Antimicrobial V-Shaped Copper(II) Pentaiodide: Insights to Bonding Pattern and Susceptibility
Abstract
:1. Introduction
2. Results and Discussion
2.1. Spectroscopical Characterization
2.1.1. UV-Vis Spectroscopy
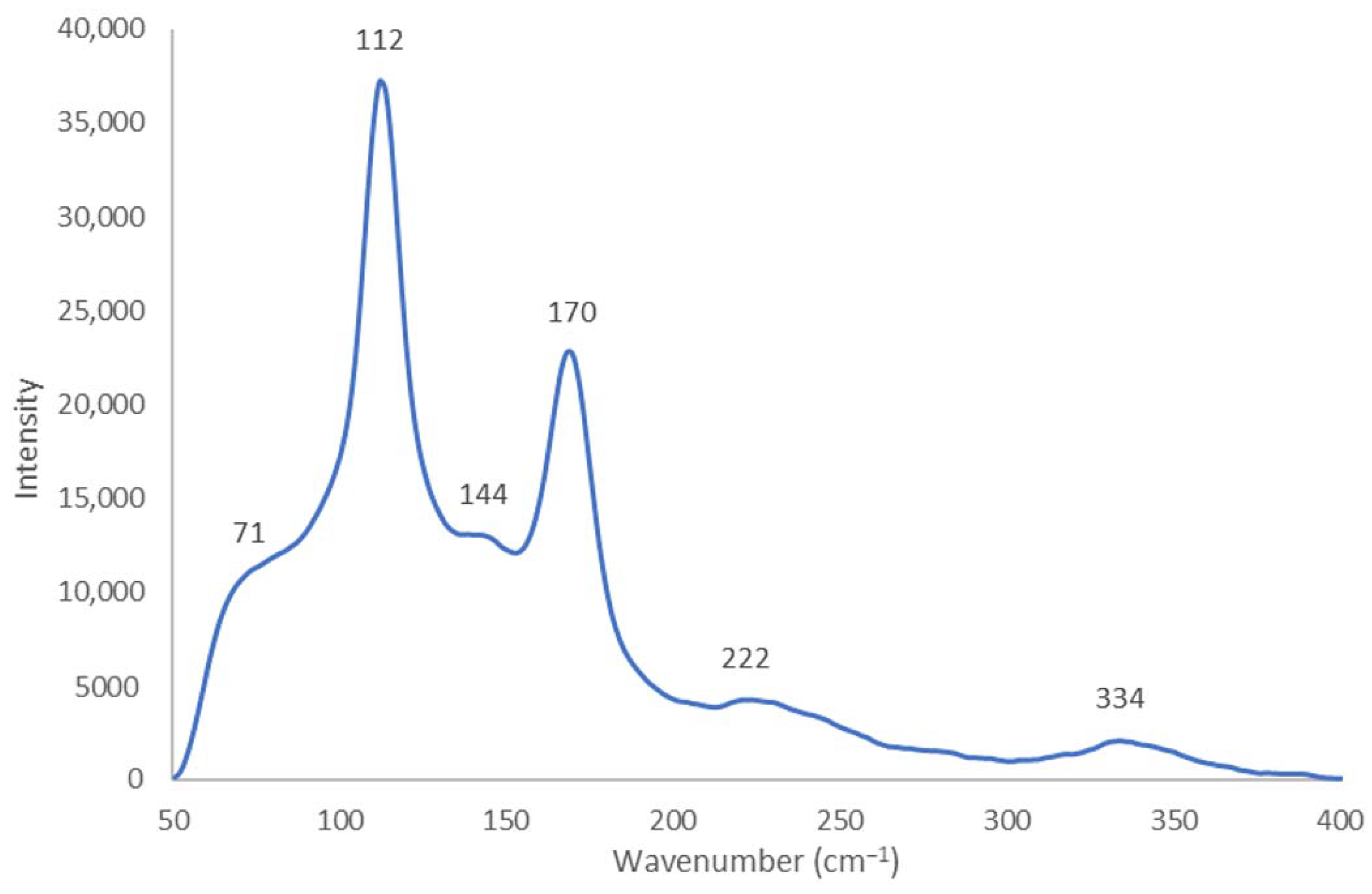
2.1.2. Raman Spectroscopy
2.1.3. Fourier-Transform Infrared (FTIR) Spectroscopy
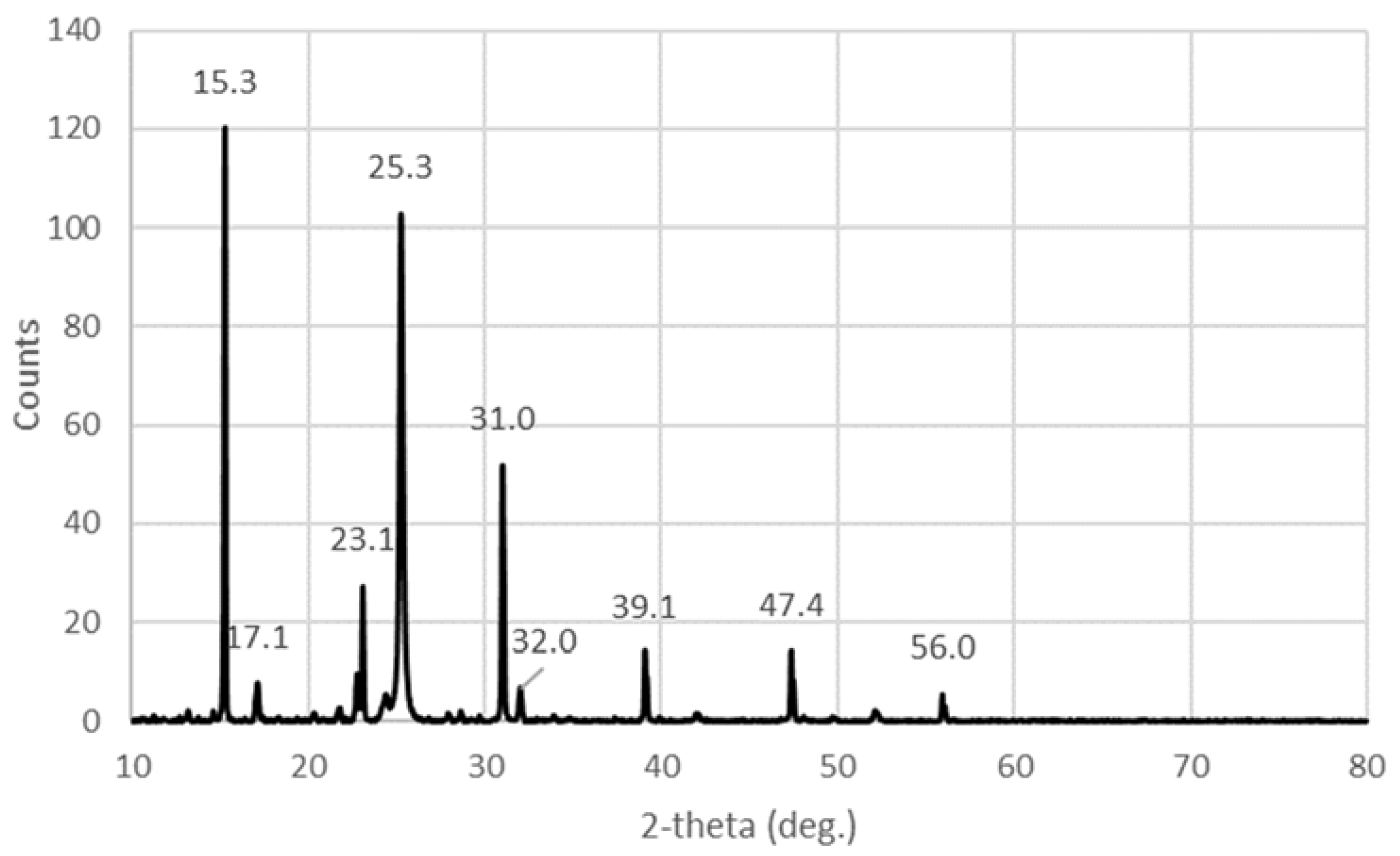
2.1.4. X-ray Diffraction (XRD)
2.2. Elemental Composition and Morphological Examination
2.2.1. Scanning Electron Microscope (SEM)
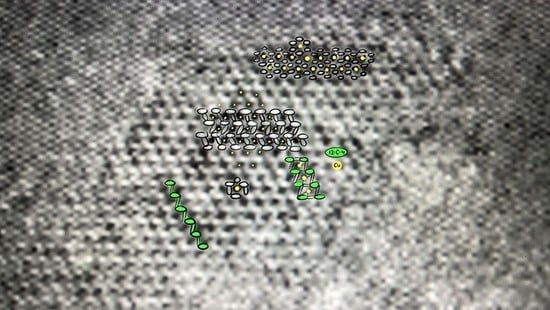
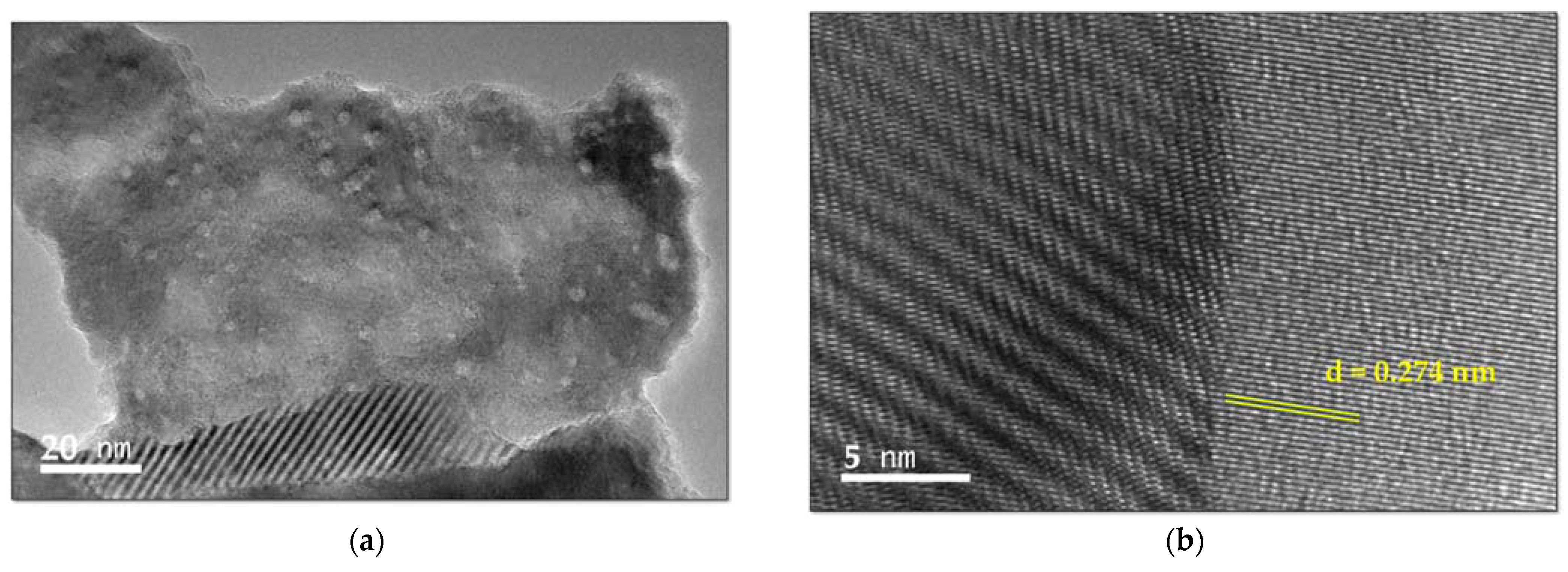
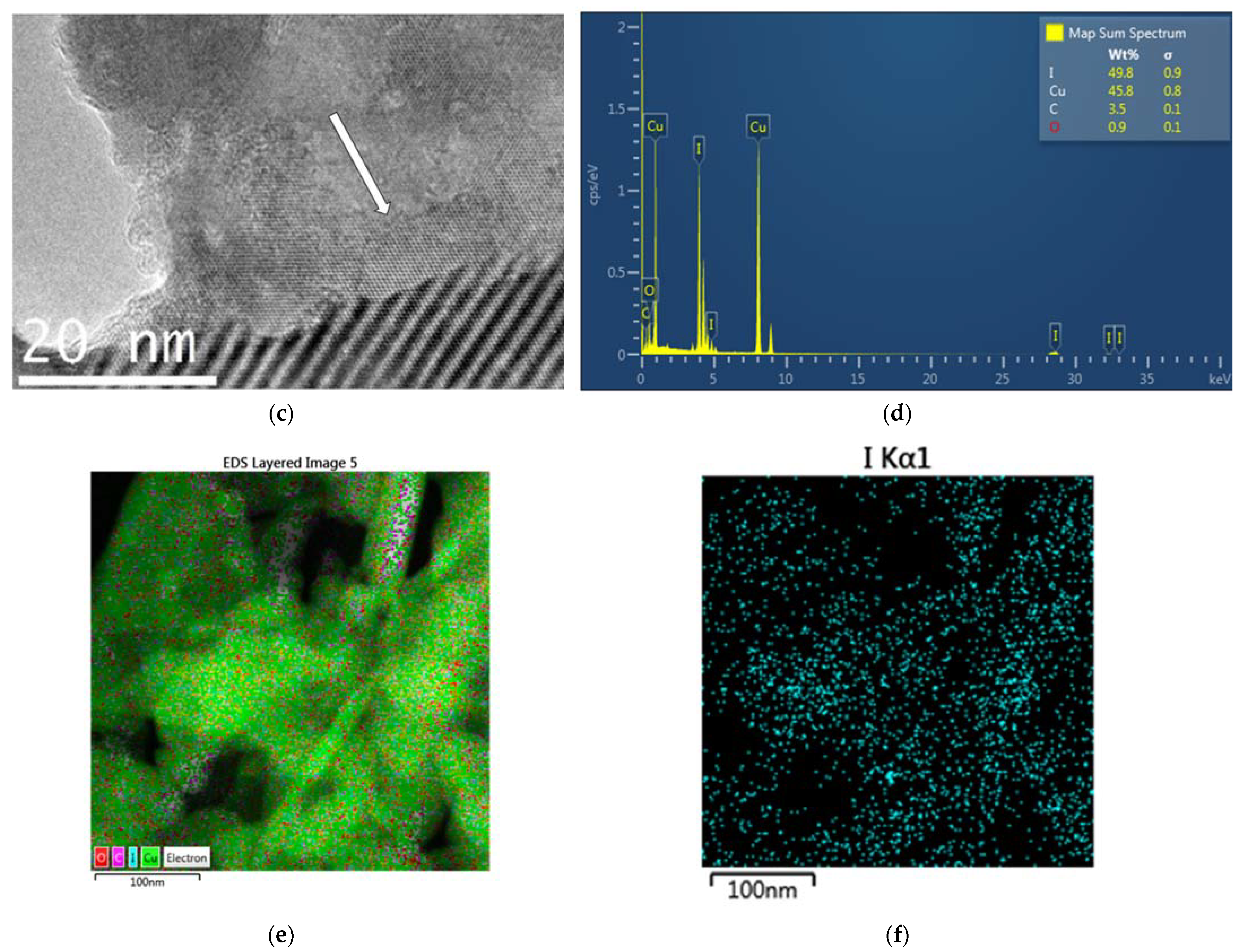
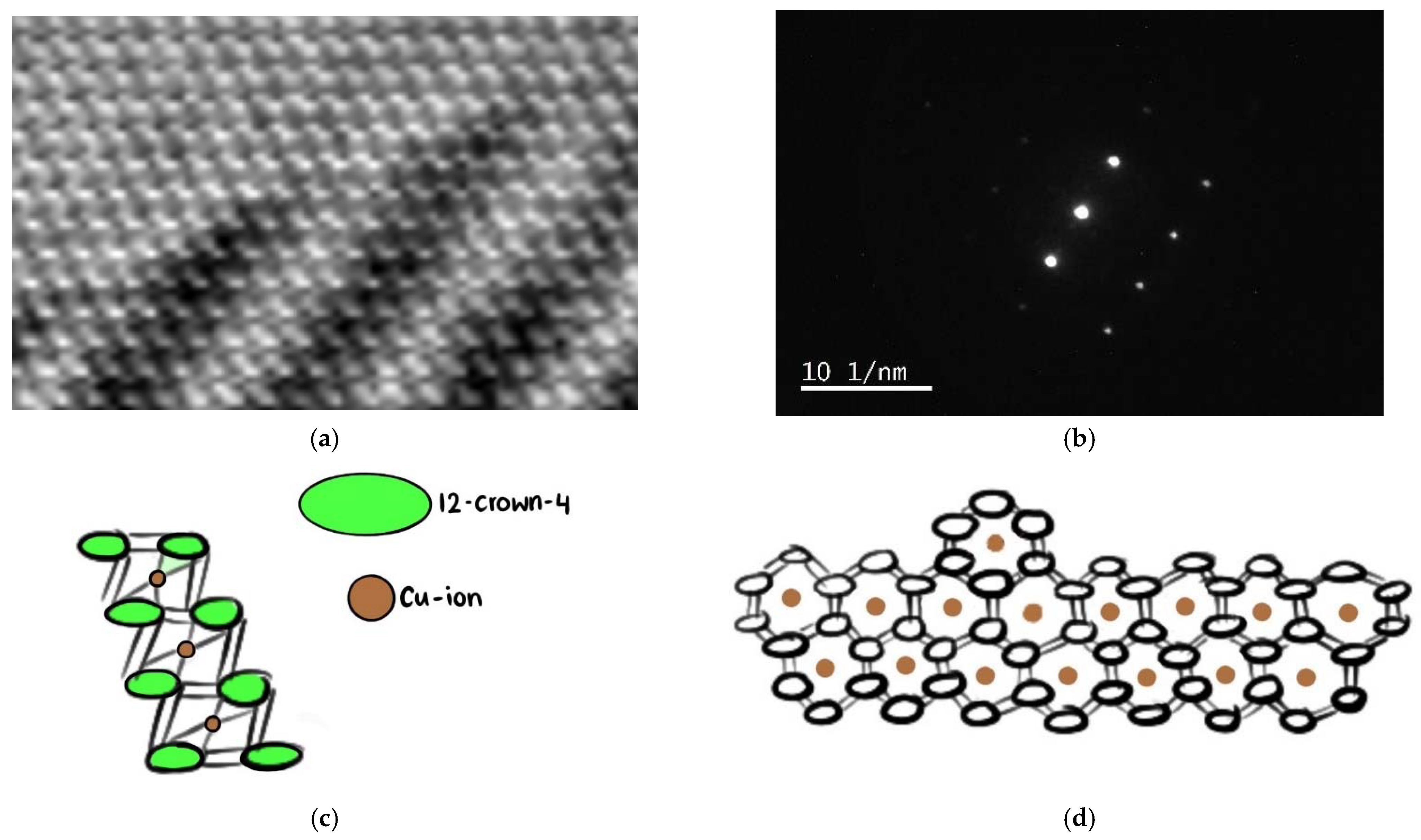
2.2.2. Transmission Electron Microscope (TEM) and Scanning Transmission Electron Microscope (STEM) Analysis with Elemental Mapping
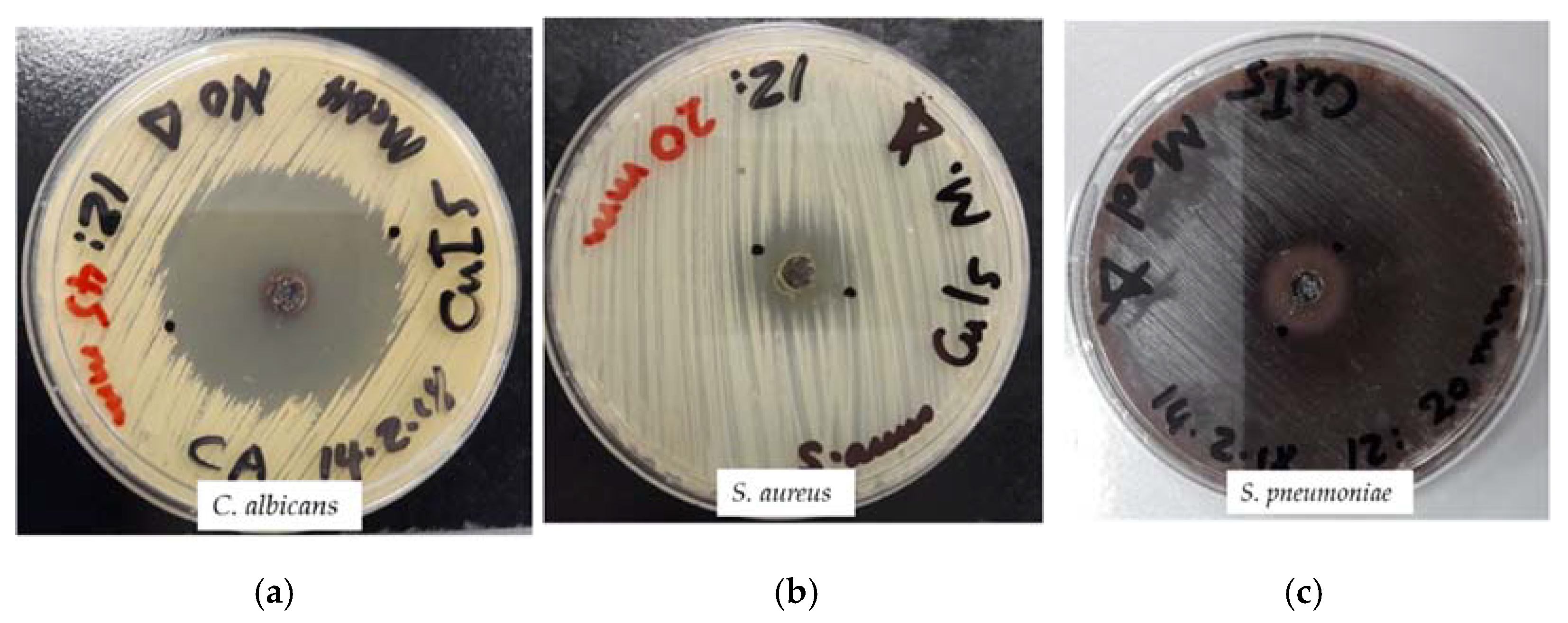
2.3. Antimicrobial Testing
3. Materials and Methods
3.1. Materials
3.2. Synthesis of [Cu(12-crown-4)2]I5
3.3. Characterization of [Cu(12-crown-4)2]I5
3.3.1. UV-Vis Spectrophotometry (UV-Vis)
3.3.2. Raman Spectroscopy
3.3.3. Fourier-Transform Infrared (FTIR) Spectroscopy
3.3.4. X-ray Diffraction (XRD)
3.3.5. Scanning Electron Microscopy (SEM) and Energy-Dispersive X-ray Spectroscopy (EDS)
3.3.6. Transmission Electron Microscopy (TEM) and Energy-Dispersive X-ray Spectroscopy (STEM-EDS)
3.4. Bacterial Strains and Culturing
3.5. Antimicrobial Testing
3.5.1. Procedure for Zone of Inhibition Plate Studies
3.5.2. Disc Diffusion Method
3.6. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e-00181-19. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Hogberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539–563. [Google Scholar] [CrossRef] [PubMed]
- Bloukh, S.H.; Edis, Z.; Shaikh, A.A.; Pathan, H.M. A Look Behind the Scenes at COVID-19: National Strategies of Infection Control and Their Impact on Mortality. Int. J. Environ. Res. Public Health 2020, 17, 5616. [Google Scholar] [CrossRef]
- Mahoney, A.R.; Safaee, M.M.; Wuest, W.M.; Furst, A.L. The silent pandemic: Emergent antibiotic resistances following the global response to SARS-CoV-2. iScience 2021, 24, 102304. [Google Scholar] [CrossRef]
- Jones, I.A.; Joshi, L.T. Biocide Use in the Antimicrobial Era: A Review. Molecules 2021, 26, 2276. [Google Scholar] [CrossRef]
- Vazquez-Munoz, R.; Lopez-Ribot, J.L. Nanotechnology as an Alternative to Reduce the Spread of COVID-19. Challenges 2020, 11, 15. [Google Scholar] [CrossRef]
- Lee, S.H.; Jun, B.-H. Silver Nanoparticles: Synthesis and Application for Nanomedicine. Int. J. Mol. Sci. 2019, 20, 865. [Google Scholar] [CrossRef]
- Fanoro, O.T.; Oluwafemi, O.S. Bactericidal Antibacterial Mechanism of Plant Synthesized Silver, Gold and Bimetallic Nanoparticles. Pharmaceutics 2020, 12, 1044. [Google Scholar] [CrossRef]
- Mamatha, G.; Rajulu, A.V.; Madhukar, K. In Situ Generation of Bimetallic Nanoparticles in Cotton Fabric Using Aloe Vera Leaf Extract, as a Reducing Agent. J. Nat. Fibers 2020, 17, 1121–1129. [Google Scholar] [CrossRef]
- Edis, Z.; Haj Bloukh, S.; Ashames, A.; Ibrahim, M. Copper-Based Nanoparticles, their chemistry and Antibacterial properties: A Review. In Chemistry for a Clean and Healthy Planet, 1st ed.; Ramasami, P., Gupta Bhowon, M., Jhaumeer Laulloo, S., Li Kam Wah, H., Eds.; Springer Nature AG: Cham, Switzerland, 2019; pp. 401–428. ISBN 13 978-3-030-20282-8. [Google Scholar]
- El-Kahky, D.; Attia, M.; Easa, S.M.; Awad, N.M.; Helmy, E.A. Interactive Effects of Biosynthesized Nanocomposites and Their Antimicrobial and Cytotoxic Potentials. Nanomaterials 2021, 11, 903. [Google Scholar] [CrossRef] [PubMed]
- Wahab, M.A.; Li, L.; Li, H.; Abdala, A. Silver Nanoparticle-Based Nanocomposites for Combating Infectious Pathogens: Recent Advances and Future Prospects. Nanomaterials 2021, 11, 581. [Google Scholar] [CrossRef]
- Reda, M.; Ashames, A.; Edis, Z.; Bloukh, S.; Bhandare, R.; Abu Sara, H. Green Synthesis of Potent Antimicrobial Silver Nanoparticles Using Different Plant Extracts and Their Mixtures. Processes 2019, 7, 510. [Google Scholar] [CrossRef]
- Bloukh, S.H.; Edis, Z.; Abu Sara, H.; Alhamaidah, M.A. Antimicrobial Properties of Lepidium sativum L. Facilitated Silver Nanoparticles. Pharmaceutics 2021, 13, 1352. [Google Scholar] [CrossRef]
- McNeilly, O.; Mann, R.; Hamidian, M.; Gunawan, C. Emerging Concern for Silver Nanoparticle Resistance in Acinetobacter Baumannii and Other Bacteria. Front. Microbiol. 2021, 12, 894. [Google Scholar] [CrossRef]
- Kaiho, T. Iodine Chemistry and Applications, 1st ed.; Kaiho, T., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 15–410. ISBN 78-1-118-46629-2. [Google Scholar]
- Baig, N.; Shetty, S.; Al-Mousawi, S.; Alameddine, B. Synthesis of conjugated Polymers via Cyclopentannulation reaction: Promising Materials for Iodine Adsorption. Polym. Chem. 2020, 11, 3066–3074. [Google Scholar] [CrossRef]
- Miensah, E.D.; Kokuloku, L.T., Jr.; Gu, A.; Chen, K.; Wang, P.; Gong, C.; Mao, P.; Chen, K.; Jiao, Y.; Yang, Y. Effects of activation parameters on Zeolitic imidazolate framework JUC-160-derived, nitrogen-doped hierarchical nanoporous carbon and its volatile iodine capture properties. Colloids Surfaces A Physicochem. Eng. Asp. 2022, 650, 129478. [Google Scholar] [CrossRef]
- Inokuchi, Y.; Nakatsuma, M.; Kida, M.; Ebata, T. Conformation of Alkali Metal Ion–Benzo-12-Crown-4 Complexes Investigated by UV Photodissociation and UV–UV Hole-Burning Spectroscopy. J. Phys. Chem. A 2016, 120, 6394–6401. [Google Scholar] [CrossRef]
- Staffeld, P.; Kaller, M.; Ehni, P.; Ebert, M.; Laschat, S.; Giesselmann, F. Improved Electronic Transport in Ion Complexes of Crown Ether Based Columnar Liquid Crystals. Crystals 2019, 9, 74. [Google Scholar] [CrossRef]
- Okuda, M.; Hiramatsu, T.; Yasuda, M.; Ishigaki, M.; Ozaki, Y.; Hayashi, M.; Tominaga, K.; Chatani, E. Theoretical Modeling of Electronic Structures of Polyiodide Species Included in α-Cyclodextrin. J. Phys. Chem. B 2020, 124, 4089–4096. [Google Scholar] [CrossRef]
- Hiramatsu, T.; Yamamoto, N.; Ha, S.; Masuda, Y.; Yasuda, M.; Ishigaki, M.; Yuzu, K.; Ozaki, Y.; Chatani, E. Iodine staining as a useful probe for distinguishing insulin amyloid polymorphs. Sci. Rep. 2020, 10, 16741. [Google Scholar] [CrossRef] [PubMed]
- Uthayaraj, S.; Karunarathne, D.G.B.C.; Kumara, G.R.A.; Murugathas, T.; Rasalingam, S.; Rajapakse, R.M.G.; Ravirajan, P.; Velauthapillai, D. Powder Pressed Cuprous Iodide (CuI) as A Hole Transporting Material for Perovskite Solar Cells. Materials 2019, 12, 2037. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wu, Z.; Liu, F.; Liu, S.; Liu, J.; Wang, Y.; Yan, T. Encapsulating a high content of iodine into an active graphene substrate as a cathode material for high-rate lithium–iodine batteries. J. Mater. Chem. A 2017, 5, 15235–15242. [Google Scholar] [CrossRef]
- Chang, J.; Zhao, G.; Zhao, X.; He, C.; Pang, S.; Shreeve, J.M. New Promises from an Old Friend: Iodine-Rich Compounds as Prospective Energetic Biocidal Agents. Accounts Chem. Res. 2021, 54, 332–343. [Google Scholar] [CrossRef]
- Makhayeva, D.N.; Irmukhametova, G.S.; Khutoryanskiy, V.V. Polymeric Iodophors: Preparation, Properties, and Biomedical Applications. Rev. J. Chem. 2020, 10, 40–57. [Google Scholar] [CrossRef]
- Zhang, S.; Kai, C.; Liu, B.; Zhang, S.; Wei, W.; Xu, X.; Zhou, Z. Facile fabrication of cellulose membrane containing polyiodides and its antibacterial properties. Appl. Surf. Sci. 2020, 500, 144046. [Google Scholar] [CrossRef]
- Sai, M.; Zhong, S.; Tang, Y.; Ma, W.; Sun, Y.; Ding, D. Research on the preparation and antibacterial properties of 2-N-thiosemicarbazide-6-O-hydroxypropyl chitosan membranes with iodine. J. Appl. Polym. Sci. 2014, 131, 1–8. [Google Scholar] [CrossRef]
- Tashiro, K.; Takahama, T.; Wang, M.F. X-ray study of Poly(vinyl Alcohol)-Iodine complex prepared from the dilute iodine solution as a hint to know the inner structure of polarizer. Polymer 2021, 233, 124180. [Google Scholar] [CrossRef]
- Borjihan, Q.; Zhu, Y.; Zhou, R.; Zhang, X.; Zhang, L.; Luan, S.; Dong, A. Controlled Engineering of Nano-Povidones for Efficient Iodine Recovery and Antibacterial Reutilization. ACS Sustain. Chem. Eng. 2020, 8, 11704–11712. [Google Scholar] [CrossRef]
- Xu, X.; Guan, Y. Investigating the Complexation and Release Behaviors of Iodine in Poly(vinylpyrrolidone)-Iodine Systems through Experimental and Computational Approaches. Ind. Eng. Chem. Res. 2020, 59, 22667–22676. [Google Scholar] [CrossRef]
- Edis, Z.; Bloukh, S.H.; Sara, H.A.; Azelee, N.I.W. Antimicrobial Biomaterial on Sutures, Bandages and Face Masks with Potential for Infection Control. Polymers 2022, 14, 1932. [Google Scholar] [CrossRef] [PubMed]
- Edis, Z.; Bloukh, S.H.; Abu Sara, H.; Bhakhoa, H.; Rhyman, L.; Ramasami, P. “Smart” triiodide compounds: Does halogen bonding influence antimicrobial activities? Pathogens 2019, 8, 182–202. [Google Scholar] [CrossRef] [PubMed]
- Bloukh, S.H.; Edis, Z.; Ibrahim, M.R.; Abu Sara, H. “Smart” antimicrobial nanocomplexes with potential to decrease surgical site infections (SSI). Pharmaceutics 2020, 12, 361. [Google Scholar] [CrossRef]
- Edis, Z.; Bloukh, S.H. Facile Synthesis of Antimicrobial Aloe Vera-“Smart” Triiodide-PVP Biomaterials. Biomimetics 2020, 5, 45. [Google Scholar] [CrossRef] [PubMed]
- Edis, Z.; Bloukh, S.H. Facile Synthesis of Bio-Antimicrobials with “Smart” Triiodides. Molecules 2021, 26, 3553. [Google Scholar] [CrossRef]
- Svensson, P.H.; Kloo, L. Synthesis, structure, and bonding in polyiodide and metal iodide−iodine systems. Chem. Rev. 2003, 103, 1649–1684. [Google Scholar] [CrossRef]
- Wang, Y.; Xue, Y.; Wang, X.; Cui, Z.; Wang, L. The stable polyiodides: Experimental and Theoretical Studies of Formation Mechanism. J. Mol. Struct. 2014, 1074, 231–239. [Google Scholar] [CrossRef]
- Bartashevich, E.V.; Grigoreva, E.A.; Yushina, I.D.; Bulatova, L.M.; Tsirelson, V.G. Modern level for the prediction of properties of iodine-containing organic compounds: Iodine forming halogen bonds. Russ. Chem. Bull. Int. Ed. 2017, 66, 1–12. [Google Scholar] [CrossRef]
- Wei, Y.J.; Liu, C.G.; Mo, L.P. Ultraviolet absorption spectra of iodine, iodide ion and triiodide ion. Guang Pu Xue Yu Guang Pu Fen Xi 2005, 25, 86–88. [Google Scholar]
- Savastano, M.; Bazzicalupi, C.; Gellini, C.; Bianchi, A. Genesis of Complex Polyiodide Networks: Insights on the Blue Box/I−/I2 Ternary System. Crystals 2020, 10, 387. [Google Scholar] [CrossRef]
- Savastano, M.; Bazzicalupi, C.; García, C.; Gellini, C.; de la Torre, M.D.L.; Mariani, P.; Pichierri, F.; Bianchi, A.; Melguizo, M. Iodide and triiodide anion complexes involving anion–π interactions with a tetrazine-based receptor. Dalton Trans. 2017, 46, 4518. [Google Scholar] [CrossRef] [PubMed]
- Shestimerova, T.A.; Golubev, N.A.; Bykov, M.A.; Mironov, A.V.; Fateev, S.A.; Tarasov, A.B.; Turkevych, I.; Wei, Z.; Dikarev, E.V.; Shevelkov, A.V. Molecular and Supramolecular Structures of Triiodides and Polyiodobismuthates of Phenylenediammonium and Its N,N-dimethyl Derivative. Molecules 2021, 26, 5712. [Google Scholar] [CrossRef] [PubMed]
- Reiss, G.J. I5––polymers with a layered arrangement: Synthesis, spectroscopy, and structure of a new polyiodide salt in the nicotine/HI/I2 system. Z. Naturforsch. B 2015, 70, 735–739. [Google Scholar] [CrossRef]
- Shestimerova, T.A.; Mironov, A.V.; Bykov, M.A.; Grigorieva, A.V.; Wei, Z.; Dikarev, E.V.; Shevelkov, A.V. Assembling Polyiodides and Iodobismuthates Using a Template Effect of a Cyclic Diammonium Cation and Formation of a Low-Gap Hybrid Iodobismuthate with High Thermal Stability. Molecules 2020, 25, 2765. [Google Scholar] [CrossRef] [PubMed]
- Shestimerova, T.A.; Bykov, A.V.; Kuznetsov, A.N.; Grishko, A.Y.; Wei, Z.; Dikarev, E.V.; Shevelkov, A.V. Pattern of covalent and non-covalent interactions within the pentaiodide anion in the structure of (3-HOC5H9NH2)I5. Z. Anorg. Allgem. Chem. 2022, 648, e202200039. [Google Scholar] [CrossRef]
- Reiss, G.J. A cyclic I102− anion in the layered crystal structure of theophyllinium pentaiodide, C7H9I5N4O2. Z. Krist. NCS 2019, 234, 737–739. [Google Scholar] [CrossRef]
- Edis, Z.; Bloukh, S. Preparation, Structural and Spectroscopical Characterization of a Pentaiodide [Rb(12-crown-4)2]I5. Z. Naturforsch. B 2013, 68b, 1340–1346. [Google Scholar] [CrossRef]
- Edis, Z.; Raheja, R.; Bloukh, S.H.; Bhandare, R.R.; Sara, H.A.; Reiss, G.J. Antimicrobial Hexaaquacopper (II) Complexes with Novel Polyiodide Chains. Polymers 2021, 13, 1005. [Google Scholar] [CrossRef]
- Haj Bloukh, S.; Edis, Z. Structure and Antimicrobial properties of bis(1,4,7,10-tetraoxacyclododecane-κ4O,O′,O′′,O′′′)cesium pentaiodide, C16H32CsI5O8. Z. Krist. NCS 2020, 235, 759–761. [Google Scholar] [CrossRef]
- Edis, Z. Polyiodides of 12-Crown-4 Complexes with Alkali Cations. Ph.D. Thesis, University of Cologne, Cologne, Germany, 1999. [Google Scholar]
- Świergiel, J.; Grembowski, J.; Jadżyn, J. Odd-even alternation in molecular structure and self-assembling of some macrocyclic polyethers. J. Mol. Liquids 2017, 229, 472–476. [Google Scholar] [CrossRef]
- Cooper, T.E.; Carl, D.R.; Oomens, J.; Steill, J.D.; Armentrout, P.B. Infrared Spectroscopy of Divalent Zinc and Cadmium Crown Ether Systems. J. Phys. Chem. A 2011, 115, 5408–5422. [Google Scholar] [CrossRef] [PubMed]
- Al-Rusaese, S.; Al-Kahtani, A.A.; El-Azhary, A.A. Experimental and Theoretical Study of the Vibrational Spectra of 12-Crown-4-Alkali Metal Cation Complexes. J. Phys. Chem. 2006, 110, 8676–8687. [Google Scholar] [CrossRef] [PubMed]
- Bloukh, S.H.; Edis, Z. Halogen bonding in Crystal structure of bis(1,4,7,10-tetraoxacyclododecane-κ4O,O′,O′′,O′′′)cesium triiodide, C16H32CsI3O8. Z. Krist. NCS 2020, 235, 717–719. [Google Scholar] [CrossRef]
- Edis, Z.; Bloukh, S.H. Preparation and structural and spectroscopic characterization of triiodides [M(12-crown-4)2]I3 with M = Na and Rb. Z. Nat. 2014, 69, 995–1002. [Google Scholar] [CrossRef]
- Andrei, A.; Öztürk, Y.; Khalfaoui-Hassani, B.; Rauch, J.; Marckmann, D.; Trasnea, P.-I.; Daldal, F.; Koch, H.-G. Cu Homeostasis in Bacteria: The Ins and Outs. Membranes 2020, 10, 242. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.W.; Perry, D.M.; Kirby, W.M.M. Single-disk antibiotic-sensitivity testing of staphylococci: An analysis of technique and results. AMA Arch. Int. Med. 1959, 104, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk Susceptibility Testing, 28th ed.; M100S; CLSI: Wayne, PA, USA, 2018; Volume 38. [Google Scholar]





| Group | 1 | 2 [(I−)2(I2)] | [18] | [19] | [23] | [22]exp | [22]calc |
|---|---|---|---|---|---|---|---|
| I2/12-crown-4 | 204 vs | 204 | 204 | 204 | |||
| I3−/12-crown-4 I3− [I3−…I2] | 228 s,br 290 m,br 360 s,br | 290 359 | 228 290 359 | 229 291 359 | 290 360 | 223 292 353 | 217 292 365/339Y |
| I− | 201 vs | 201 | 201 | ||||
| I5− | 445 vw,br | 455 | 450 | 460 |
| Group | 1. | 2 | [42] | [45] | [39] | [47] | [22] | [23] | [28] |
|---|---|---|---|---|---|---|---|---|---|
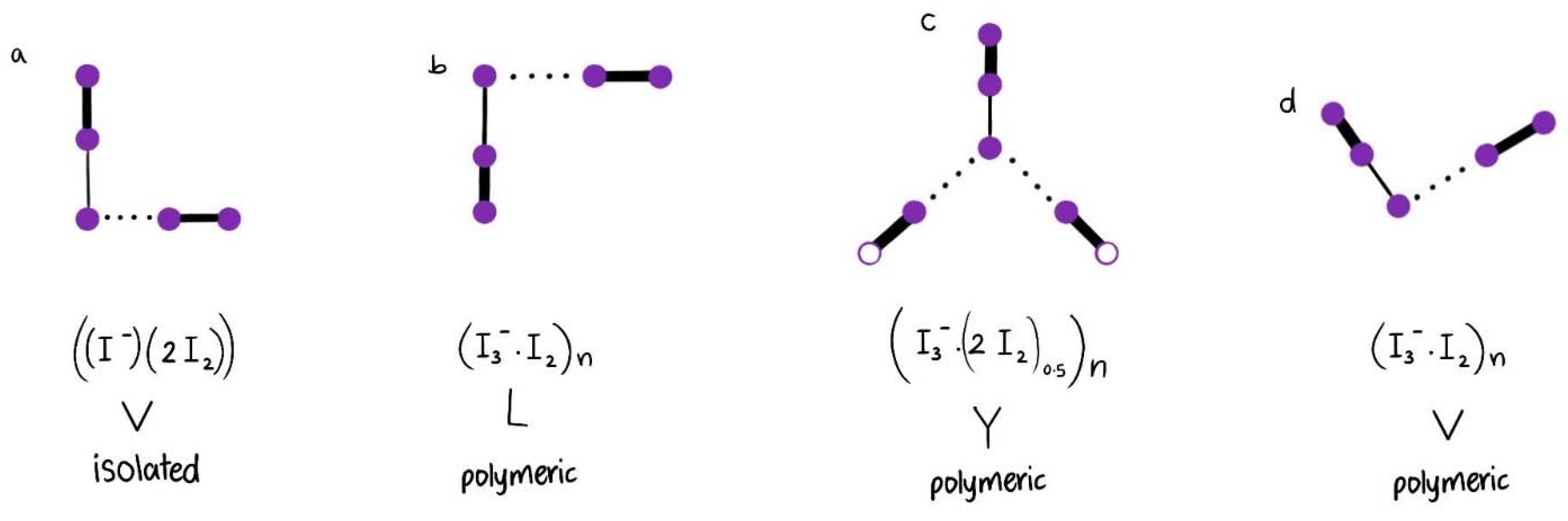
| I5− Type | V | Y | V | V | V | L | Linear | Linear | |
| Structure | [(I3−)(I2)] | [(I3−)2(I2)0.5] | [(I−)2(I2)] | [(I3−)(I2)] | [(I3−)(I2)] | [(I3−)(I2)] | [(I5−)]n | [(I5−)]n | |
| I2 as [I2….I−] or [I2….I3−] | s 170 νs | vs 172 νs | s 170 νs w 165L νas | 177 νs | 178 νs 163 νas | m 164 νs | 160 νas | 163 νas | |
| I3- | m, sh [I-I-I−] 71 ν2bend vs [I-I-I−] 112 νs w, sh [I-I….I−] 222 νas | 71 ν2bend vs 111 νs w 222 νas | 67 ν2bend 114 νs | 108 νs 211 νas 423 | 80 ν2bend vs 111 νs s 127 νas | 70 ν2bend 110 νs 220 νas | 108 νs 218 νas | 110 νs 217 νas | |
| I5- | m,sh 144 νas | m,sh 142 νas | w 144 νas | 151 νas | 147 νas | ||||
| as [I2….I3−] | vw, sh 334 νas | 334 νas | 334 νas | 318 νas | 337 νas | 331 νas | |||
| 12-crown-4 | vw 2926 | vw 2855 |
| Compound | Type | [I2] (Å) | [I3−] (Å) | [I2….I3−] (Å) | [I3−] Angle (°) | [I2] (cm−1) | [I3−] (cm−1) | [I5−]/[I2….I3−] (cm−1) |
|---|---|---|---|---|---|---|---|---|
| [47] Polymeric Chain [(I3−)(I2)]n | L | 2.75 | 2.83 3.05 2.94 * | 3.38 3.40 3.39 * | 177.45 | m 164 νs | 80 ν2bend vs 111 νs s 127 νas | |
| [45] Polymeric [(I3−)(I2)]n | V | 2.74 | 2.90 2.93 2.92 * | 3.41 | 176.43 | m 177 νs | 67 ν2bend vs 114 νs | vs 151 νas |
| [Cu(H2O)6(12-crown-4)5]I6 x 2I2 Polymeric chain | Y | 2.76 2.77 | 2.80 3.06 | 3.38 3.38 | 176.46 | vs 172 νs | m,sh 142 νas | |
| [(I3−)2(I2)0.5]n | 2.77* | 2.93* | 3.38* | |||||
| [Rb(12-crown-4)2]I5 Isolated [(I−)2(I2)] | V | 2.79 | 2.82 3.09 2.96 * | 3.09 | 177.30 | m 163 νs w,sh 180 νs | 72 ν2bend m,br 96 νs w,br 190-230 νas | s,br 142 νas |
| [Cu(12-crown-4)2]I5 Chain | V | ≥2.77 ≤2.75 | ≥2.92 ≤2.93 | ≥3.39 ≤3.41 | ~180 ≥176.43 + ≤177.45 + | s 170 νs | m,sh [I-I-I−] 71 ν2bend vs [I-I-I−] 112 νs w,sh [I-I….I−] 222 νas | m,sh 144 νas vw,sh 334 νas |
| ν1 (O–H)*s ν2 (O–H)*as | ν (C–H)as | ν (CH2)as,s | ν (C–H)s ν (O–H)* | δ (C–H)as ν3 (O–H)* | δ (C–C) | δ (C–H) δ (C–C) | ν (C–O) ν (C–O-C) | ν (CH–CH) ν (O–H)* | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | w 2950 | w 2900 | w 2850 | w 1450 | w 1365 | w 1290 | w 1250 vs 1125 s 1100 m 1075 m 1025 w 975 m 919 | m 848 vw 815 | |
| 2 | w,sh 3393 * w 3240 * w 3179 * | w 2953 | w 2905 | w 2859 | w 1445 vw 1638 * | w 1362 | w 1287 | w 1248 m 1124 vs 1090 s 1069 s 1022 m 916 | s 841 m 810 ν56 m 588 m 548 ν57/ν16 m 521 |
| 3 | 3746 *s 3418 *as | 2951 | 2905 | 2863 2777 * 2733 * | 1443 1642 * | vw 1360 | 1287 | 1243 1133 1092 m 1022 911 | m 844 549 628 * |
| 4 | 2954 | 2906 | 2865 | 1444 | 1363 | 1289 | 1245 1134 1095 1023 915 | 849 |
| Group | 1 | [25] | [33] | [29] | [30] |
|---|---|---|---|---|---|
| I2 | 23.1 w 25.3 s 31 m 39.1 vw 47.4 vw | 24.5 s 25 s 28 vs 37 w 38 w 43 w 46 m | 22.8 m 23.8 m 25.5 m 27 m 30 m 37 vw 46 w | 25 29 36 | |
| I5− | 15.3 vs | 8 m 10 m 13.4 vs |
| Strain | Antibiotic | A | AWB | AWC | 1+B | 2+B | 3+B | 1+C | 2+C | 3+C |
|---|---|---|---|---|---|---|---|---|---|---|
| S. pneumoniae ATCC 49619 | G | 18 | 20 | 20 | 25 | 0 | 0 | 19 | 0 | 0 |
| S. aureus ATCC 25923 | G | 28 | 20 | 23 | 11 | 0 | 0 | 35 | 14 | 0 |
| S. pyogenes ATCC 19615 | C | 25 | 25 | 20 | 13 | 0 | 0 | 21 | 0 | 0 |
| E. faecalis ATCC 29212 | CTX | 25 | 20 | 19 | 16 | 0 | 0 | 18 | 0 | 0 |
| B. subtilis WDCM 00003 | S | 21 | 19 | 21 | 18 | 12 | 0 | 33 | 11 | 0 |
| P. mirabilis ATCC 29906 | G | 30 | 0 | 0 | 20 | 0 | 0 | 15 | 0 | 0 |
| P. aeruginosa WDCM 00026 | CTX | 23 | 17 | 16 | 12 | 0 | 0 | 12 | 0 | 0 |
| E. coli WDCM 00013 | A | 23 | 15 | 15 | 15 | 9 | 0 | 25 | 8 | 0 |
| K. pneumoniae WDCM 00097 | CTX | 30 | NA | NA | 16 | 8 | 0 | 24 | 7 | 0 |
| C. albicans WDCM 00054 | NY | 16 | 45 | 51 | 25 | 12 | 0 | 53 | 14 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Edis, Z.; Bloukh, S.H. Antimicrobial V-Shaped Copper(II) Pentaiodide: Insights to Bonding Pattern and Susceptibility. Molecules 2022, 27, 6437. https://doi.org/10.3390/molecules27196437
Edis Z, Bloukh SH. Antimicrobial V-Shaped Copper(II) Pentaiodide: Insights to Bonding Pattern and Susceptibility. Molecules. 2022; 27(19):6437. https://doi.org/10.3390/molecules27196437
Chicago/Turabian StyleEdis, Zehra, and Samir Haj Bloukh. 2022. "Antimicrobial V-Shaped Copper(II) Pentaiodide: Insights to Bonding Pattern and Susceptibility" Molecules 27, no. 19: 6437. https://doi.org/10.3390/molecules27196437