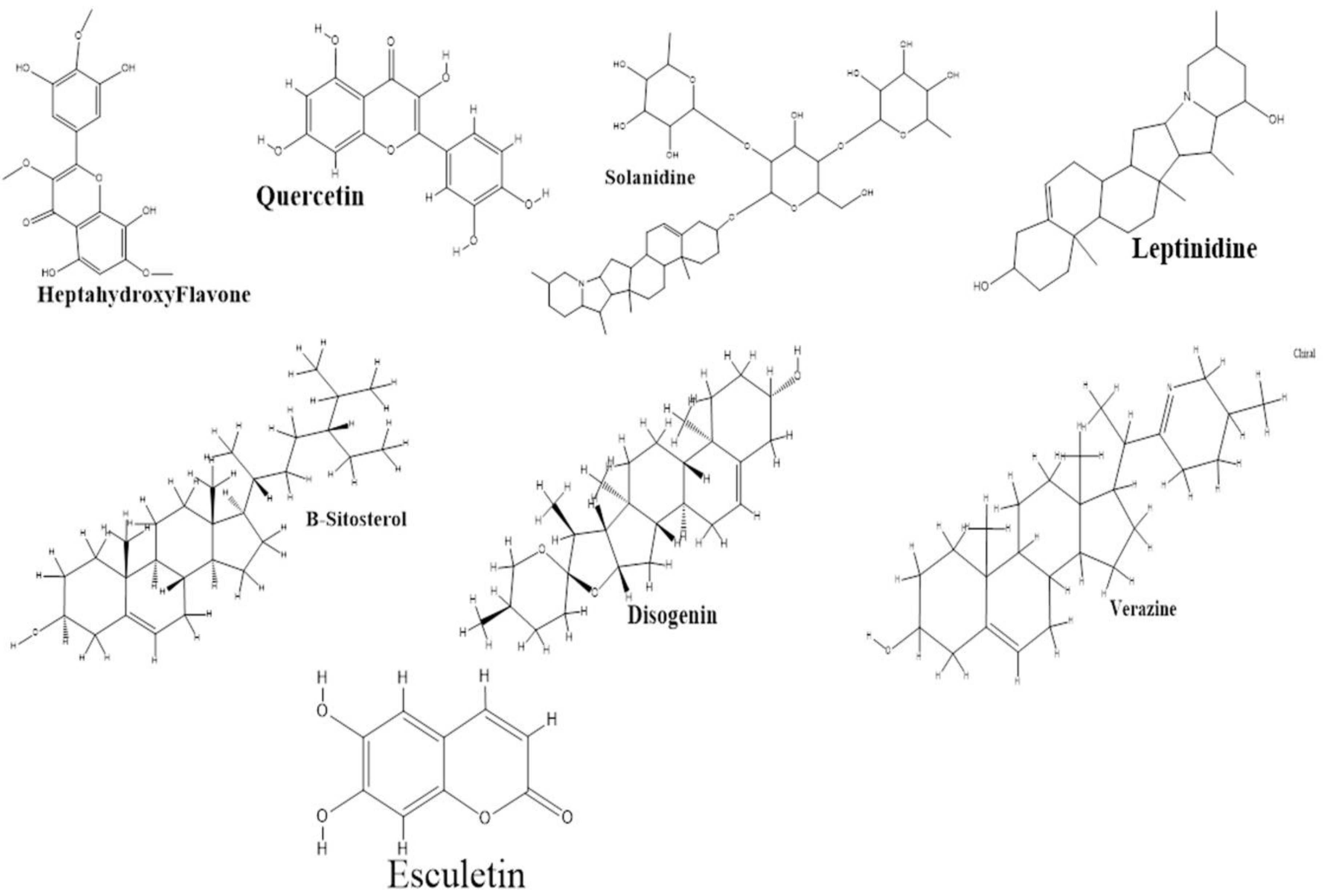
2.2. Active Compounds and Target Screening
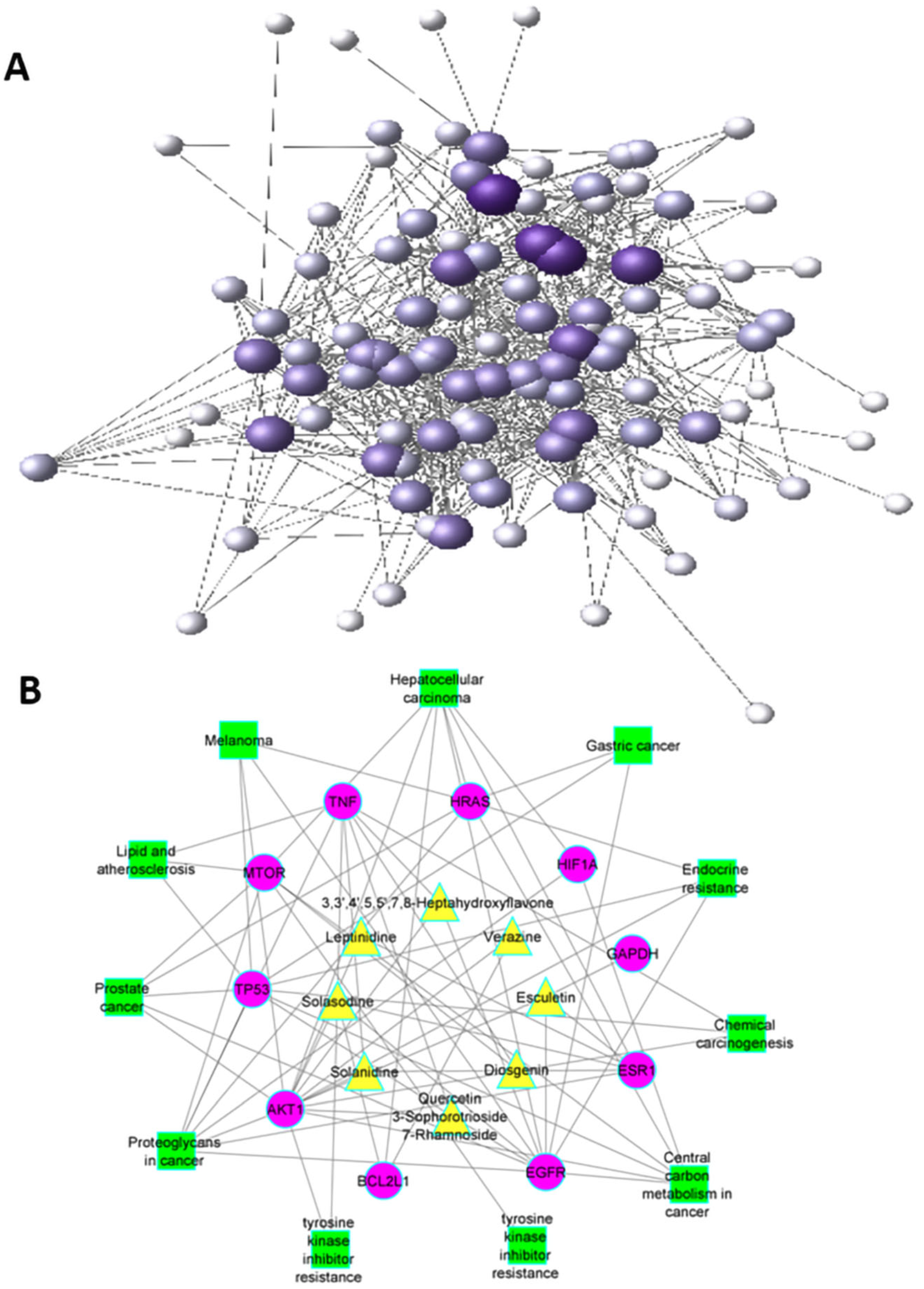
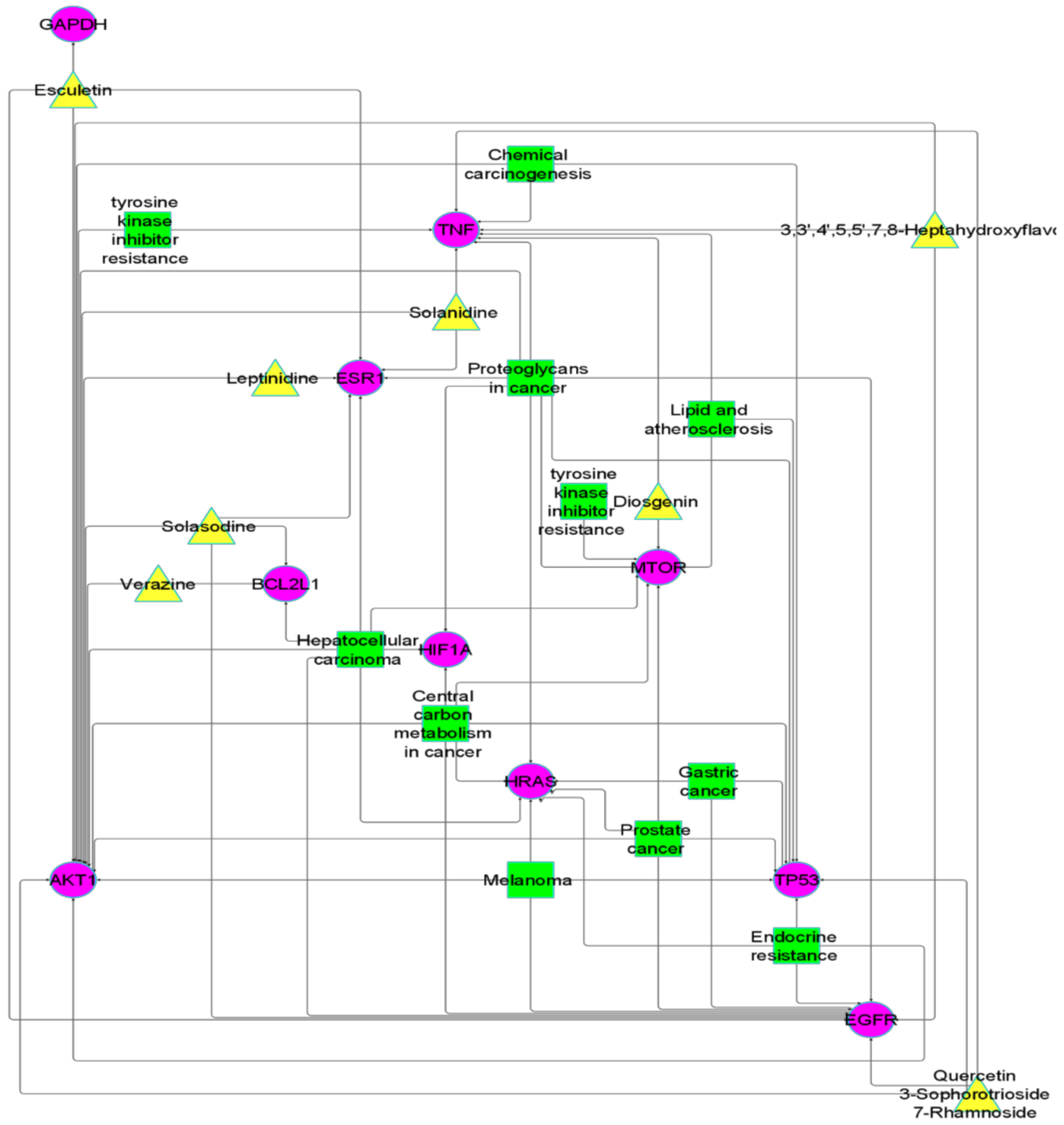
2.3. Protein–Protein Network and Hub Genes
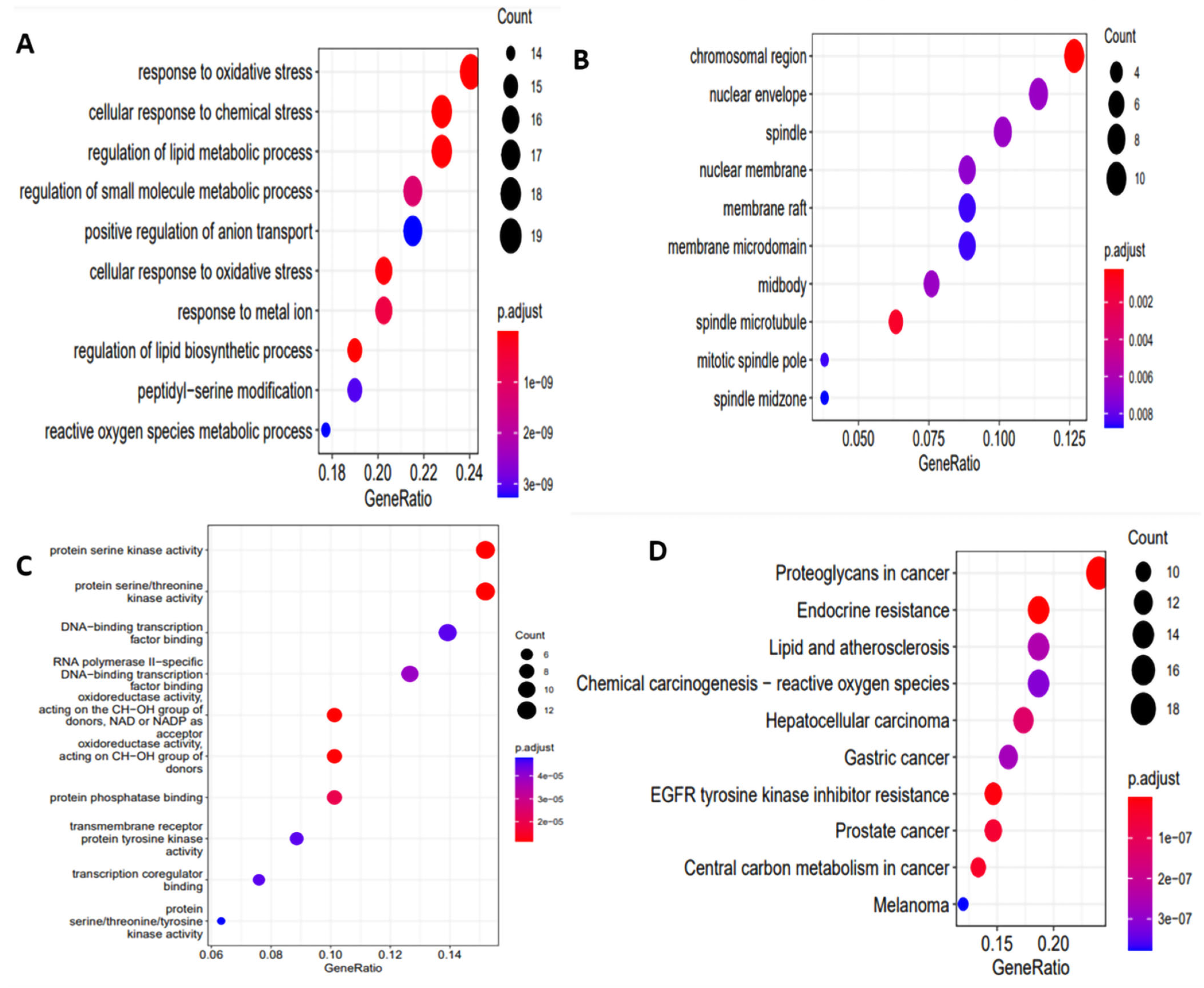
2.4. GO Enrichment Analysis
2.5. Pathway Network Construction
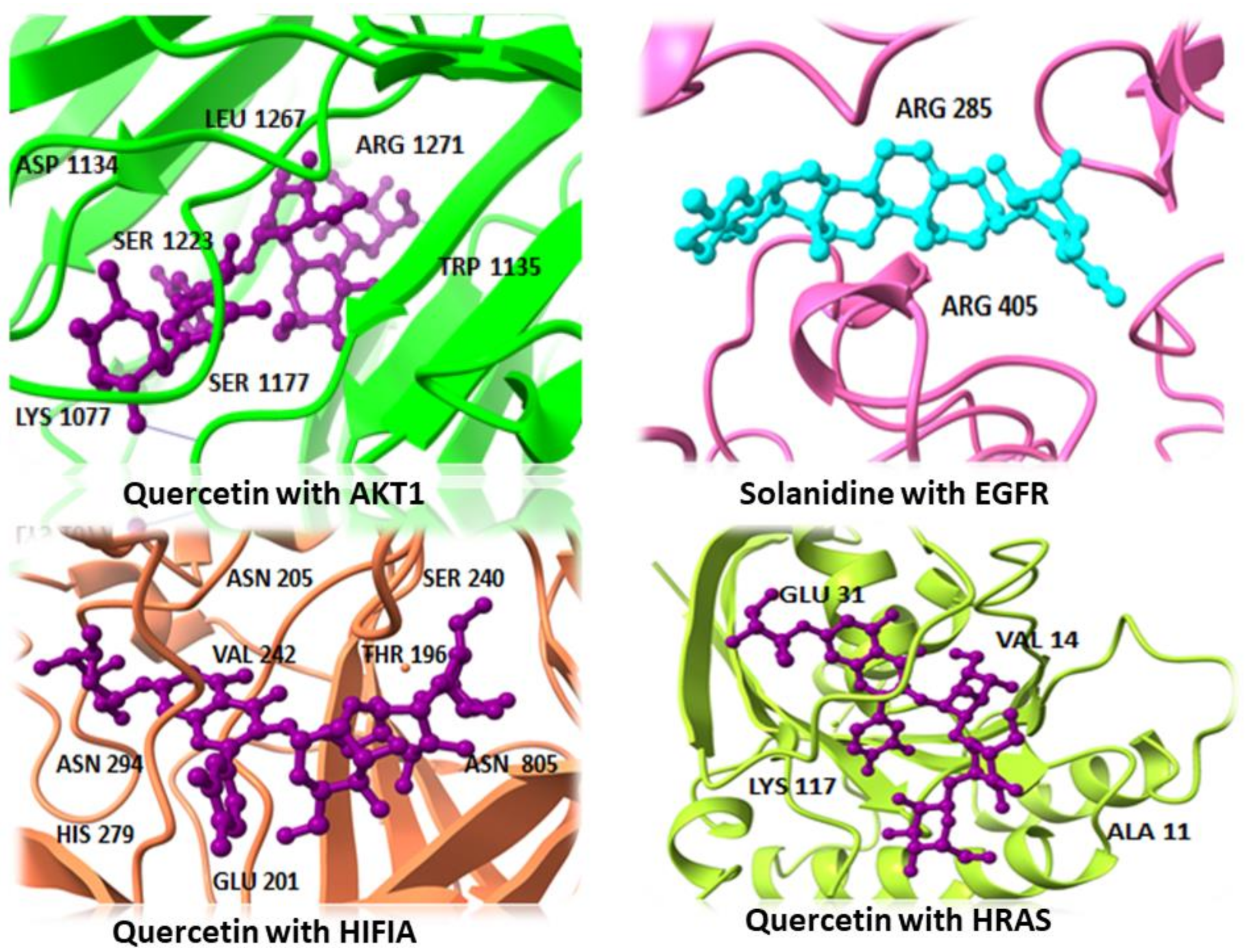
2.6. Molecular Docking
3. Discussion
4. Material and Methods
4.1. Collection and Identification of S. surattense
4.2. Extraction of Plant Material
4.3. Preparation of Stock Solutions
4.4. In Vitro Assay and Cell Culture Medium
4.5. Cell Proliferation Assay/Toxicological Analysis of Plant Extract Using WST-8 Assay
Screening of Active Compounds and Targets
4.6. Prediction of HCC Targets
4.7. Screening of Key Targets
4.8. Analysis of Functional Enrichment
4.9. Network Construction
4.10. Molecular Docking
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005, 55, 74–108. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Montal, R.; Sia, D.; Finn, R.S. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2018, 15, 599–616. [Google Scholar] [CrossRef] [PubMed]
- Balogh, J.; Victor, D., 3rd; Asham, E.H.; Burroughs, S.G.; Boktour, M.; Saharia, A.; Li, X.; Ghobrial, R.M.; Monsour, H.P., Jr. Hepatocellular carcinoma: A review. J. Hepatocell. Carcinoma 2016, 3, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Dutta, R.; Mahato, R.I. Recent advances in hepatocellular carcinoma therapy. Pharmacol. Ther. 2017, 173, 106–117. [Google Scholar] [CrossRef]
- Forner, A.; Da Fonseca, L.G.; Díaz-González, Á.; Sanduzzi-Zamparelli, M.; Reig, M.; Bruix, J. Controversies in the management of hepatocellular carcinoma. JHEP Rep. 2019, 1, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Plenge, R.M. Disciplined approach to drug discovery and early development. Sci. Transl. Med. 2016, 8, 349ps15. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Wang, X.-D.; Niu, Y.-Y.; Duan, D.-D.; Yang, X.; Hao, J.; Zhu, C.-H.; Chen, D.; Wang, K.-X.; Qin, X.-M. Molecular targets of Chinese herbs: A clinical study of hepatoma based on network pharmacology. Sci. Rep. 2016, 6, 24944. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, T.R.; Akerele, O. Medicinal Plants: Their Role in Health and Biodiversity; University of Pennsylvania Press: Philadelphia, PA, USA, 2015. [Google Scholar]
- Li, X.; Yang, G.; Li, X.; Zhang, Y.; Yang, J.; Chang, J.; Sun, X.; Zhou, X.; Guo, Y.; Xu, Y. Traditional Chinese medicine in cancer care: A review of controlled clinical studies published in Chinese. PLoS ONE 2013, 8, e60338. [Google Scholar]
- Verma, S.; Singh, S. Current and future status of herbal medicines. Vet. World 2008, 1, 347. [Google Scholar] [CrossRef]
- Sudhanshu, N.R.; Mittal, S.; Menghani, E. Antioxidant activity of Solanum surratense and Solanum nigrum methanolic extract: An in vitro evaluation. JAPR 2012, 3, 9–12. [Google Scholar]
- Dey, P.; Kundu, A.; Chakraborty, H.J.; Kar, B.; Choi, W.S.; Lee, B.M.; Bhakta, T.; Atanasov, A.G.; Kim, H.S. Therapeutic value of steroidal alkaloids in cancer: Current trends and future perspectives. Int. J. Cancer 2019, 145, 1731–1744. [Google Scholar] [CrossRef]
- Hopkins, A.L. Network pharmacology. Nat. Biotechnol. 2007, 25, 1110–1111. [Google Scholar] [CrossRef]
- Zhang, G.-B.; Li, Q.-Y.; Chen, Q.-L.; Su, S.-B. Network pharmacology: A new approach for Chinese herbal medicine research. Evid.-Based Complement. Altern. Med. 2013, 2013, 621423. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Li, R.; Ma, T.; Gu, J.; Liang, X.; Li, S. Imbalanced network biomarkers for traditional Chinese medicine Syndrome in gastritis patients. Sci. Rep. 2013, 3, 1543. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Wang, X.; Liu, Y.; Ye, J.; Liu, Q.; Li, Y.; Li, S. Integrating network pharmacology and metabolomics study on anti-rheumatic mechanisms and antagonistic effects against methotrexate-induced toxicity of Qing-Luo-Yin. Front. Pharmacol. 2018, 9, 1472. [Google Scholar] [CrossRef] [PubMed]
- Regassa, H.; Sourirajan, A.; Kumar, V.; Pandey, S.; Kumar, D.; Dev, K. A Review of Medicinal Plants of the Himalayas with Anti-Proliferative Activity for the Treatment of Various Cancers. Cancers 2022, 14, 3898. [Google Scholar] [CrossRef] [PubMed]
- Ni, T.; Wang, H.; Li, D.; Tao, L.; Lv, M.; Jin, F.; Wang, W.; Feng, J.; Qian, Y.; Sunagawa, M. Huachansu Capsule inhibits the proliferation of human gastric cancer cells via Akt/mTOR pathway. Biomed. Pharmacother. 2019, 118, 109241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Mo, Z.; Zhang, S.; Li, X. A network pharmacology approach to reveal the underlying mechanisms of Artemisia annua on the treatment of hepatocellular carcinoma. Evid.-Based Complement. Altern. Med. 2021, 2021, 8947304. [Google Scholar] [CrossRef]
- Noor, F.; Noor, A.; Ishaq, A.R.; Farzeen, I.; Saleem, M.H.; Ghaffar, K.; Aslam, M.F.; Aslam, S.; Chen, J.-T. Recent advances in diagnostic and therapeutic approaches for breast cancer: A comprehensive review. Curr. Pharm. Des. 2021, 27, 2344–2365. [Google Scholar] [CrossRef] [PubMed]
- Noor, F.; Saleem, M.H.; Chen, J.-T.; Javed, M.R.; Al-Megrin, W.A.; Aslam, S. Integrative bioinformatics approaches to map key biological markers and therapeutic drugs in Extramammary Paget’s disease of the scrotum. PLoS ONE 2021, 16, e0254678. [Google Scholar]
- Fujiwara, N.; Friedman, S.L.; Goossens, N.; Hoshida, Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J. Hepatol. 2018, 68, 526–549. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Xu, X.; Wang, L.; Zhu, B.; Wang, X.; Xia, J. The role of EGF-EGFR signalling pathway in hepatocellular carcinoma inflammatory microenvironment. J. Cell. Mol. Med. 2014, 18, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Villanueva, A.; Lachenmayer, A.; Finn, R.S. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat. Rev. Clin. Oncol. 2015, 12, 408–424. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, Z.; Sun, Y.; Geng, X.; Hu, Y.; Meng, H.; Ge, J.; Qu, L. Synthesis of luminescent carbon dots with ultrahigh quantum yield and inherent folate receptor-positive cancer cell targetability. Sci. Rep. 2018, 8, 1086. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Ijaz, S.; Mohammad, I.S.; Muhammad, K.S.; Akhtar, N.; Khan, H.M.S. Aglycone solanidine and solasodine derivatives: A natural approach towards cancer. Biomed. Pharmacother. 2017, 94, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Morillo, M.; Rojas, J.; Lequart, V.; Lamarti, A.; Martin, P. Natural and synthetic derivatives of the steroidal glycoalkaloids of Solanum genus and biological activity. Nat. Prod. Chem. Res. 2020, 8, 1–14. [Google Scholar]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Pan, W.; Lu, Z.; Xia, C. Pharmacological mechanisms underlying the hepatoprotective effects of Ecliptae herba on hepatocellular carcinoma. Evid.-Based Complement. Altern. Med. 2021, 2021, 5591402. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Chen, F.; Zhong, Z.; Tan, H.Y.; Wang, N.; Liu, Y.; Fang, X.; Yang, T.; Feng, Y. Interpreting the pharmacological mechanisms of huachansu capsules on hepatocellular carcinoma through combining network pharmacology and experimental evaluation. Front. Pharmacol. 2020, 11, 414. [Google Scholar] [CrossRef]
- Citri, A.; Yarden, Y. EGF–ERBB signalling: Towards the systems level. Nat. Rev. Mol. Cell Biol. 2006, 7, 505–516. [Google Scholar] [CrossRef]
- Xu, X.; Sakon, M.; Nagano, H.; Hiraoka, N.; Yamamoto, H.; Hayashi, N.; Dono, K.; Nakamori, S.; Umeshita, K.; Ito, Y. Akt2 expression correlates with prognosis of human hepatocellular carcinoma. Oncol. Rep. 2004, 11, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Manning, B.D.; Cantley, L.C. AKT/PKB signaling: Navigating downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Li, L.; Jin, Y.; Chen, Z.; Duan, L.; Cao, M.; Ma, M.; Wu, Z. A network pharmacology approach to reveal the underlying mechanisms of Paeonia lactiflora Pall. on the treatment of Alzheimer’s disease. Evid.-Based Complement. Altern. Med. 2019, 2019, 8706589. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Tripathi, S. Concept of standardization, extraction and pre phytochemical screening strategies for herbal drug. J. Pharmacogn. Phytochem. 2014, 2, 115–119. [Google Scholar]
- Rasul, M.G. Conventional extraction methods use in medicinal plants, their advantages and disadvantages. Int. J. Basic Sci. Appl. Comput. 2018, 2, 10–14. [Google Scholar]
- Rehman, S.; Ashfaq, U.A.; Riaz, S.; Javed, T.; Riazuddin, S. Antiviral activity of Acacia nilotica against Hepatitis C Virus in liver infected cells. Virol. J. 2011, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Bokhari, M.; Carnachan, R.J.; Cameron, N.R.; Przyborski, S.A. Culture of HepG2 liver cells on three dimensional polystyrene scaffolds enhances cell structure and function during toxicological challenge. J. Anat. 2007, 211, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Ammerman, N.C.; Beier-Sexton, M.; Azad, A.F. Growth and maintenance of Vero cell lines. Curr. Protoc. Microbiol. 2008, 11, A.4E.1–A.4E.7. [Google Scholar] [CrossRef] [PubMed]
- Khalid, H.; Landry, K.B.; Ijaz, B.; Ashfaq, U.A.; Ahmed, M.; Kanwal, A.; Froeyen, M.; Mirza, M.U. Discovery of novel Hepatitis C virus inhibitor targeting multiple allosteric sites of NS5B polymerase. Infect. Genet. Evol. 2020, 84, 104371. [Google Scholar] [CrossRef] [PubMed]
- Gies, V.; Lopinski, G.; Augustine, J.; Cheung, T.; Kodra, O.; Zou, S. The impact of processing on the cytotoxicity of graphene oxide. Nanoscale Adv. 2019, 1, 817–826. [Google Scholar] [CrossRef]
- Araki, T.; Yamada, M.; Ohnishi, H.; Sano, S.; Uetsuki, T.; Hatanaka, H. Shp-2 specifically regulates several tyrosine-phosphorylated proteins in brain-derived neurotrophic factor signaling in cultured cerebral cortical neurons. J. Neurochem. 2000, 74, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, M.; Wispriyono, B.; Igisu, H. Increased cytotoxicity of cadmium in fibroblasts lacking c-fos. Biochem. Pharmacol. 2000, 59, 1573–1576. [Google Scholar] [CrossRef]
- Rehman, S.; Ijaz, B.; Fatima, N.; Muhammad, S.A.; Riazuddin, S. Therapeutic potential of Taraxacum officinale against HCV NS5B polymerase: In-vitro and In silico study. Biomed. Pharmacother. 2016, 83, 881–891. [Google Scholar] [CrossRef]
- Sesang, W.; Punyanitya, S.; Pitchuanchom, S.; Udomputtimekakul, P.; Nuntasaen, N.; Banjerdpongchai, R.; Wudtiwai, B.; Pompimon, W. Cytotoxic aporphine alkaloids from leaves and twigs of Pseuduvaria trimera (Craib). Molecules 2014, 19, 8762–8772. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, S.; Khalid, H.R.; Haq, W.U.; Aslam, S.; Alshammari, A.; Alharbi, M.; Riaz Rajoka, M.S.; Khurshid, M.; Ashfaq, U.A. Implementation of System Pharmacology and Molecular Docking Approaches to Explore Active Compounds and Mechanism of Ocimum sanctum against Tuberculosis. Processes 2022, 10, 298. [Google Scholar] [CrossRef]


| Compound Name | MW | nHBD | nHBA | LogP | Lipinksi Rule |
|---|---|---|---|---|---|
| 3,3′,4′,5,5′,7,8-Heptahydroxyflavone | 390.1 | 3 | 9 | 2.6 | Accepted |
| Leptinidine | 413.33 | 2 | 3 | 4.6 | Accepted |
| 3′,4′,5,5′,7-Pentahydroxy-3-methoxy flavone | 332.05 | 5 | 8 | 2.11 | Accepted |
| Rishitin | 222.16 | 2 | 2 | 2.38 | Accepted |
| Spirosolan-3-ol | 415.35 | 2 | 3 | 4.9 | Accepted |
| Spirosol-5-en-3-ol | 413.33 | 2 | 3 | 5.0 | Accepted |
| Spirost-5-ene-3,25-diol | 430.31 | 2 | 4 | 4.4 | Accepted |
| Spirost-5-en-3-ol | 414.31 | 1 | 3 | 5.5 | Accepted |
| 11-Spirovetivene-2,14-diol | 236.18 | 1 | 2 | 2.2 | Accepted |
| Verazine | 413.33 | 2 | 3 | 5.1 | Accepted |
| Campesterol | 400.37 | 1 | 1 | 5.5 | Accepted |
| Coumarin | 146.04 | 0 | 1 | 1.6 | Accepted |
| Diosgenin | 414.31 | 1 | 3 | 5.5 | Accepted |
| Esculetin | 178.03 | 2 | 4 | 0.9 | Accepted |
| Esculin | 340.08 | 5 | 9 | −0.6 | Accepted |
| Methyl caffeate | 194.06 | 2 | 4 | 1.9 | Accepted |
| Solanidine | 397.33 | 1 | 2 | 5.6 | Accepted |
| Solanocapsine | 430.36 | 4 | 4 | 5.0 | Accepted |
| Solasodine | 413.33 | 2 | 3 | 5.2 | Accepted |
| Tomatidinol | 413.33 | 2 | 3 | 5.3 | Accepted |
| Solanine | 867.5 | 9 | 16 | 2.0 | Rejected |
| Quercetin 3-Galactoside 7-Rhamnoside | 610.15 | 10 | 16 | −0.8 | Rejected |
| Quercetin 3-Sophorotrioside 7-Rhamnoside | 934.26 | 16 | 26 | −2.9 | Rejected |
| Target Proteins | Compounds | Binding Affinity (kcal/mol) | RMSD | Interacting Residues |
|---|---|---|---|---|
| AKT1 | Quercetin | −15.833 | 1.54 | SER:1177, ASP:1134, TRP:1135, SER:1223, LYS:1077, LEU:1267, ARG:1271 |
| Solanidine | −14.353 | 1.62 | SER:1177, VAL:1181, ASP:1229 | |
| HeptahydroxyFlavone | −13.48 | 0.81 | SER:1177, VAL:1220, THR:1265 | |
| BCL2L1 | Esculetin | −7.054 | 1.81 | GLN:26, GLN:160 |
| HeptahydroxyFlavone | −9.353 | 1.71 | GLU:158, GLN:160 | |
| Quercetin | −7.396 | 2.79 | GLN:26, GLU:158, SER:25, SER:23 | |
| EGFR | Solanidine | −15.81 | 2.37 | ARG B:285, ARG B:405 |
| Quercetin | −15.154 | 2.09 | ARG B:285, SER B:342, ARG B:405 | |
| HeptahydroxyFlavone | −13.245 | 1.38 | SER B:11, ARG B:285, ARG B:405 | |
| ESR | Quercetin | −7.167 | 2.44 | ASP 351 |
| HeptahydroxyFlavone | −12.73 | 1.27 | ASP:351 | |
| Leptinidine | −12.532 | 0.71 | ASP:351 | |
| H1F1A | Quercetin | −22.917 | 2.68 | ASN A:205, ASN A:294, THR A:196, SER A:240, VAL A:242, GLU A:201, HIS A:279ASN B:805 |
| Solanidine | −17.346 | 1.31 | GLU A:201, GLN A:203, ASN A:205, TRP A:296, PRO A:274, TRP A:277 | |
| HeptahydroxyFlavone | −17.46 | 0.81 | LEU A:101, ASN B:803 | |
| HRAS | Verazine | −13.267 | 1.07 | LYS:16, THR:35, GLN:61, LYS:117 |
| Quercetin | −20.618 | 1.91 | ALA:11, VAL:14, GLU:31, LYS:117 | |
| HeptahydroxyFlavone | −17.543 | 1.64 | VAL:14, GLY:15, LYS:16, SER:17, LYS:117 | |
| MTOR | Quercetin | −12.382 | 1.81 | LYS B:2046, TYR B:2089, GLY B:2093, ARG B:2043 |
| Solanidine | −8.277 | 1.15 | ARG B:2043, ASN B:2044 | |
| HeptahydroxyFlavone | −8.719 | 1.99 | ARG B:2043, | |
| TNF | Esculetin | −9.637 | 0.95 | GLY B:122, GLY C:121 |
| HeptahydroxyFlavone | −5.584 | 1.94 | TYR A:119, TYR B:119, GLY C:121 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalid, H.R.; Aamir, M.; Tabassum, S.; Alghamdi, Y.S.; Alzamami, A.; Ashfaq, U.A. Integrated System Pharmacology Approaches to Elucidate Multi-Target Mechanism of Solanum surattense against Hepatocellular Carcinoma. Molecules 2022, 27, 6220. https://doi.org/10.3390/molecules27196220
Khalid HR, Aamir M, Tabassum S, Alghamdi YS, Alzamami A, Ashfaq UA. Integrated System Pharmacology Approaches to Elucidate Multi-Target Mechanism of Solanum surattense against Hepatocellular Carcinoma. Molecules. 2022; 27(19):6220. https://doi.org/10.3390/molecules27196220
Chicago/Turabian StyleKhalid, Hafiz Rameez, Muhammad Aamir, Sana Tabassum, Youssef Saeed Alghamdi, Ahmad Alzamami, and Usman Ali Ashfaq. 2022. "Integrated System Pharmacology Approaches to Elucidate Multi-Target Mechanism of Solanum surattense against Hepatocellular Carcinoma" Molecules 27, no. 19: 6220. https://doi.org/10.3390/molecules27196220
APA StyleKhalid, H. R., Aamir, M., Tabassum, S., Alghamdi, Y. S., Alzamami, A., & Ashfaq, U. A. (2022). Integrated System Pharmacology Approaches to Elucidate Multi-Target Mechanism of Solanum surattense against Hepatocellular Carcinoma. Molecules, 27(19), 6220. https://doi.org/10.3390/molecules27196220









