Optimization and Identification of Single Mutation in Hemoglobin Variants with 2,2,2 Trifluoroethanol Modified Digestion Method and Nano−LC Coupled MALDI MS/MS
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Experimental Design of the Study
2.3. Phase I: Strategies Used for Screening and Optimization of Sample Processing Methods for Complete Sequence Coverage in Recombinant Hemoglobins
2.3.1. Recombinant Hemoglobin
2.3.2. Conventional Digestion of rHb
2.3.3. 10% ACN Treatment Prior Trypsin Digestion
2.3.4. Digestion of rHbs by Various Proteases like Trypsin, Glu−C and Lys−C
2.3.5. TFE Treatment for rHbs
2.4. Phase II: Identification of Hb Variants by TFE Method
2.4.1. Ethical Approval and Blood Sample Collection
2.4.2. Sample Processing and Isolation of Pure Hemoglobin
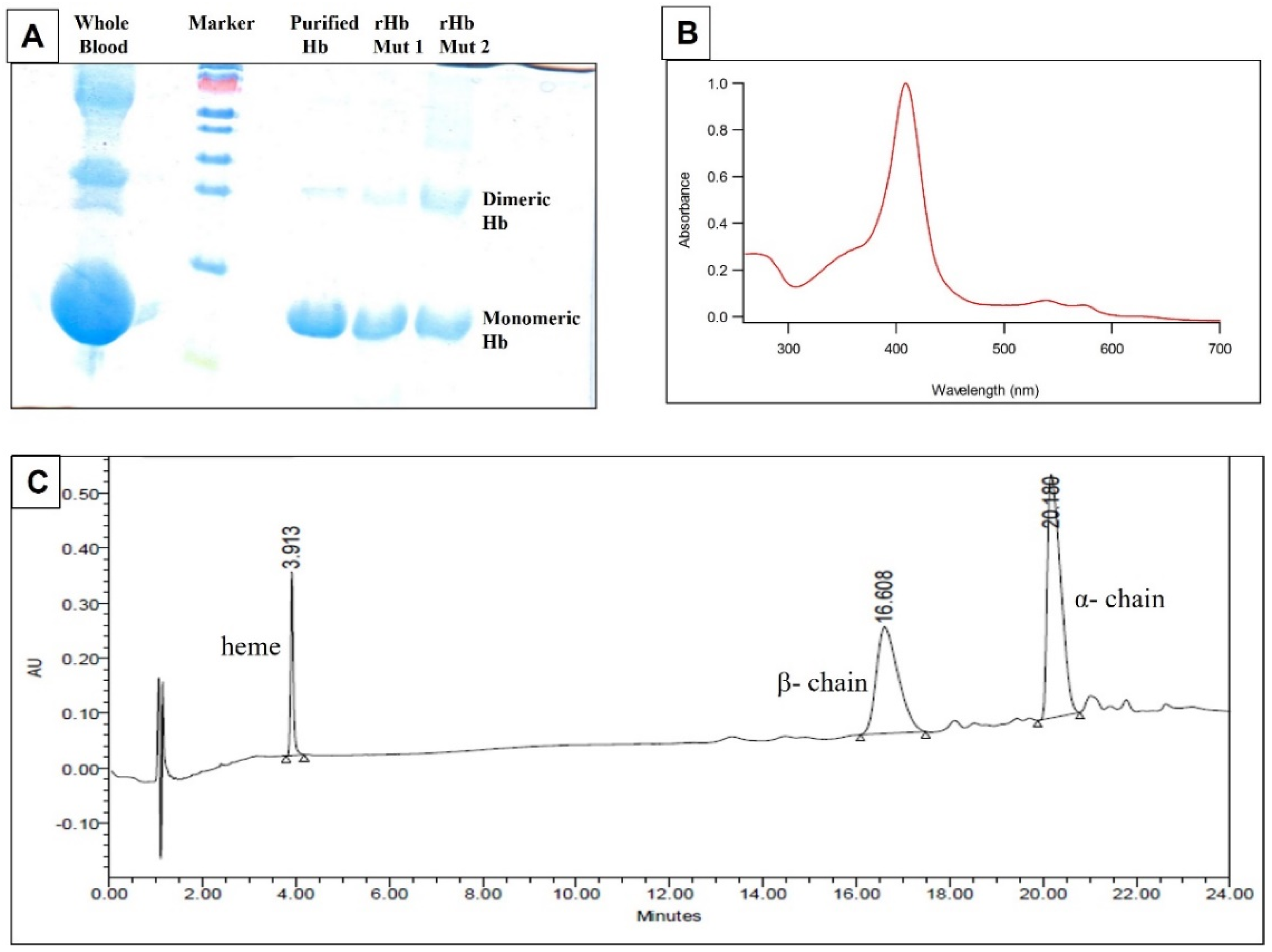
2.4.3. Assessment of Quality of Pure Hb by SDS−PAGE, Absorption Spectroscopy and UPLC
2.4.4. TFE Treatment of Human Hb Variant Samples
2.5. Nano−liquid Chromatography for Separation of Peptides
2.6. MALDI TOF/TOF Analysis
2.7. Database Generation and Searching
3. Results
3.1. Quality and Purity Assessment of Isolated Pure Hb and Recombinant Mutants
3.2. Low Sequence Coverage Was Obtained with the Conventional Method in a Single Spot
3.3. Acetonitrile (10%) Treatment and LC−MALDI Improved the Sequence Coverage Significantly
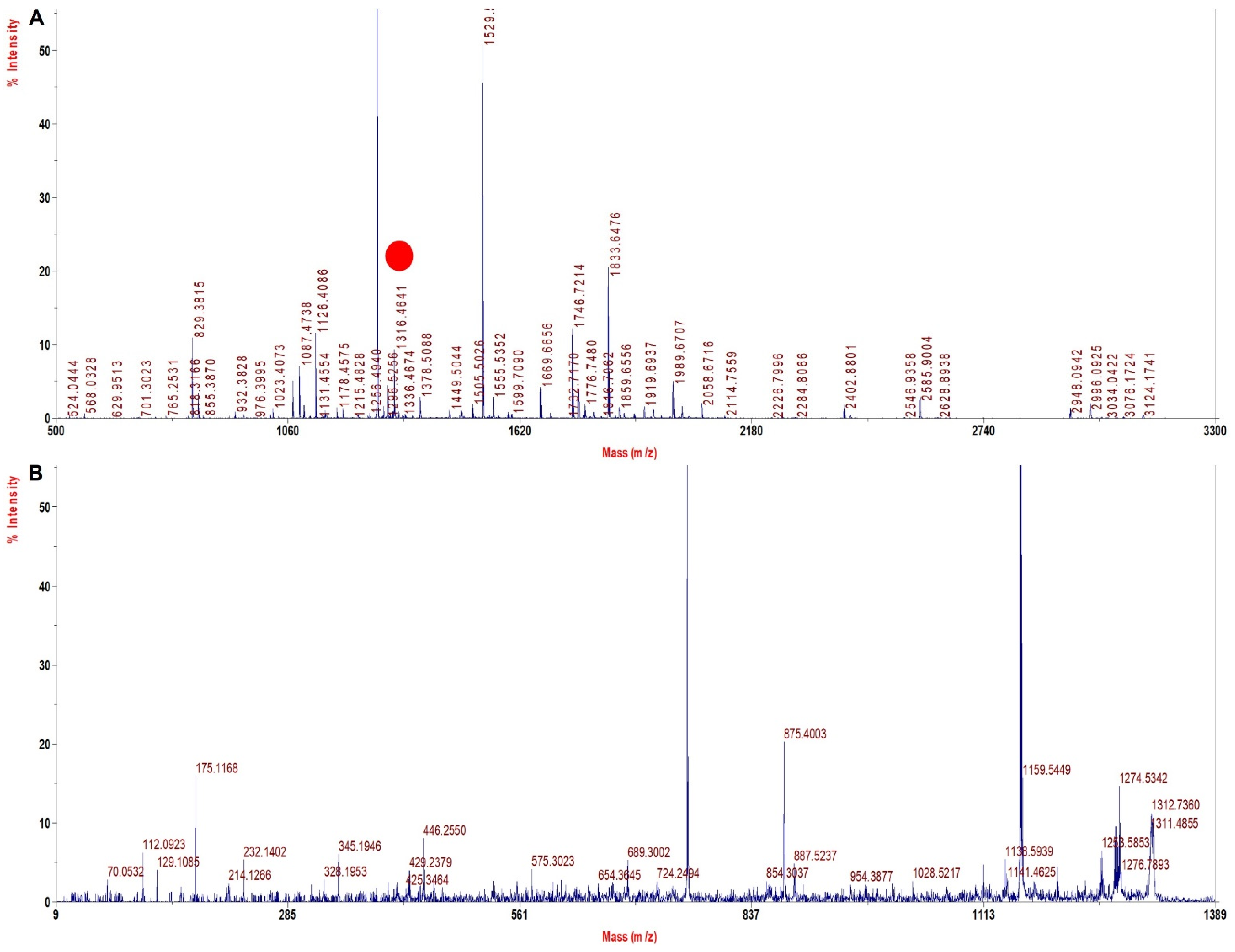
3.4. Digestion by Multiple Proteases Increased the Sequence Coverage up to 100%
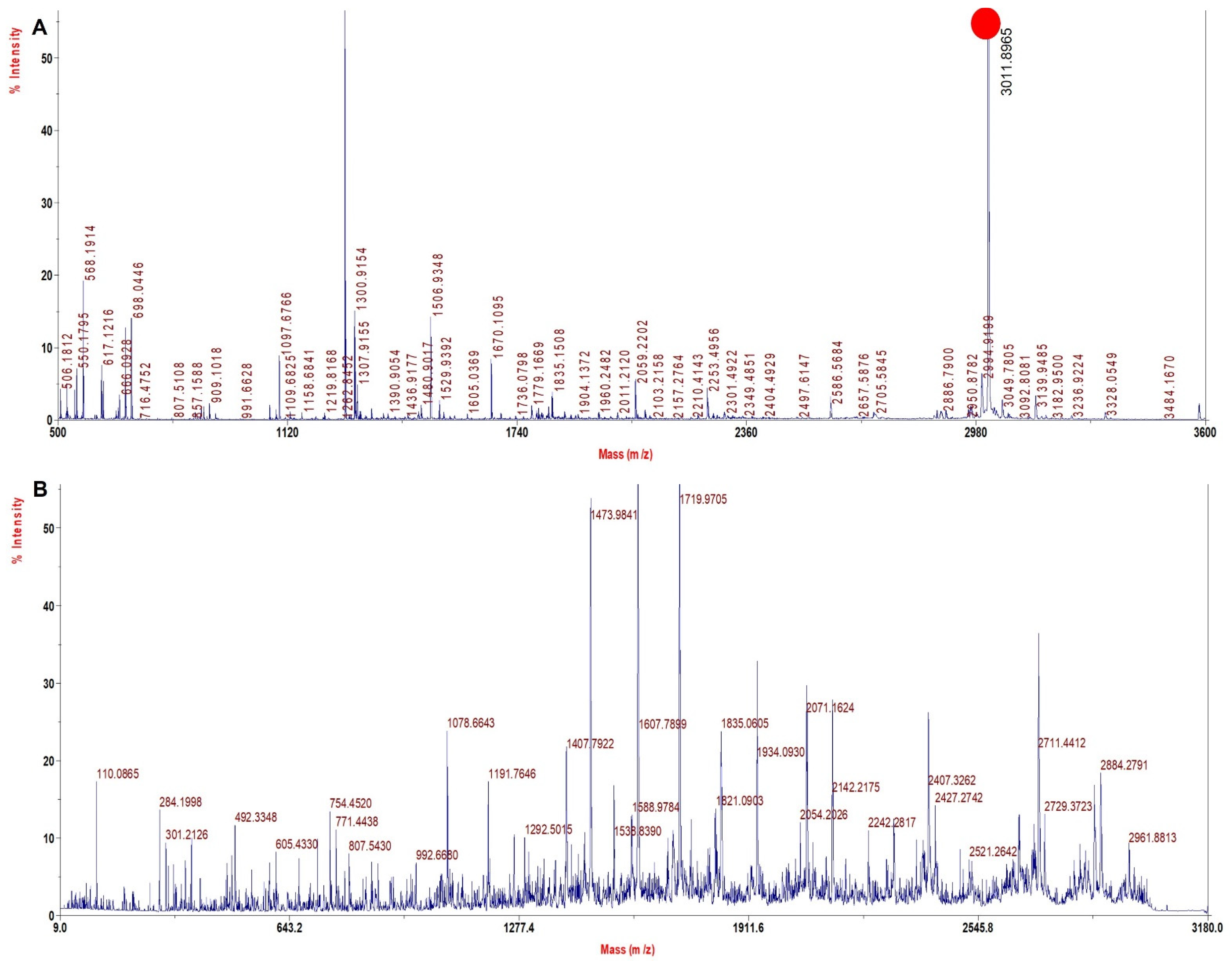
3.5. TFE Treatment Digestion Method Improved Sequence Coverage Significantly in rHb
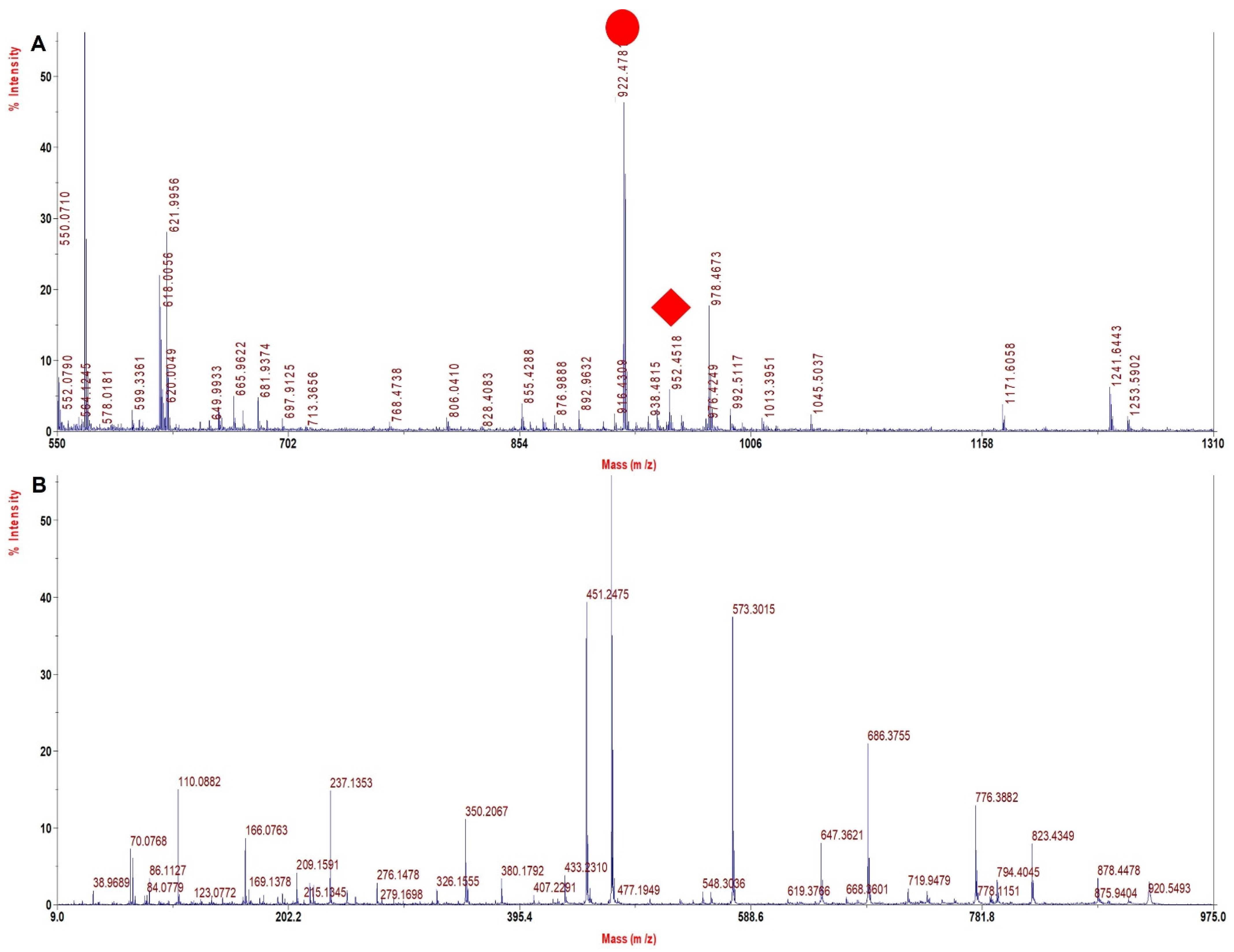
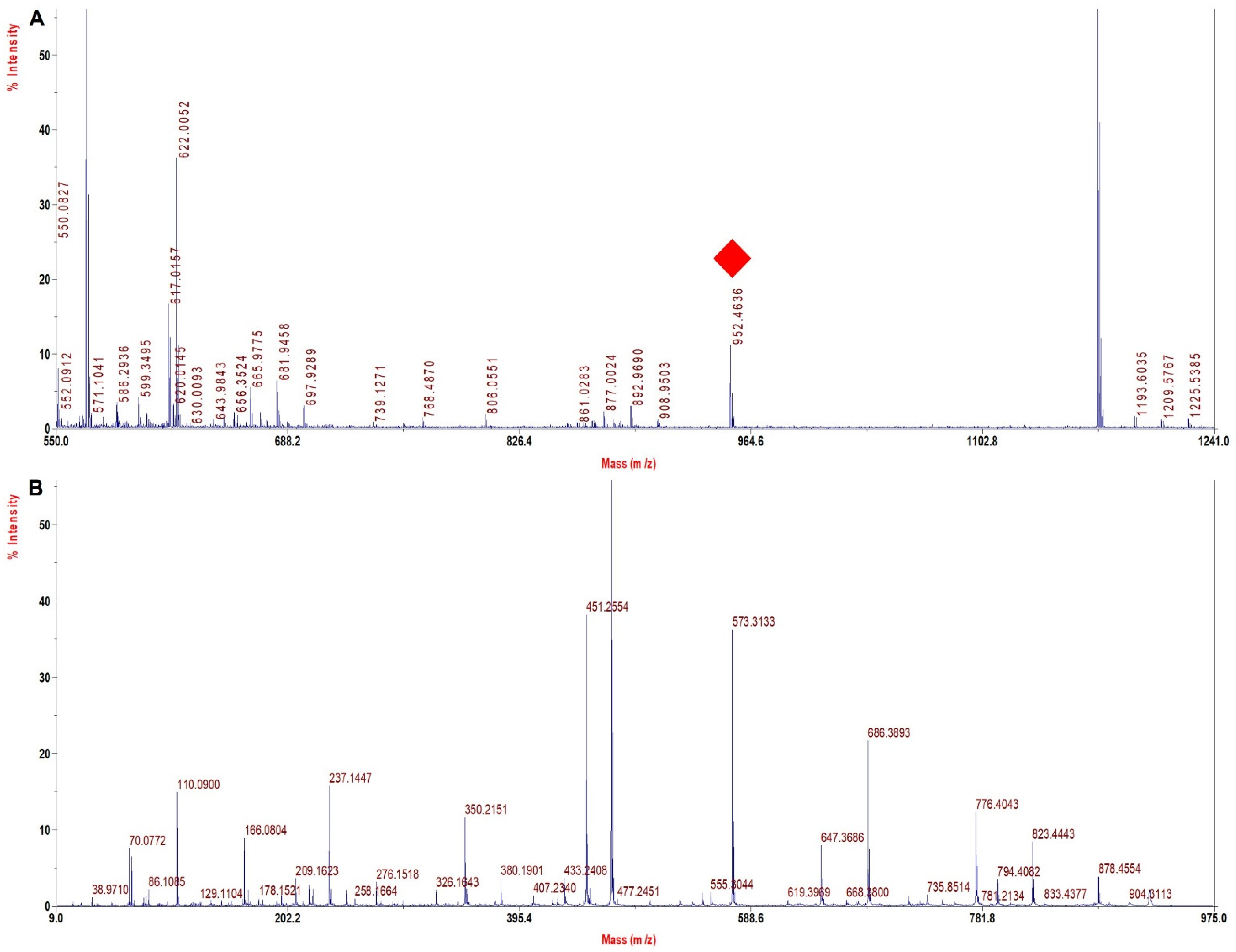
3.6. TFE Modified Digestion Method Improved Sequence Coverage Significantly in Human Hb Variants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Balgir, R.S. The burden of hemoglobinopathies in India and the challenges ahead. Curr. Sci. 2000, 79, 1536–1547. [Google Scholar]
- Chakrabarti, S.; Mandal, K.; Pathak, S.; Patra, A.; Pal, S. Haemoglobinopathies among the tribal and non-tribal antenatal mothers in a tertiary care hospital of rural West Bengal, India. Bangladesh J. Med Sci. 2016, 15, 90–94. [Google Scholar] [CrossRef]
- Weatherall, D.J.; Clegg, J.B. Inherited haemoglobin disorders: An increasing global health problem. Bull. World Health Organ. 2001, 79, 704–712. [Google Scholar]
- Weatherall, D.J. The inherited diseases of hemoglobin are an emerging global health burden. Blood 2010, 115, 4331–4336. [Google Scholar] [CrossRef]
- Ryan, K.; Bain, B.J.; Worthington, D.; James, J.; Plews, D.; Mason, A.; Roper, D.; Rees, D.C.; de la Salle, B.; Streetly, A.; et al. Significant haemoglobinopathies: Guidelines for screening and diagnosis. Br. J. Haematol. 2010, 149, 35–49. [Google Scholar] [CrossRef]
- Weatherall, D.; Akinyanju, O.; Fucharoen, S.; Olivieri, N.; Musgrove, P. Inherited Disorders of Hemoglobin. In Disease Control Prioritites in Developing Countries, 2nd ed.; The International Bank for Reconstruction and Development, The World Bank: Washington, DC, USA, 2006; Chapter 34. [Google Scholar]
- Das, R.; Mitra, G.; Mathew, B.; Bhat, V.; Ross, C.; Pal, D.; Mandal, A.K. Mass Spectrometry-Based Diagnosis of Hemoglobinopa-thies: A Potential Tool for the Screening of Genetic Disorder. Biochem. Genet. 2016, 54, 816–825. [Google Scholar] [CrossRef]
- Ou, C.-N.; Rognerud, C.L. Diagnosis of hemoglobinopathies: Electrophoresis vs. HPLC. Clin. Chim. Acta 2001, 313, 187–194. [Google Scholar] [CrossRef]
- Joutovsky, A.; Hadzi-Nesic, J.; Nardi, M.A. HPLC retention time as a diagnostic tool for hemoglobin variants and hemoglobi-nopathies: A study of 60,000 samples in a clinical diagnostic laboratory. Clin. Chem. 2004, 50, 1736–1747. [Google Scholar] [CrossRef]
- Kleinert, P.; Schmid, M.; Zurbriggen, K.; Speer, O.; Schmugge, M.; Roschitzki, B.; Durka, S.S.; Leopold, U.; Kuster, T.; Heizmann, C.W.; et al. Mass Spectrometry: A Tool for Enhanced Detection of Hemoglobin Variants. Clin. Chem. 2008, 54, 69–76. [Google Scholar] [CrossRef]
- Wada, Y.; Hayashi, A.; Masanori, F.; Katakuse, I.; Ichihara, T.; Nakabushi, H.; Matsuo, T.; Sakurai, T.; Matsuda, H. Characterization of a new fetal hemoglobin variant, Hb F Izumi Aγ6GluåGly, by molecular secondary ion mass spectrometry. Biochim. Biophys. Acta 1983, 749, 244–248. [Google Scholar] [CrossRef]
- Das, R.; Mitra, G.; Mathew, B.; Ross, C.; Bhat, V.; Mandal, A.K. Automated analysis of hemoglobin variants using nanoLC-MS and customized databases. J. Proteome Res. 2013, 12, 3215–3222. [Google Scholar] [CrossRef]
- Tang, N.; Miller, C. An Integrated Approach to Improve Sequence Coverage and Protein Identification by Combining LC-MALDI MS/MS and Nano-LC/MS/MS. In Proceedings of the Agilent Technologies, 53 rd ASMS Conference on Mass Spectrometry, San Antonio, TX, USA, 5–9 June 2005. [Google Scholar]
- Biringer, R.G.; Amato, H.; Harrington, M.G.; Fonteh, A.N.; Riggins, J.N.; Hühmer, A.F.R. Enhanced sequence coverage of proteins in human cerebrospinal fluid using multiple enzymatic digestion and linear ion trap LC-MS/MS. Briefings Funct. Genom. Proteom. 2006, 5, 144–153. [Google Scholar] [CrossRef]
- Giansanti, P.; Tsiatsiani, L.; Low, T.Y.; Heck, A.J.R. Six alternative proteases for mass spectrometry–based proteomics beyond trypsin. Nat. Protoc. 2016, 11, 993–1006. [Google Scholar] [CrossRef]
- Proc, J.L.; Kuzyk, M.A.; Hardie, D.B.; Yang, J.; Smith, D.S.; Jackson, A.M.; Parker, C.E.; Borchers, C.H. A Quantitative Study of the Effects of Chaotropic Agents, Surfactants, and Solvents on the Digestion Efficiency of Human Plasma Proteins by Trypsin. J. Proteome Res. 2010, 9, 5422–5437. [Google Scholar] [CrossRef]
- Meza, J.E.; Miller, C.A.; Fischer, S.M. Improved Tryptic Digestion of Proteins Using 2,2,2-Trifluoroethanol (TFE) Poster–ABRF; Agilent Technologies, Inc.: Santa Clara, CA, USA, 2004. [Google Scholar]
- Strader, M.B.; Tabb, D.L.; Hervey, W.J.; Pan, C.; Hurst, G.B. Efficient and Specific Trypsin Digestion of Microgram to Nanogram Quantities of Proteins in Organic−Aqueous Solvent Systems. Anal. Chem. 2005, 78, 125–134. [Google Scholar] [CrossRef]
- Russell, W.K.; Park, Z.-Y.; Russell, D.H. Proteolysis in Mixed Organic−Aqueous Solvent Systems: Applications for Peptide Mass Mapping Using Mass Spectrometry. Anal. Chem. 2001, 73, 2682–2685. [Google Scholar] [CrossRef]
- Alam, A.; Mataj, A.; Yang, Y.; Boysen, R.I.; Bowden, D.K.; Hearn, M.T.W. Rapid Microwave-Assisted Chemical Cleavage—Mass Spectrometric Method for the Identification of Hemoglobin Variants in Blood. Anal. Chem. 2010, 82, 8922–8930. [Google Scholar] [CrossRef]
- Cole, R.D.; Stein, W.H.; Moore, S. On the Cysteine Content of Human Hemoglobin. J. Biol. Chem. 1958, 233, 1359–1363. [Google Scholar] [CrossRef]
- Liu, K.Z.; Tsang, K.S.; Li, C.K.; Shaw, R.A.; Mantsch, H.H. Infrared spectroscopic identification of beta-thalassemia. Clin. Chem. 2003, 49, 1125–1132. [Google Scholar] [CrossRef]
- Saraswathi, M.; Nakanishi, T.; Shimizu, A. Relative quantification of glycated Cu-Zn superoxide dismutase in erythrocytes by electrospray ionization mass spectrometry. Biochim. Biophys. Acta 1999, 1426, 483–490. [Google Scholar] [CrossRef]
- Bhargava, M.; Viken, K.J.; Dey, S.; Steinbach, M.S.; Wu, B.; Jagtap, P.D.; Higgins, L.; Panoskaltsis-Mortari, A.; Weisdorf, D.J.; Kumar, V.; et al. Proteome Profiling in Lung Injury after Hematopoietic Stem Cell Transplantation. Biol. Blood Marrow Transplant. 2016, 22, 1383–1390. [Google Scholar] [CrossRef]
- Laing, R.W.; Bhogal, R.H.; Wallace, L.; Boteon, Y.; Neil, D.A.H.; Smith, A.; Stephenson, B.T.F.; Schlegel, A.; Hübscher, S.G.; Mirza, D.F.; et al. The Use of an Acellular Oxygen Carrier in a Human Liver Model of Normothermic Machine Perfusion. Transplantation 2017, 101, 2746–2756. [Google Scholar] [CrossRef]
- Carvalho, A.S.; Rodriguez, M.S.; Matthiesen, R. Red Blood Cells in Clinical Proteomics. Methods Mol. Biol. 2017, 1619, 173–181. [Google Scholar] [CrossRef]
- Wajcman, H. Analysis of Hemoglobins and Globin Chains by High-Performance Liquid Chromatography. Methods Mol. Biol. 2003, 82, 21–29. [Google Scholar] [CrossRef]
- Hardison, R.C.; Chui, D.H.; Giardine, B.; Riemer, C.; Patrinos, G.P.; Anagnou, N.; Miller, W.; Wajcman, H. HbVar: A relational database of human hemoglobin variants and thalassemia mutations at the globin gene server. Hum. Mutat. 2002, 19, 225–233. [Google Scholar] [CrossRef]
- Drapeau, G.R.; Boily, Y.; Houmard, J. Purification and Properties of an Extracellular Protease of Staphylococcus aureus. J. Biol. Chem. 1972, 247, 6720–6726. [Google Scholar] [CrossRef]
- Edwards, R.L.; Creese, A.J.; Baumert, M.; Griffiths, P.; Bunch, J.; Cooper, H.J. Hemoglobin Variant Analysis via Direct Surface Sampling of Dried Blood Spots Coupled with High-Resolution Mass Spectrometry. Anal. Chem. 2011, 83, 2265–2270. [Google Scholar] [CrossRef]
- Hachani, J.; Duban-Deweer, S.; Pottiez, G.; Renom, G.; Flahaut, C.; Périni, J.-M. MALDI-TOF MS profiling as the first-tier screen for sickle cell disease in neonates: Matching throughput to objectives. Proteom. Clin. Appl. 2011, 5, 405–414. [Google Scholar] [CrossRef]
- Chen, L.; Wang, N.; Li, L. Development of microwave-assisted acid hydrolysis of proteins using a commercial microwave reactor and its combination with LC–MS for protein full-sequence analysis. Talanta 2014, 129, 290–295. [Google Scholar] [CrossRef]
- Zubarev, R.A.; Horn, D.M.; Fridriksson, E.K.; Kelleher, N.L.; Kruger, N.A.; Lewis, M.A.; Carpenter, B.K.; McLafferty, F.W. Electron Capture Dissociation for Structural Characterization of Multiply Charged Protein Cations. Anal. Chem. 2000, 72, 563–573. [Google Scholar] [CrossRef]
- Syka, J.E.P.; Coon, J.J.; Schroeder, M.J.; Shabanowitz, J.; Hunt, D.F. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA 2004, 101, 9528–9533. [Google Scholar] [CrossRef]
- Edwards, R.L.; Martin, N.J.; Cooper, H.J. Hemoglobin variant analysis of whole blood and dried blood spots by MS. Bioanalysis 2013, 5, 2043–2052. [Google Scholar] [CrossRef]
- Edwards, R.L.; Griffiths, P.; Bunch, J.; Cooper, H.J. Top-Down Proteomics and Direct Surface Sampling of Neonatal Dried Blood Spots: Diagnosis of Unknown Hemoglobin Variants. J. Am. Soc. Mass Spectrom. 2012, 23, 1921–1930. [Google Scholar] [CrossRef]
- Graça, D.C.; Hartmer, R.; Jabs, W.; Beris, P.; Clerici, L.; Stoermer, C.; Samii, K.; Hochstrasser, D.; Tsybin, Y.O.; Scherl, A.; et al. Identification of hemoglobin variants by top-down mass spectrometry using selected diagnostic product ions. Anal. Bioanal. Chem. 2015, 407, 2837–2845. [Google Scholar] [CrossRef]
- Daniel, Y.A.; Turner, C.; Haynes, R.M.; Hunt, B.J.; Dalton, R.N. Rapid and specific detection of clinically significant haemoglo-binopathies using electrospray mass spectrometry-mass spectrometry. Br. J. Haematol. 2005, 130, 635–643. [Google Scholar] [CrossRef]
- Boemer, F.; Cornet, Y.; Libioulle, C.; Segers, K.; Bours, V.; Schoos, R. 3-years experience review of neonatal screening for hemo-globin disorders using tandem mass spectrometry. Clin. Chim. Acta 2011, 412, 1476–1479. [Google Scholar] [CrossRef]
- Blackburn, K.; Goshe, M.B. Challenges and strategies for targeted phosphorylation site identification and quantification using mass spectrometry analysis. Briefings Funct. Genom. Proteom. 2008, 8, 90–103. [Google Scholar] [CrossRef]
- Grote, E.; Fu, Q.; Ji, W.; Liu, X.; Van Eyk, J.E. Using Pure Protein to Build a Multiple Reaction Monitoring Mass Spectrometry Assay for Targeted Detection and Quantitation. Methods Mol. Biol. 2013, 1005, 199–213. [Google Scholar] [CrossRef]
- Bird, G.H.; Shin, J.A. MALDI-TOF mass spectrometry characterization of recombinant hydrophobic mutants containing the GCN4 basic region/leucine zipper motif. Biochim. Biophys. Acta 2002, 1597, 252–259. [Google Scholar] [CrossRef]
- Wheeler, A.R.; Moon, H.; Bird, C.A.; Loo, R.R.; Kim, C.J.; Loo, J.A.; Garrell, R.L. Digital microfluidics with in-line sample purifi-cation for proteomics analyses with MALDI-MS. Anal. Chem. 2005, 77, 534–540. [Google Scholar] [CrossRef]
- Xu, S.; Ye, M.; Xu, D.; Li, X.; Pan, A.C.; Zou, H. Matrix with High Salt Tolerance for the Analysis of Peptide and Protein Samples by Desorption/Ionization Time-of-Flight Mass Spectrometry. Anal. Chem. 2006, 78, 2593–2599. [Google Scholar] [CrossRef]
- Gekko, K.; Ohmae, E.; Kameyama, K.; Takagi, T. Acetonitrile-protein interactions: Amino acid solubility and preferential solva-tion. Biochim. Biophys. Acta 1998, 1387, 195–205. [Google Scholar] [CrossRef]
- Waas, M.; Bhattacharya, S.; Chuppa, S.; Wu, X.; Jensen, D.R.; Omasits, U.; Wollscheid, B.; Volkman, B.F.; Noon, K.R.; Gundry, R.L. Combine and conquer: Surfactants, solvents, and chaotropes for robust mass spectrometry based analyses of membrane pro-teins. Anal. Chem. 2014, 86, 1551–1559. [Google Scholar] [CrossRef]
- Meza, J.E.; Miller, C.A.; Fischer, S.M. Excellence in Microfluidics: The Effect of Denaturing Agents on Protein Identification by Mass Spectrometry; ABRF: Savannah, GA, USA; Palo Alto, CA, USA, 2005; p. P142-S. [Google Scholar]
| Sample Name | Sequence Coverage (%) | |
|---|---|---|
| Alpha | Beta | |
| rHb Control | 88.0 | 96.6 |
| rHb Mut 1 β 63 (H > G) | 82.4 | 97.3 |
| rHb Mut 2 β 28 (L > F) | 70.4 | 93.9 |
| rHb Mut 3 β 67 (V > A) | 69.0 | 79.6 |
| rHb Mut 4 α 62 (V > N) | 79.6 | 82.3 |
| Mutation Name | Sequence Coverage LC−MALDI (ACN) (%) | Mutation Identified Trypsin/Glu−C/Lys−C | |||||
|---|---|---|---|---|---|---|---|
| Trypsin | Glu−C | Lys−C | |||||
| Alpha | Beta | Alpha | Beta | Alpha | Beta | ||
| rHb Control | 89.4 | 99.3 | 31.7 | 91.2 | 96.5 | 93.9 | |
| rHb Mut 1 β 63 (H > G) | 85.9 | 100 | 85.9 | 100 | 91.5 | 95.9 | √/−/− |
| rHb Mut 2 β 28 (L > F) | 54.9 | 86.4 | 74.6 | 100 | 96.5 | 98.6 | √/√/− |
| rHb Mut 3 β 67 (V > A) | 69.0 | 79.6 | 92.3 | 87.8 | 90.8 | 98.6 | √/−/− |
| rHb Mut 4 α 62 (V > N) | 79.6 | 82.3 | 66.2 | 85.7 | 96.5 | 95.9 | √/−/− |
| Human Hb Control | 99.3 | 99.3 | 76.8 | 99.3 | 88.7 | 98.0 | |
| Sample Name | Sequence Coverage (%) | |||||
|---|---|---|---|---|---|---|
| Trypsin + Glu−C Combined | Glu−C + Lys−C Combined | Lys−C + Trypsin Combined | ||||
| Chain | α | β | α | β | α | β |
| Human Hb Control | 100 | 100 | 100 | 100 | 100 | 100 |
| rHb Control | 80.3 | 96.6 | 96.5 | 97.3 | 100 | 95.2 |
| rHb Mutant 1 | 94.4 | 100 | 100 | 100 | 91.5 | 97.3 |
| rHb Mutant 2 | 93.0 | 100 | 100 | 100 | 100 | 100 |
| rHb Mutant 3 | 92.3 | 100 | 100 | 100 | 100 | 100 |
| rHb Mutant 4 | 88.7 | 100 | 100 | 100 | 100 | 100 |
| Mutation Name | Sequence Coverage (%) | Mutation Identified | |
|---|---|---|---|
| Alpha | Beta | ||
| rHb Control | 97.3 | 99.3 | − |
| rHb Mut 1 β 63 (H > G) | 97.9 | 100 | √ |
| rHb Mut 2 β 28 (L > F) | 99.3 | 99.3 | √ |
| rHb Mut 3 β 67 (V > A) | 97.9 | 99.3 | √ |
| rHb Mut 4 α 62 (V > N) | 99.3 | 99.3 | √ |
| Human Hb Control | 99.3 | 99.3 | − |
| HbS Heterozygous β 6 (E > V) (1908) | 96.6 | 100 | √ |
| HbS Homozygous β 6 (E > V) (1909) | 70.4 | 100 | √ |
| HbE β 26 (E > K) (1963) | 99.3 | 100 | √ |
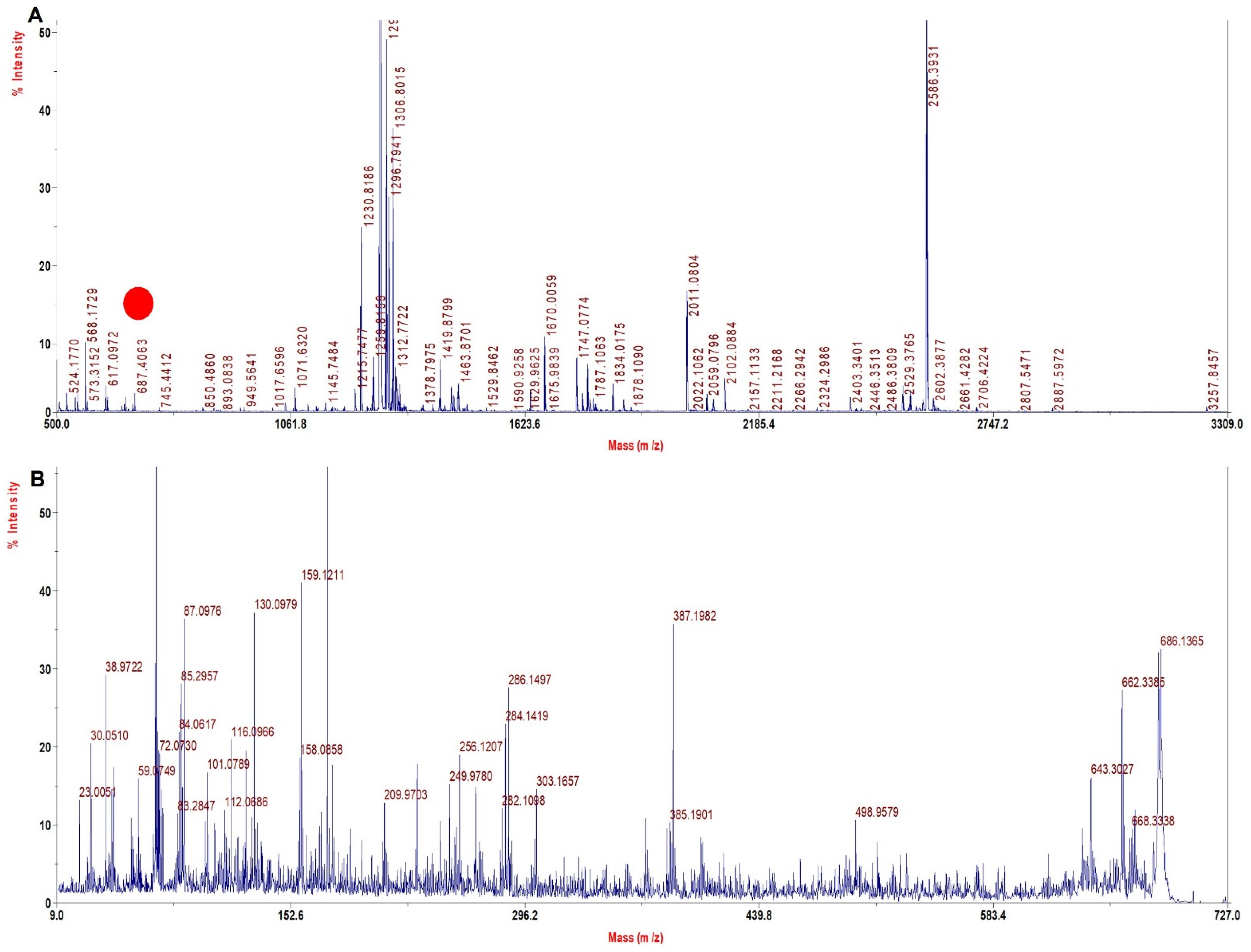
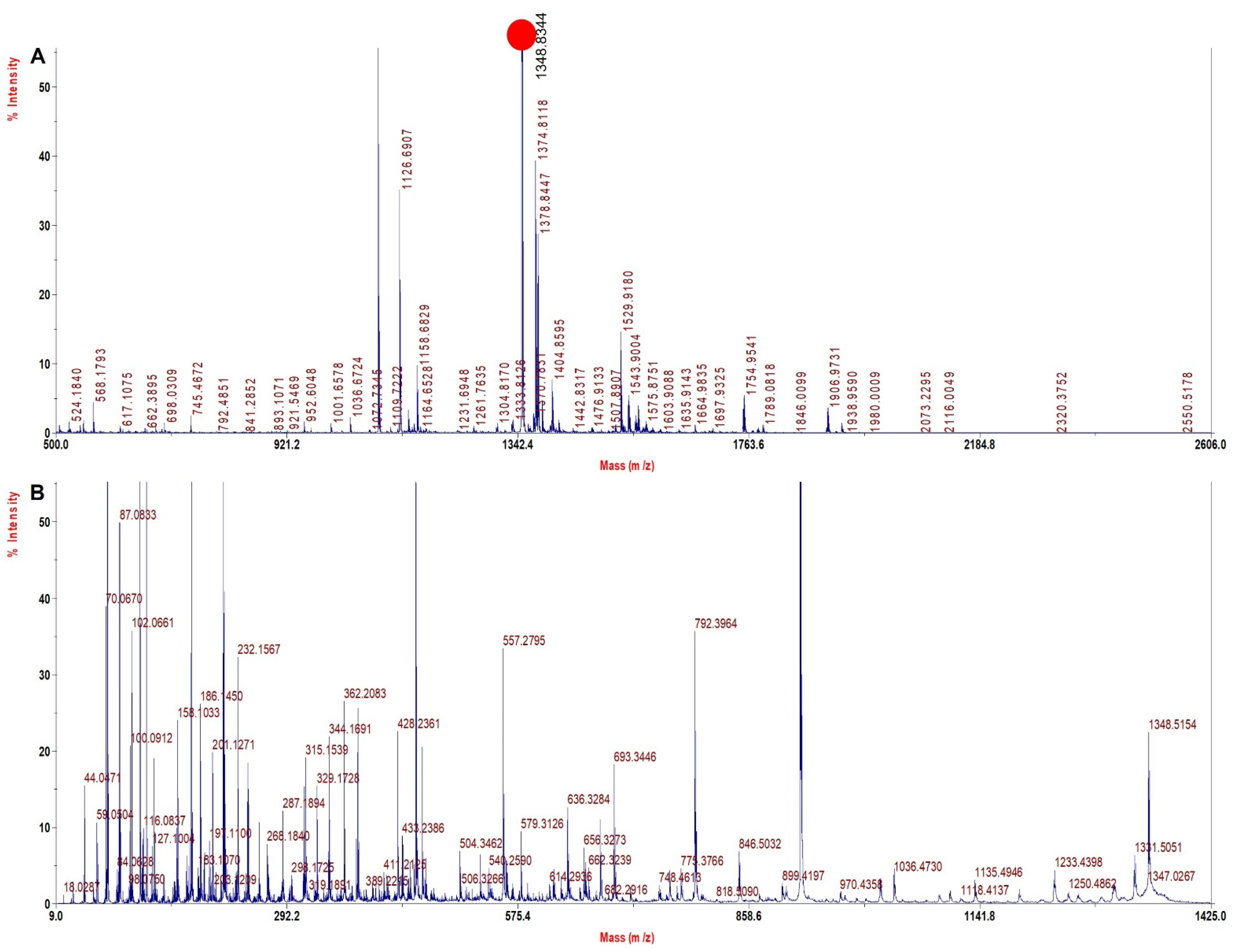
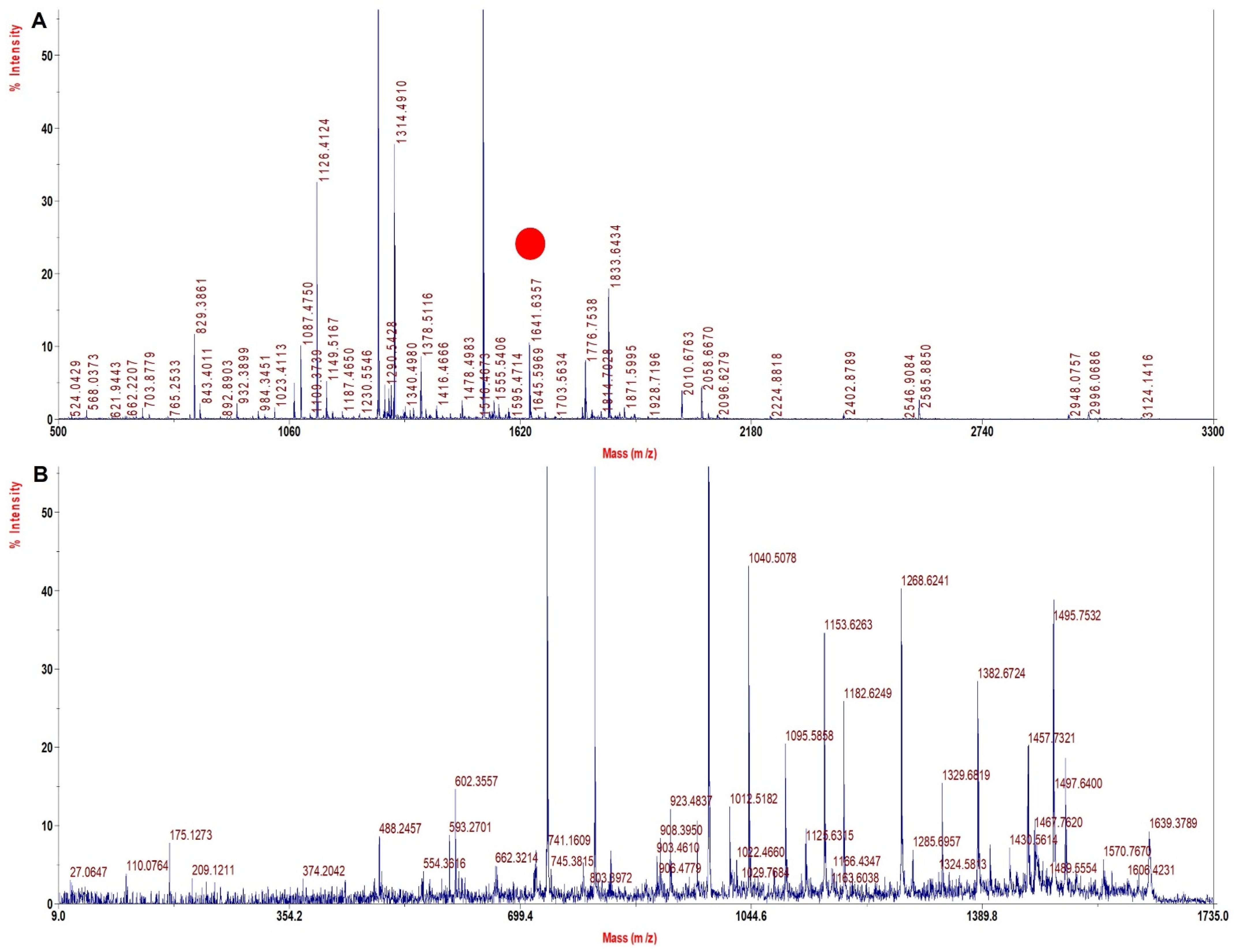
| Sample | Wild Type Precursor Ion MH+ | Mutated Precursor Ion MH+ | Mass Difference (Dalton) | Tryptic Signature Peptide | Inference |
|---|---|---|---|---|---|
| rHb Mut 1 | 765.2 | 687.9 | −80 | VKAGGKK | β 63 His > Gly |
| rHb Mut 2 | 1314.4 | 1348.4 | +34 | VNVDEVGGEAFGR | β 28 Leu > Phe |
| rHb Mut 3 | 1669.6 | 1641.6 | −28 | ALGAFSDGLAHLDNLK | β 67 Val > Ala |
| rHb Mut 4 | 2995.5 | 3010.5 | +15 | NADALTNAVAHVDDMPNALSALSDLHAHK | α 62 Val > Asn |
| HbS hetero | 952.4 | 922.4 | −30 | VHLTPVEK | β 6 Glu > Val |
| HbS homo | 952.4 | 922.4 | −30 | VHLTPVEK | β 6 Glu > Val |
| HbE | 1316.4 | 1315.5 | −1 | VNVDEVGGKALGR | β 26 Glu > Lys |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dasauni, P.; Singh, N.; Chhabra, V.; Mahapatra, M.; Saxena, R.; Kundu, S. Optimization and Identification of Single Mutation in Hemoglobin Variants with 2,2,2 Trifluoroethanol Modified Digestion Method and Nano−LC Coupled MALDI MS/MS. Molecules 2022, 27, 6357. https://doi.org/10.3390/molecules27196357
Dasauni P, Singh N, Chhabra V, Mahapatra M, Saxena R, Kundu S. Optimization and Identification of Single Mutation in Hemoglobin Variants with 2,2,2 Trifluoroethanol Modified Digestion Method and Nano−LC Coupled MALDI MS/MS. Molecules. 2022; 27(19):6357. https://doi.org/10.3390/molecules27196357
Chicago/Turabian StyleDasauni, Pushpanjali, Nirpendra Singh, Varun Chhabra, Manoranjan Mahapatra, Renu Saxena, and Suman Kundu. 2022. "Optimization and Identification of Single Mutation in Hemoglobin Variants with 2,2,2 Trifluoroethanol Modified Digestion Method and Nano−LC Coupled MALDI MS/MS" Molecules 27, no. 19: 6357. https://doi.org/10.3390/molecules27196357
APA StyleDasauni, P., Singh, N., Chhabra, V., Mahapatra, M., Saxena, R., & Kundu, S. (2022). Optimization and Identification of Single Mutation in Hemoglobin Variants with 2,2,2 Trifluoroethanol Modified Digestion Method and Nano−LC Coupled MALDI MS/MS. Molecules, 27(19), 6357. https://doi.org/10.3390/molecules27196357







