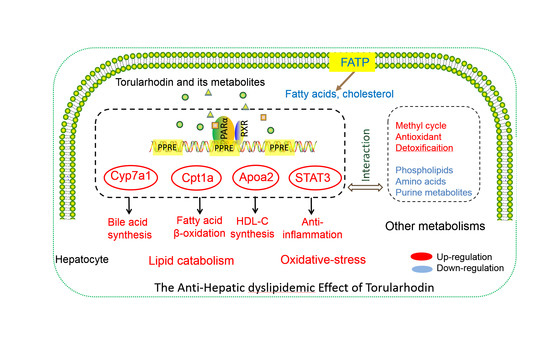
Torularhodin Alleviates Hepatic Dyslipidemia and Inflammations in High-Fat Diet-Induced Obese Mice via PPARα Signaling Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Animal and Treatment
2.3. Phenotype Parameter Analysis
2.4. Proteomics Experiment
2.4.1. Hepatic Protein Sample Preparation
2.4.2. Peptide Desalination
2.4.3. HPLC-Mass Spectrometry
2.4.4. Mascot Qualitative-Quantitative Analysis
2.5. Hepatic Metabolite Analysis
2.6. Western Blotting Analysis
2.7. Statistical and Bioinformatics Analysis
3. Results
3.1. Anti-Obesity and Anti-Hepatic Dyslipidemia Effect of Torularhodin
3.2. Proteomics and Metabolite Profiling
3.2.1. Lipid Metabolism-Related Hepatic DEPs
3.2.2. Other Metabolic Processes-Related Hepatic DEPs
3.2.3. Hepatic Metabolite Profiling
3.3. The Correlation between Phenotypic Parameters and Hepatic Proteins/Metabolites
3.4. Anti-Hepatic Inflammation by Torularhodin
3.5. Hepatic Protein-Protein Interaction
3.6. KEGG Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Méndez-Sánchez, N.; Bugianesi, E.; Gish, R.G.; Lammert, F.; Tilg, H.; Nguyen, M.H.; Sarin, S.K.; Fabrellas, N.; Zelber-Sagi, S.; Fan, J.G.; et al. Global multi-stakeholder endorsement of the MAFLD definition. Lancet. Gastroenterol. Hepatol. 2022, 7, 388–390. [Google Scholar] [CrossRef]
- Sommerauer, C.; Kutter, C. Noncoding RNAs and RNA binding proteins—Emerging governors of liver physiology and metabolic diseases. Am. J. Physiol. Cell Physiol. 2022. [Google Scholar] [CrossRef]
- Patil, R.; Sood, G.K. Non-alcoholic fatty liver disease and cardiovascular risk. World J. Gastrointest. Pathophysiol. 2017, 8, 51–58. [Google Scholar] [CrossRef]
- Eggersdorfer, M.; Wyss, A. Carotenoids in human nutrition and health. Arch. Biochem. Biophys. 2018, 652, 18–26. [Google Scholar] [CrossRef]
- Yao, N.; Yan, S.; Guo, Y.; Wang, H.; Li, X.; Wang, L.; Hu, W.; Li, B.; Cui, W. The association between carotenoids and subjects with overweight or obesity: A systematic review and meta-analysis. Food Funct 2021, 12, 4768–4782. [Google Scholar] [CrossRef]
- Clugston, R.D. Carotenoids and fatty liver disease: Current knowledge and research gaps. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2020, 1865, 158597. [Google Scholar] [CrossRef]
- Kot, A.M.; Błażejak, S.; Gientka, I.; Kieliszek, M.; Bryś, J. Torulene and torularhodin: “new” fungal carotenoids for industry? Microb. Cell Factories 2018, 17, 49. [Google Scholar] [CrossRef]
- Sakaki, H.; Nakanishi, T.; Komemushi, S.; Namikawa, K.; Miki, W. Torularhodin as a Potent Scavenger against Peroxyl Radicals Isolated from a Soil Yeast, Rhodotorula glutinis. J. Clin. Biochem. Nutr. 2001, 30, 1–10. [Google Scholar] [CrossRef]
- Zhang, W.; Hua, H.; Guo, Y.; Cheng, Y.; Pi, F.; Yao, W.; Xie, Y.; Qian, H. Torularhodin from Sporidiobolus pararoseus Attenuates d-galactose/AlCl(3)-Induced Cognitive Impairment, Oxidative Stress, and Neuroinflammation via the Nrf2/NF-κB Pathway. J. Agric. Food Chem. 2020, 68, 6604–6614. [Google Scholar] [CrossRef]
- Du, C.; Li, Y.; Guo, Y.; Han, M.; Zhang, W.; Qian, H. Torularhodin, isolated from Sporidiobolus pararoseus, inhibits human prostate cancer LNCaP and PC-3 cell growth through Bcl-2/Bax mediated apoptosis and AR down-regulation. RSC Adv. 2015, 5, 106387–106395. [Google Scholar] [CrossRef]
- Li, J.; Liu, C.; Guo, Y.; Pi, F.; Yao, W.; Xie, Y.; Cheng, Y.; Qian, H. Determination of the effects of torularhodin against alcoholic liver diseases by transcriptome analysis. Free Radic. Biol. Med. 2019, 143, 47–54. [Google Scholar] [CrossRef]
- Liu, C.; Cui, Y.; Pi, F.; Guo, Y.; Cheng, Y.; Qian, H. Torularhodin Ameliorates Oxidative Activity in Vitro and d-Galactose-Induced Liver Injury via the Nrf2/HO-1 Signaling Pathway in Vivo. J. Agric. Food Chem. 2019, 67, 10059–10068. [Google Scholar] [CrossRef]
- Zhang, D.; Zheng, W.; Li, X.; Liang, G.; Ye, N.; Liu, Y.; Li, A.; Liu, X.; Zhang, R.; Cheng, J.; et al. Investigation of Obesity-Alleviation Effect of Eurycoma longifolia on Mice Fed with a High-Fat Diet through Metabolomics Revealed Enhanced Decomposition and Inhibition of Accumulation of Lipids. J. Proteome Res. 2021, 20, 2714–2724. [Google Scholar] [CrossRef]
- Hu, S.; Li, P.; Zhang, R.; Liu, X.; Wei, S. Integrated metabolomics and proteomics analysis reveals energy metabolism disorders in the livers of sleep-deprived mice. J. Proteom. 2021, 245, 104290. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Tsai, T.H.; Chen, E.; Li, L.; Saha, P.; Lee, H.J.; Huang, L.S.; Shelness, G.S.; Chan, L.; Chang, B.H. The constitutive lipid droplet protein PLIN2 regulates autophagy in liver. Autophagy 2017, 13, 1130–1144. [Google Scholar] [CrossRef]
- Feng, Q.; Kalari, K.; Fridley, B.L.; Jenkins, G.; Ji, Y.; Abo, R.; Hebbring, S.; Zhang, J.; Nye, M.D.; Leeder, J.S.; et al. Betaine-homocysteine methyltransferase: Human liver genotype-phenotype correlation. Mol. Genet. Metab. 2011, 102, 126–133. [Google Scholar] [CrossRef]
- Nicolaou, M.; Andress, E.J.; Zolnerciks, J.K.; Dixon, P.H.; Williamson, C.; Linton, K.J. Canalicular ABC transporters and liver disease. J. Pathol. 2012, 226, 300–315. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, J.-K.; Kim, H. ABCB7 simultaneously regulates apoptotic and non-apoptotic cell death by modulating mitochondrial ROS and HIF1α-driven NFκB signaling. Oncogene 2020, 39, 1969–1982. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, J.H.; Noh, S.; Hur, H.J.; Sung, M.J.; Hwang, J.T.; Park, J.H.; Yang, H.J.; Kim, M.S.; Kwon, D.Y.; et al. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. J. Proteome Res. 2011, 10, 722–731. [Google Scholar] [CrossRef]
- Yuan, W.; Yu, B.; Yu, M.; Kuai, R.; Morin, E.E.; Wang, H.; Hu, D.; Zhang, J.; Moon, J.J.; Chen, Y.E.; et al. Synthetic high-density lipoproteins delivering liver X receptor agonist prevent atherogenesis by enhancing reverse cholesterol transport. J. Control. Release Off. J. Control. Release Soc. 2021, 329, 361–371. [Google Scholar] [CrossRef]
- Soran, H.; Schofield, J.D.; Durrington, P.N. Antioxidant properties of HDL. Front Pharm. 2015, 6, 222. [Google Scholar] [CrossRef]
- Xu, Y.; Li, Y.; Jadhav, K.; Pan, X.; Zhu, Y.; Hu, S.; Chen, S.; Chen, L.; Tang, Y.; Wang, H.H.; et al. Hepatocyte ATF3 protects against atherosclerosis by regulating HDL and bile acid metabolism. Nat. Metab. 2021, 3, 59–74. [Google Scholar] [CrossRef]
- Lu, M.; Lu, Q.; Zhang, Y.; Tian, G. ApoB/apoA1 is an effective predictor of coronary heart disease risk in overweight and obesity. J. Biomed. Res. 2011, 25, 266–273. [Google Scholar] [CrossRef]
- Hrzenjak, A.; Frank, S.; Wo, X.; Zhou, Y.; Van Berkel, T.; Kostner, G.M. Galactose-specific asialoglycoprotein receptor is involved in lipoprotein (a) catabolism. Biochem. J. 2003, 376 Pt 3, 765–771. [Google Scholar] [CrossRef]
- Narvekar, P.; Berriel Diaz, M.; Krones-Herzig, A.; Hardeland, U.; Strzoda, D.; Stöhr, S.; Frohme, M.; Herzig, S. Liver-Specific Loss of Lipolysis-Stimulated Lipoprotein Receptor Triggers Systemic Hyperlipidemia in Mice. Diabetes 2009, 58, 1040–1049. [Google Scholar] [CrossRef]
- Zhang, Y.-N.; Wang, Q.-Q.; Chen, Y.-S.; Shen, C.; Xu, C.-F. Association between Serum Uric Acid to HDL-Cholesterol Ratio and Nonalcoholic Fatty Liver Disease in Lean Chinese Adults. Int. J. Endocrinol. 2020, 2020, 5953461. [Google Scholar] [CrossRef]
- Reilly, S.M.; Saltiel, A.R. Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 2017, 13, 633–643. [Google Scholar] [CrossRef]
- Gross, B.; Pawlak, M.; Lefebvre, P.; Staels, B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat. Rev. Endocrinol. 2017, 13, 36–49. [Google Scholar] [CrossRef]
- Montaigne, D.; Butruille, L.; Staels, B. PPAR control of metabolism and cardiovascular functions. Nat. Rev. Cardiol. 2021, 18, 809–823. [Google Scholar] [CrossRef]
- Pawlak, M.; Lefebvre, P.; Staels, B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2015, 62, 720–733. [Google Scholar] [CrossRef]
- Janani, C.; Ranjitha Kumari, B.D. PPAR gamma gene--a review. Diabetes Metab. Syndr. 2015, 9, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Gervois, P.; Torra, I.P.; Fruchart, J.C.; Staels, B. Regulation of lipid and lipoprotein metabolism by PPAR activators. Clin. Chem. Lab. Med. 2000, 38, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Rakhshandehroo, M.; Knoch, B.; Müller, M.; Kersten, S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res 2010, 2010, 612089. [Google Scholar] [CrossRef]
- Duval, C.; Müller, M.; Kersten, S. PPARalpha and dyslipidemia. Biochim. Biophys. Acta 2007, 1771, 961–971. [Google Scholar] [CrossRef] [PubMed]
- Régnier, M.; Polizzi, A.; Smati, S.; Lukowicz, C.; Fougerat, A.; Lippi, Y.; Fouché, E.; Lasserre, F.; Naylies, C.; Bétoulières, C.; et al. Hepatocyte-specific deletion of Pparα promotes NAFLD in the context of obesity. Sci. Rep. 2020, 10, 6489. [Google Scholar] [CrossRef]
- Daniels, T.F.; Killinger, K.M.; Michal, J.J.; Wright, R.W., Jr.; Jiang, Z. Lipoproteins, cholesterol homeostasis and cardiac health. Int. J. Biol. Sci. 2009, 5, 474–488. [Google Scholar] [CrossRef] [PubMed]
- Darabi, M.; Kontush, A. High-density lipoproteins (HDL): Novel function and therapeutic applications. Biochim. Et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2022, 1867, 159058. [Google Scholar] [CrossRef]
- Zhou, L.; Li, C.; Gao, L.; Wang, A. High-density lipoprotein synthesis and metabolism (Review). Mol. Med. Rep. 2015, 12, 4015–4021. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.M.; Wiesolek, H.L.; Sumagin, R. ICAM-1: A master regulator of cellular responses in inflammation, injury resolution, and tumorigenesis. J. Leukoc. Biol. 2020, 108, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, I.; Suryawanshi, A.; Hong, Y.; Ranganathan, P.; Shanmugam, A.; Ahmad, S.; Swafford, D.; Manicassamy, B.; Ramesh, G.; Koni, P.A.; et al. Homeostatic PPARα Signaling Limits Inflammatory Responses to Commensal Microbiota in the Intestine. J. Immunol. 2016, 196, 4739–4749. [Google Scholar] [CrossRef] [PubMed]













Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Cheng, Y.; Li, J.; Liu, C.; Qian, H.; Zhang, G. Torularhodin Alleviates Hepatic Dyslipidemia and Inflammations in High-Fat Diet-Induced Obese Mice via PPARα Signaling Pathway. Molecules 2022, 27, 6398. https://doi.org/10.3390/molecules27196398
Li X, Cheng Y, Li J, Liu C, Qian H, Zhang G. Torularhodin Alleviates Hepatic Dyslipidemia and Inflammations in High-Fat Diet-Induced Obese Mice via PPARα Signaling Pathway. Molecules. 2022; 27(19):6398. https://doi.org/10.3390/molecules27196398
Chicago/Turabian StyleLi, Xingming, Yuliang Cheng, Jiayi Li, Chang Liu, He Qian, and Genyi Zhang. 2022. "Torularhodin Alleviates Hepatic Dyslipidemia and Inflammations in High-Fat Diet-Induced Obese Mice via PPARα Signaling Pathway" Molecules 27, no. 19: 6398. https://doi.org/10.3390/molecules27196398
APA StyleLi, X., Cheng, Y., Li, J., Liu, C., Qian, H., & Zhang, G. (2022). Torularhodin Alleviates Hepatic Dyslipidemia and Inflammations in High-Fat Diet-Induced Obese Mice via PPARα Signaling Pathway. Molecules, 27(19), 6398. https://doi.org/10.3390/molecules27196398