Engineering Peptide Inhibitors of the HFE–Transferrin Receptor 1 Complex
Abstract
1. Introduction
2. Results and Discussion
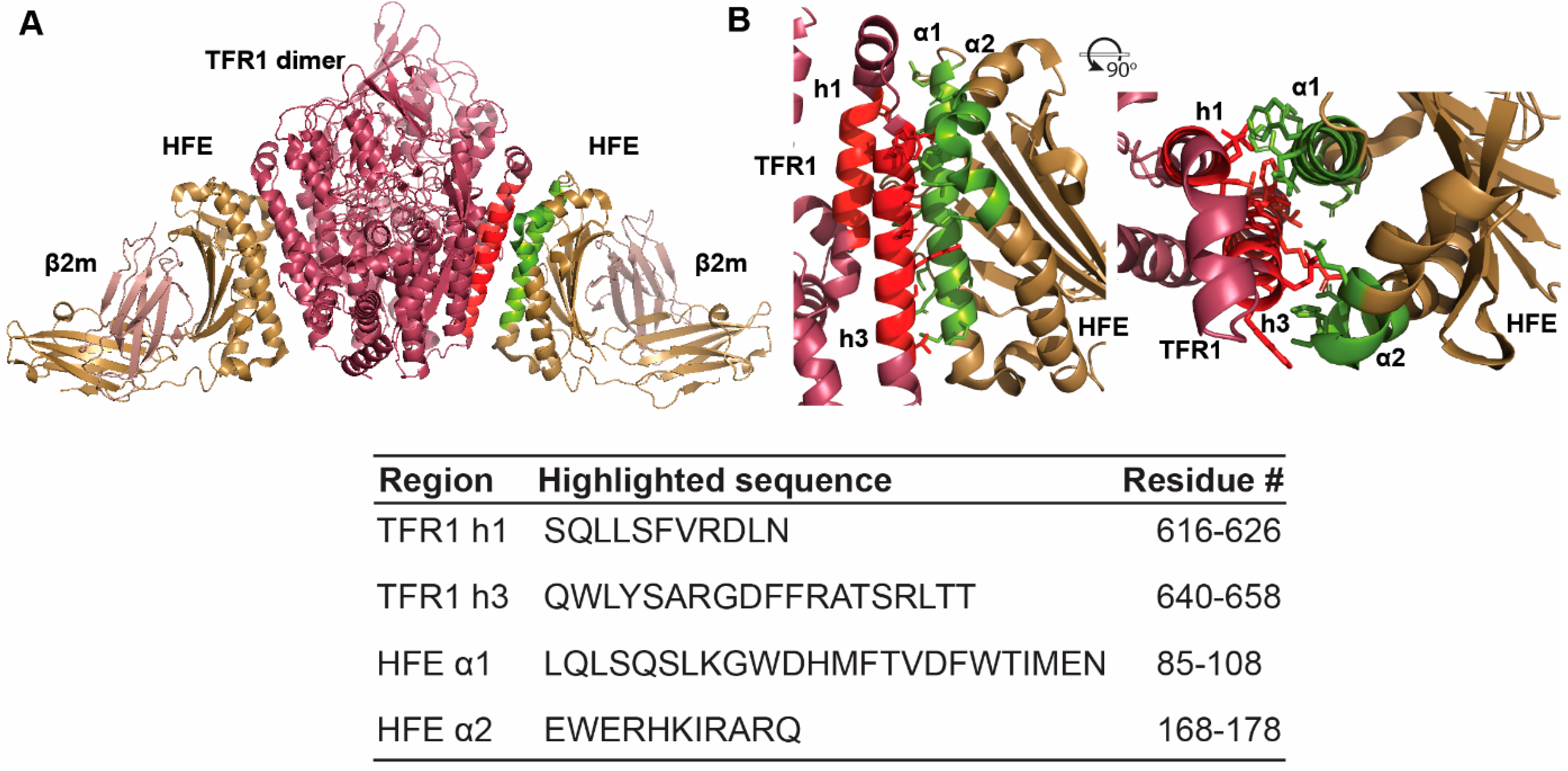
2.1. Rational Design of Peptide Inhibitors of the HFE/TFR1 Complex
2.2. Structural Features of HFE- and TFR1-like Peptides
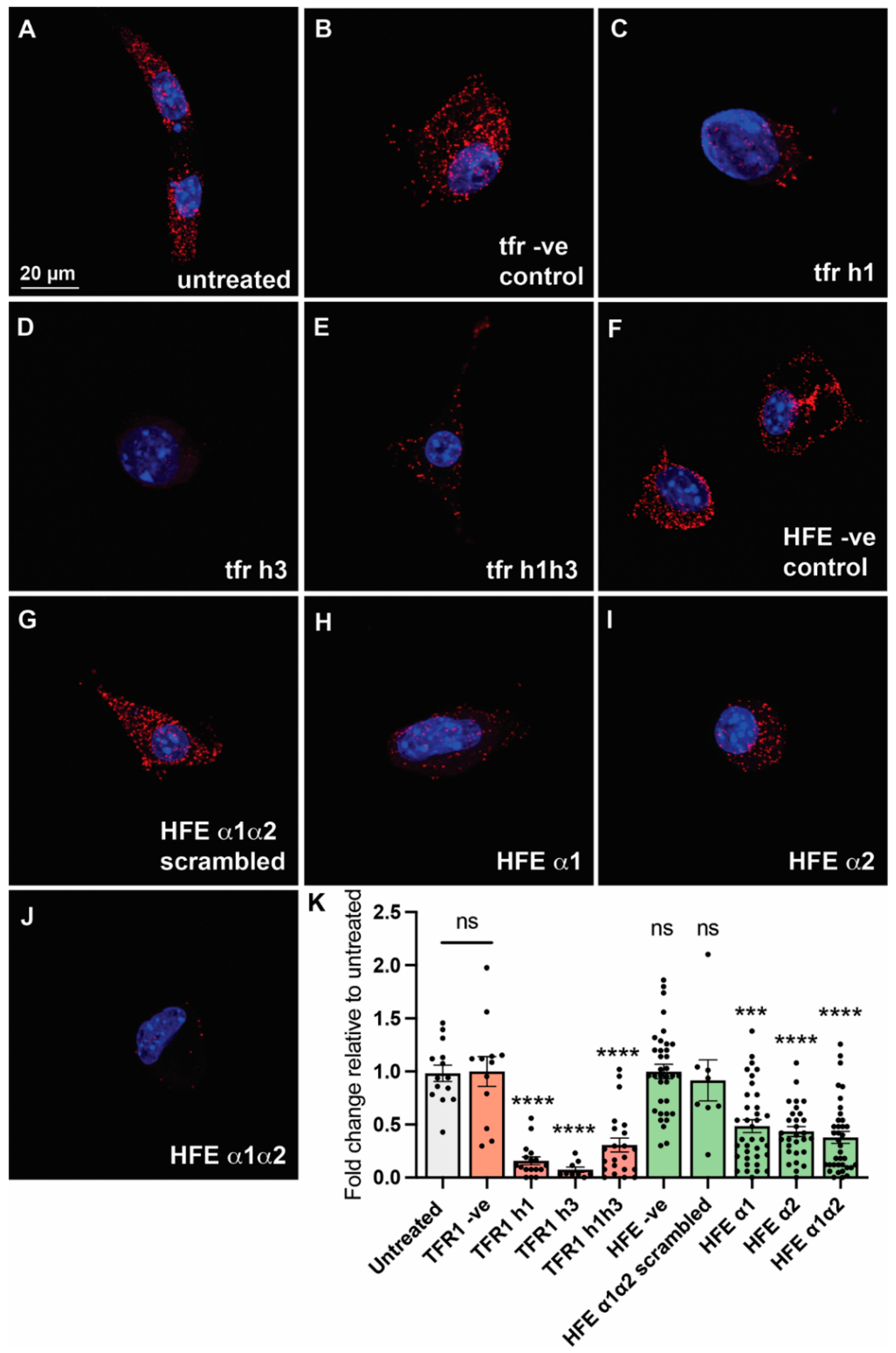
2.3. HFE- and TFR1-like Peptides Inhibit HFE/TFR1 Complex Formation in an In Situ Proximity Ligation Assay
2.4. Optimising HFE α1α2 by Helical Stapling
2.5. Structural Analysis of the Stapled HFE α1α2 Analogues
2.6. HFE α1α2-Single Stapled Analogues Are of Similar Potency as HFE α1α2 at Disrupting HFE/TFR1 Complex Formation in an In Situ Proximity Ligation Assay
3. Conclusions
4. Materials and Methods
4.1. General Methods
4.2. Peptide Synthesis
4.3. In Vitro Immunofluorescence and Proximity Ligation Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jordan, J.B.; Poppe, L.; Haniu, M.; Arvedson, T.; Syed, R.; Li, V.; Kohno, H.; Kim, H.; Schnier, P.D.; Harvey, T.S.; et al. Hepcidin revisited, disulfide connectivity, dynamics, and structure. J. Biol. Chem. 2009, 284, 24155–24167. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Systemic Iron Homeostasis. Physiol. Rev. 2013, 93, 1721–1741. [Google Scholar] [CrossRef]
- Pietrangelo, A. Hepcidin in human iron disorders: Therapeutic implications. J. Hepatol. 2011, 54, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Andrews, N.C. Disorders of iron metabolism. N. Engl. J. Med. 1999, 341, 1986–1995. [Google Scholar] [CrossRef]
- Beutler, E. Hemochromatosis: Genetics and pathophysiology. Annu. Rev. Med. 2006, 57, 331–347. [Google Scholar] [CrossRef]
- Bennett, M.J.; Lebron, J.A.; Bjorkman, P.J. Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature 2000, 403, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Lebron, J.A.; Bennett, M.J.; Vaughn, D.E.; Chirino, A.J.; Snow, P.M.; Mintier, G.A.; Feder, J.N.; Bjorkman, P.J. Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell 1998, 93, 111–123. [Google Scholar] [CrossRef]
- West, A.P.; Giannetti, A.M.; Herr, A.B.; Bennett, M.J.; Nangiana, J.S.; Pierce, J.R.; Weiner, L.P.; Snow, P.M.; Bjorkman, P.J. Mutational analysis of the transferrin receptor reveals overlapping HFE and transferrin binding sites. J. Mol. Biol. 2001, 313, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, P.J.; Toran, P.T.; Giannetti, A.M.; Bjorkman, P.J.; Andrews, N.C. The transferrin receptor modulates HFE-dependent regulation of hepcidin expression. Cell Metab. 2008, 7, 205–214. [Google Scholar] [CrossRef]
- Gao, J.; Chen, J.; Kramer, M.; Tsukamoto, H.; Zhang, A.-S.; Enns, C.A. Interaction of the Hereditary Hemochromatosis Protein HFE with Transferrin Receptor 2 Is Required for Transferrin-Induced Hepcidin Expression. Cell Metab. 2009, 9, 217–227. [Google Scholar] [CrossRef]
- D’Alessio, F.; Hentze, M.W.; Muckenthaler, M.U. The hemochromatosis proteins HFE, TfR2, and HJV form a membrane-associated protein complex for hepcidin regulation. J. Hepatol. 2012, 57, 1052–1060. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, P.J.; Fleming, M.D. Transgenic HFE-dependent induction of hepcidin in mice does not require transferrin receptor-2. Am. J. Hematol. 2012, 87, 588–595. [Google Scholar] [CrossRef]
- McDonald, C.J.; Wallace, D.F.; Ostini, L.; Subramaniam, V.N. Parenteral vs. oral iron: Influence on hepcidin signaling pathways through analysis of Hfe/Tfr2-null mice. Am. J. Physiol.-Gastrointest. Liver Physiol. 2014, 306, G132–G139. [Google Scholar] [CrossRef] [PubMed]
- Rishi, G.; Crampton, E.M.; Wallace, D.F.; Subramaniam, V.N. In situ proximity ligation assays indicate that hemochromatosis proteins HFE and transferrin receptor 2 (Tfr2) do not interact. PLoS ONE 2013, 8, e77267. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.G.; Wang, Y.; Wu, Q.; Cheng, W.H.; Liu, W.; Zhao, Y.; Mayeur, C.; Schmidt, P.J.; Yu, P.B.; Wang, F.; et al. HFE interacts with the BMP type I receptor ALK3 to regulate hepcidin expression. Blood 2014, 124, 1335–1343. [Google Scholar] [CrossRef] [PubMed]
- Jochim, A.L.; Arora, P.S. Systematic analysis of helical protein interfaces reveals targets for synthetic inhibitors. ACS Chem. Biol. 2010, 5, 919–923. [Google Scholar] [CrossRef]
- de Araujo, A.D.; Hoang, H.N.; Kok, W.M.; Diness, F.; Gupta, P.; Hill, T.A.; Driver, R.W.; Price, D.A.; Liras, S.; Fairlie, D.P. Comparative α-helicity of cyclic pentapeptides in water. Angew. Chem. Int. Ed. 2014, 53, 6965–6969. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, S.; Zhang, W.D.; Hu, H.G. Stapled Helical Peptides Bearing Different Anchoring Residues. Chem. Rev. 2020, 120, 10079–10144. [Google Scholar] [CrossRef] [PubMed]
- Barton, J.C.; Sawada-Hirai, R.; Rothenberg, B.E.; Acton, R.T. Two novel missense mutations of the HFE gene (I105T and G93R) and identification of the S65C mutation in Alabama hemochromatosis probands. Blood Cells Mol. Dis. 1999, 25, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Bigam, C.G.; Holm, A.; Hodges, R.S.; Sykes, B.D. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR 1995, 5, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 1992, 31, 1647–1651. [Google Scholar] [CrossRef] [PubMed]
- Skinner, S.P.; Fogh, R.H.; Boucher, W.; Ragan, T.J.; Mureddu, L.G.; Vuister, G.W. CcpNmr AnalysisAssign: A flexible platform for integrated NMR analysis. J. Biomol. NMR 2016, 66, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Dawber, R.S.; Zhang, P.; Walko, M.; Wilson, A.J.; Wang, X. Peptide-based inhibitors of protein-protein interactions: Biophysical, structural and cellular consequences of introducing a constraint. Chem. Sci. 2021, 12, 5977–5993. [Google Scholar] [CrossRef] [PubMed]
- Mól, A.R.; Castro, M.S.; Fontes, W. NetWheels: A web application to create high quality peptide helical wheel and net projections. bioRxiv 2018. [CrossRef]
- Schroeder, C.I.; Rosengren, K.J. Three-Dimensional Structure Determination of Peptides Using Solution Nuclear Magnetic Resonance Spectroscopy. Methods Mol. Biol. 2020, 2068, 129–162. [Google Scholar] [CrossRef] [PubMed]
- Shelton, P.T.; Jensen, K.J. Linkers, resins, and general procedures for solid-phase peptide synthesis. In Peptide Synthesis and Applications, 2nd ed.; Jensen, K.J., Shelton, P.T., Pedersen, S.L., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 1047, pp. 23–41. [Google Scholar]
- Yue, C.W.; Thierry, J.; Potier, P. 2-Phenyl Isopropyl Esters as Carboxyl Terminus Protecting Groups in the Fast Synthesis of Peptide-Fragments. Tetrahedron Lett. 1993, 34, 323–326. [Google Scholar] [CrossRef]
- Li, D.; Elbert, D.L. The kinetics of the removal of the N-methyltrityl (Mtt) group during the synthesis of branched peptides. J. Pept. Res. 2002, 60, 300–303. [Google Scholar] [CrossRef]



| Peptide | Sequence | # Residues | Residue # in Protein Sequence and Comments |
|---|---|---|---|
| TFR1 h1 | SKLLSFMKDLN | 11 | 619–629 |
| TFR1 h3 | QWLYSARGDYFRATSRLTT | 19 | 643–661 |
| TFR1 h1h3 | SKLLSFMKDLNFKTQWLYSARGDYF | 25 | 619–629 F K632 T 643–653 |
| TFR1 negative control | LKLAQVFSDMISKD | 14 | 430–443 |
| HFE α1 | LHLSQSLKGWDYMFIVDFWTIMGN | 24 | 89–112 |
| HFE α2 | EWDEHKIRAKQ | 11 | 180–190 |
| HFE α1α2 | YMFIVDFWTIMGNGGEWDEHKIRAKQ | 26 | 100–112 GG 180–190 |
| HFE α1α2-s1 | YMFIVDFWTIMGNGGEDDEHKIRAKQ | 26 | Lactam staple linking D17 to K21 |
| HFE α1α2-s2 | YMFIVDFWTKMGNGGEWDEHKIRAKQ | 26 | Lactam staple linking D6 to K10 |
| HFE α1α2-s1/2 | YMFIVDFWTKMGNGGEDDEHKIRAKQ | 26 | Lactam staple linking D6 to K10 and D17 to K21 |
| HFE α1α2 scrambled | WDNRKAYMIGIETQDKFHIVMEWGFG | 26 | Random scrambling of the sequence of HFE α1α2 |
| HFE negative control | EDNSTSGFWRYGYDG | 15 | 140–153 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goncalves Monteiro, D.; Rishi, G.; Gorman, D.M.; Burnet, G.; Aliyanto, R.; Rosengren, K.J.; Frazer, D.M.; Subramaniam, V.N.; Clark, R.J. Engineering Peptide Inhibitors of the HFE–Transferrin Receptor 1 Complex. Molecules 2022, 27, 6581. https://doi.org/10.3390/molecules27196581
Goncalves Monteiro D, Rishi G, Gorman DM, Burnet G, Aliyanto R, Rosengren KJ, Frazer DM, Subramaniam VN, Clark RJ. Engineering Peptide Inhibitors of the HFE–Transferrin Receptor 1 Complex. Molecules. 2022; 27(19):6581. https://doi.org/10.3390/molecules27196581
Chicago/Turabian StyleGoncalves Monteiro, Daniela, Gautam Rishi, Declan M. Gorman, Guillaume Burnet, Randy Aliyanto, K. Johan Rosengren, David M. Frazer, V. Nathan Subramaniam, and Richard J. Clark. 2022. "Engineering Peptide Inhibitors of the HFE–Transferrin Receptor 1 Complex" Molecules 27, no. 19: 6581. https://doi.org/10.3390/molecules27196581
APA StyleGoncalves Monteiro, D., Rishi, G., Gorman, D. M., Burnet, G., Aliyanto, R., Rosengren, K. J., Frazer, D. M., Subramaniam, V. N., & Clark, R. J. (2022). Engineering Peptide Inhibitors of the HFE–Transferrin Receptor 1 Complex. Molecules, 27(19), 6581. https://doi.org/10.3390/molecules27196581







