Effects of Donepezil Treatment on Brain Metabolites, Gut Microbiota, and Gut Metabolites in an Amyloid Beta-Induced Cognitive Impairment Mouse Pilot Model
Abstract
:1. Introduction
2. Results
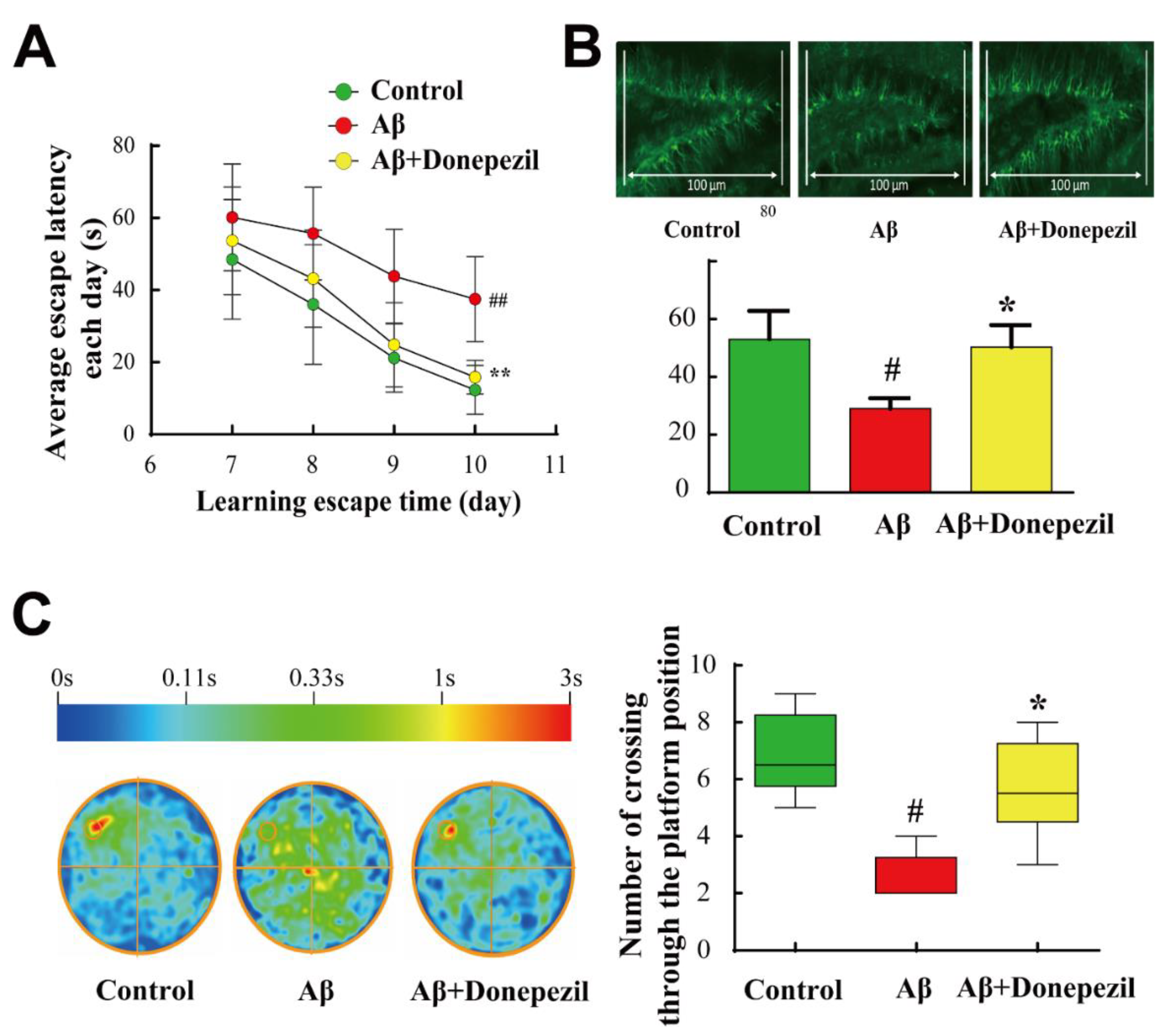
2.1. Effect of Donepezil on Aβ25–35 Memory Impairment
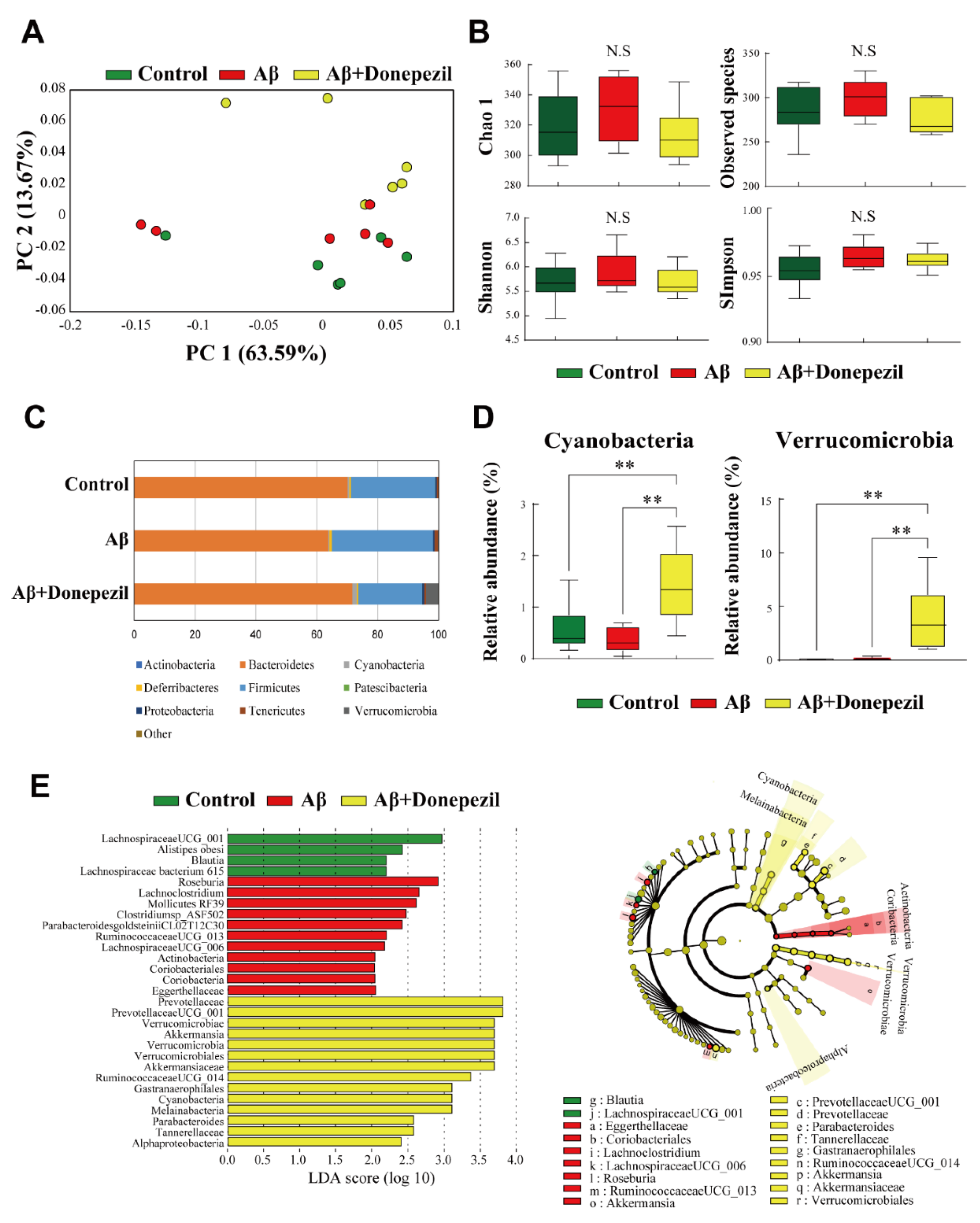
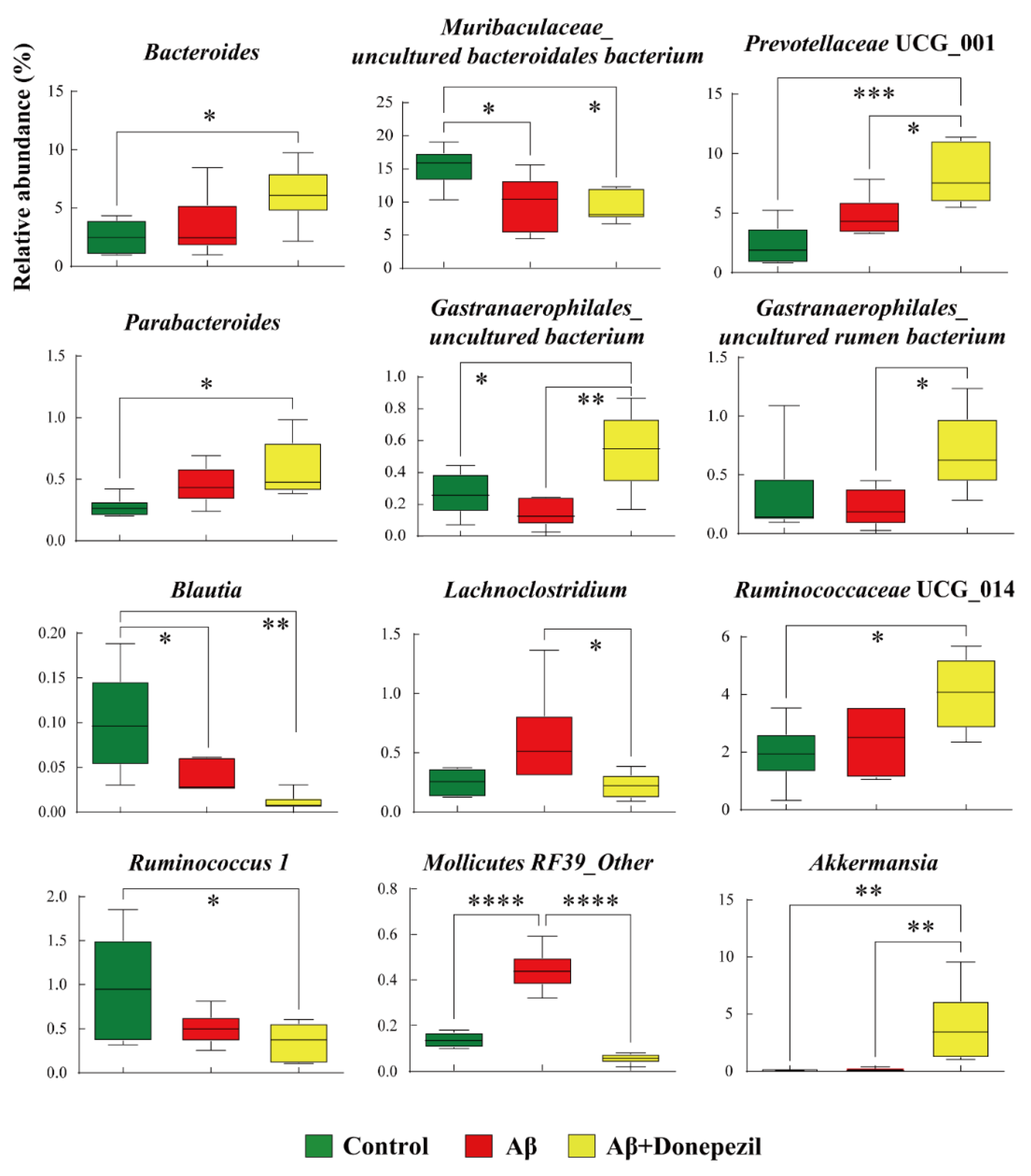
2.2. Composition of the Feces Microbiome
2.3. Profiling of Feces Metabolites
2.4. Profiling of Brain Tissue Metabolites
2.5. Profiling of HT-22 Cell Metabolites
2.6. Metabolic Pathway Analysis
2.7. Correlation between Microorganisms and Metabolites
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. In Vivo Administration
4.3. MWM Test
4.4. Collection of Animal Brain Tissue and Feces
4.5. Doublecortin Immunofluorescence Staining
4.6. Cell Culturing and Donepezil Treatment
4.7. DNA Extraction and 16S rRNA Gene Amplicon Sequencing
4.8. Metabolites Extraction from Cells, Brain Tissue, and Feces
4.9. Sample Derivatization and Metabolite Analysis
4.10. Data Processing and Multivariate Analysis
4.11. Statistical Analysis
4.12. Metabolic Pathway Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ausó, E.; Gómez-Vicente, V.; Esquiva, G. Biomarkers for Alzheimer’s disease early diagnosis. J. Pers. Med. 2020, 10, 114. [Google Scholar] [CrossRef] [PubMed]
- Worley, B.; Powers, R. Multivariate analysis in metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar]
- Mahajan, U.V.; Varma, V.R.; Griswold, M.E.; Blackshear, C.T.; An, Y.; Oommen, A.M.; Varma, S.; Troncoso, J.C.; Pletnikova, O.; O’Brien, R. Dysregulation of multiple metabolic networks related to brain transmethylation and polyamine pathways in Alzheimer disease: A targeted metabolomic and transcriptomic study. PLoS Med. 2020, 17, e1003012. [Google Scholar]
- Kim, E.; Jung, Y.S.; Kim, H.; Kim, J.S.; Park, M.; Jeong, J.; Lee, S.K.; Yoon, H.G.; Hwang, G.S.; Namkoong, K. Metabolomic signatures in peripheral blood associated with Alzheimer’s disease amyloid-β-induced neuroinflammation. J. Alzheimers Dis. 2014, 42, 421–433. [Google Scholar] [CrossRef]
- Wang, J.; Gu, B.J.; Masters, C.L.; Wang, Y.J. A systemic view of Alzheimer disease—Insights from amyloid-β metabolism beyond the brain. Nat. Rev. Neurol. 2017, 13, 612–623. [Google Scholar] [CrossRef]
- Valles-Colomer, M.; Falony, G.; Darzi, Y.; Tigchelaar, E.F.; Wang, J.; Tito, R.Y.; Schiweck, C.; Kurilshikov, A.; Joossens, M.; Wijmenga, C.; et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019, 4, 623–632. [Google Scholar] [CrossRef]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K.; et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017, 7, 13537. [Google Scholar] [CrossRef] [Green Version]
- Minter, M.R.; Hinterleitner, R.; Meisel, M.; Zhang, C.; Leone, V.; Zhang, X.; Oyler-Castrillo, P.; Zhang, X.; Much, M.W.; Shen, X.; et al. Antibiotic-induced perturbations in microbial diversity during post-natal development alters amyloid pathology in an aged APPSWE/PS1ΔE9 murine model of Alzheimer’s disease. Sci. Rep. 2017, 7, 10411. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Mo, X.; Huang, H.; Chen, X.; Liu, H.; Peng, Z.; Chen, L.; Rong, S.; Yang, W.; Xu, S.; et al. Yeast β-glucan alleviates cognitive deficit by regulating gut microbiota and metabolites in Aβ1–42-induced AD-like mice. Int. J. Biol. Macromol. 2020, 161, 258–270. [Google Scholar] [CrossRef]
- Zhuang, Z.Q.; Shen, L.L.; Li, W.W.; Fu, X.; Zeng, F.; Gui, L.; Lü, Y.; Cai, M.; Zhu, C.; Tan, Y.L.; et al. Gut microbiota is altered in patients with Alzheimer’s disease. J. Alzheimers. Dis. 2018, 63, 1337–1346. [Google Scholar] [CrossRef] [Green Version]
- Strandwitz, P.; Kim, K.H.; Terekhova, D.; Liu, J.K.; Sharma, A.; Levering, J.; McDonald, D.; Dietrich, D.; Ramadhar, T.R.; Lekbua, A.; et al. GABA-modulating bacteria of the human gut microbiota. Nat. Microbiol. 2019, 4, 396–403. [Google Scholar] [CrossRef]
- Bäuerl, C.; Collado, M.C.; Diaz Cuevas, A.; Viña, J.; Pérez Martínez, G. Shifts in gut microbiota composition in an APP/PSS 1 transgenic mouse model of Alzheimer’s disease during lifespan. Lett. Appl. Microbiol. 2018, 66, 464–471. [Google Scholar] [CrossRef]
- Cuervo-Zanatta, D.; Garcia-Mena, J.; Perez-Cruz, C. Gut microbiota alterations and cognitive impairment are sexually dissociated in a transgenic mice model of Alzheimer’s disease. J. Alzheimer’s Dis. 2021, 82, S195–S214. [Google Scholar] [CrossRef]
- Harach, T.; Marungruang, N.; Duthilleul, N.; Cheatham, V.; Mc Coy, K.; Frisoni, G.; Neher, J.; Fåk, F.; Jucker, M.; Lasser, T.; et al. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci. Rep. 2017, 7, 41802. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.Y.; Lee, D.K.; Chung, B.R.; Kim, H.V.; Kim, Y. Intracerebroventricular injection of amyloid-β peptides in normal mice to acutely induce Alzheimer-like cognitive deficits. J. Vis. Exp. 2016, 109, 53308. [Google Scholar] [CrossRef] [Green Version]
- Ohno, M. Alzheimer’s therapy targeting the β-secretase enzyme BACE1: Benefits and potential limitations from the perspective of animal model studies. Brain Res. Bull. 2016, 126, 183–198. [Google Scholar] [CrossRef]
- Zhang, N.; Gordon, M.L. Clinical efficacy and safety of donepezil in the treatment of Alzheimer’s disease in Chinese patients. Clin. Interv. Aging 2018, 13, 1963–1970. [Google Scholar] [CrossRef] [Green Version]
- D’Hooge, R.; De Deyn, P.P. Applications of the Morris water maze in the study of learning and memory. Brain Res. Rev. 2001, 36, 60–90. [Google Scholar] [CrossRef]
- Hainmueller, T.; Bartos, M. Dentate gyrus circuits for encoding, retrieval and discrimination of episodic memories. Nat. Rev. Neurosci. 2020, 21, 153–168. [Google Scholar] [CrossRef]
- Bernier, B.E.; Lacagnina, A.F.; Ayoub, A.; Shue, F.; Zemelman, B.V.; Krasne, F.B.; Drew, M.R. Dentate gyrus contributes to retrieval as well as encoding: Evidence from context fear conditioning, recall, and extinction. J. Neurosci. 2017, 37, 6359–6371. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Wissmann, P.; Geisler, S.; Leblhuber, F.; Fuchs, D. Immune activation in patients with Alzheimer’s disease is associated with high serum phenylalanine concentrations. J. Neurol. Sci. 2013, 329, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Han, Y.; Du, J.; Liu, R.; Jin, K.; Yi, W. Microbiota-gut-brain axis and the central nervous system. Oncotarget 2017, 8, 53829–53838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain. Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finegold, S.M.; Dowd, S.E.; Gontcharova, V.; Liu, C.; Henley, K.E.; Wolcott, R.D.; Youn, E.; Summanen, P.H.; Granpeesheh, D.; Dixon, D.; et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 2010, 16, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Arnoriaga-Rodríguez, M.; Fernández-Real, J.M. Microbiota impacts on chronic inflammation and metabolic syndrome-related cognitive dysfunction. Rev. Endocr. Metab. Disord. 2019, 20, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Dao, M.C.; Everard, A.; Aron-Wisnewsky, J.; Sokolovska, N.; Prifti, E.; Verger, E.O.; Kayser, B.D.; Levenez, F.; Chilloux, J.; Hoyles, L.; et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: Relationship with gut microbiome richness and ecology. Gut 2016, 65, 426–436. [Google Scholar] [CrossRef] [Green Version]
- Grander, C.; Adolph, T.E.; Wieser, V.; Lowe, P.; Wrzosek, L.; Gyongyosi, B.; Ward, D.V.; Grabherr, F.; Gerner, R.R.; Pfister, A.; et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut 2018, 67, 891–901. [Google Scholar] [CrossRef]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef] [Green Version]
- Ou, Z.; Deng, L.; Lu, Z.; Wu, F.; Liu, W.; Huang, D.; Peng, Y. Protective effects of Akkermansia muciniphila on cognitive deficits and amyloid pathology in a mouse model of Alzheimer’s disease. Nutr. Diabetes 2020, 10, 12. [Google Scholar] [CrossRef] [Green Version]
- Gershon, M.D. 5-Hydroxytryptamine (serotonin) in the gastrointestinal tract. Curr. Opin. Endocrinol. Diabetes Obes. 2013, 20, 14–21. [Google Scholar] [CrossRef] [Green Version]
- Margolis, K.G.; Stevanovic, K.; Li, Z.; Yang, Q.M.; Oravecz, T.; Zambrowicz, B.; Jhaver, K.G.; Diacou, A.; Gershon, M.D. Pharmacological reduction of mucosal but not neuronal serotonin opposes inflammation in mouse intestine. Gut 2014, 63, 928–937. [Google Scholar] [CrossRef]
- Yaghoubfar, R.; Behrouzi, A.; Ashrafian, F.; Shahryari, A.; Moradi, H.R.; Choopani, S.; Hadifar, S.; Vaziri, F.; Nojoumi, S.A.; Fateh, A.; et al. Modulation of serotonin signaling/metabolism by Akkermansia muciniphila and its extracellular vesicles through the gut-brain axis in mice. Sci. Rep. 2020, 10, 22119. [Google Scholar] [CrossRef]
- Shearman, E.; Rossi, S.; Szasz, B.; Juranyi, Z.; Fallon, S.; Pomara, N.; Sershen, H.; Lajtha, A. Changes in cerebral neurotransmitters and metabolites induced by acute donepezil and memantine administrations: A microdialysis study. Brain Res. Bull. 2006, 69, 204–213. [Google Scholar] [CrossRef]
- Vascellari, S.; Palmas, V.; Melis, M.; Pisanu, S.; Cusano, R.; Uva, P.; Perra, D.; Madau, V.; Sarchioto, M.; Oppo, V.; et al. Gut microbiota and metabolome alterations associated with Parkinson’s disease. mSystems 2020, 5, e00561-20. [Google Scholar] [CrossRef]
- O’Bryant, S.E.; Xiao, G.; Barber, R.; Huebinger, R.; Wilhelmsen, K.; Edwards, M.; Graff-Radford, N.; Doody, R.; Diaz-Arrastia, R. A blood-based screening tool for Alzheimer’s disease that spans serum and plasma: Findings from TARC and ADNI. PLoS ONE 2011, 6, e28092. [Google Scholar] [CrossRef]
- Tyagi, E.; Agrawal, R.; Nath, C.; Shukla, R. Cholinergic protection via α7 nicotinic acetylcholine receptors and PI3K-Akt pathway in LPS-induced neuroinflammation. Neurochem. Int. 2010, 56, 135–142. [Google Scholar] [CrossRef]
- Zhang, Z.; Tan, X.; Sun, X.; Wei, J.; Li, Q.X.; Wu, Z. Isoorientin affects markers of Alzheimer’s Disease via effects on the oral and gut microbiota in APP/PS1 mice. J. Nutr. 2022, 152, 140–152. [Google Scholar] [CrossRef]
- Li, W.; Zhao, Q.; Wang, J.; Wang, Y.; Wen, T. Dcf1 deletion presents alterations in gut microbiota of mice similar to Parkinson’s disease. Biochem. Biophys. Res. Commun. 2020, 529, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Bangsgaard Bendtsen, K.M.; Krych, L.; Sørensen, D.B.; Pang, W.; Nielsen, D.S.; Josefsen, K.; Hansen, L.H.; Sørensen, S.J.; Hansen, A.K. Gut microbiota composition is correlated to grid floor induced stress and behavior in the BALB/c mouse. PLoS ONE 2012, 7, e46231. [Google Scholar] [CrossRef] [Green Version]
- Darnaud, M.; Dos Santos, A.; Gonzalez, P.; Augui, S.; Lacoste, C.; Desterke, C.; De Hertogh, G.; Valentino, E.; Braun, E.; Zheng, J.; et al. Enteric delivery of regenerating family member 3 alpha alters the intestinal microbiota and controls inflammation in mice with colitis. Gastroenterology 2018, 154, 1009–1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadovnikova, I.S.; Gureev, A.P.; Ignatyeva, D.A.; Gryaznova, M.V.; Chernyshova, E.V.; Krutskikh, E.P.; Novikova, A.G.; Popov, V.N. Nrf2/ARE activators improve memory in aged mice via maintaining of mitochondrial quality control of brain and the modulation of gut microbiome. Pharmaceuticals 2021, 14, 607. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Diling, C.; Jian, Y.; Ting, L.; Guoyan, H.; Hualun, L.; Xiaocui, T.; Guoxiao, L.; Ou, S.; Chaoqun, Z.; et al. Effects of oligosaccharides from Morinda officinalis on gut microbiota and metabolome of APP/PS1 transgenic mice. Front. Neurol. 2018, 9, 412. [Google Scholar] [CrossRef]
- Park, J.Y.; Choi, J.; Lee, Y.; Lee, J.E.; Lee, E.H.; Kwon, H.J.; Yang, J.; Jeong, B.R.; Kim, Y.K.; Han, P.L. Metagenome analysis of bodily microbiota in a mouse model of Alzheimer disease using bacteria-derived membrane vesicles in blood. Exp. Neurobiol. 2017, 26, 369–379. [Google Scholar] [CrossRef]
- Gueli, M.C.; Taibi, G. Alzheimer’s disease: Amino acid levels and brain metabolic status. Neurol. Sci. 2013, 34, 1575–1579. [Google Scholar] [CrossRef] [Green Version]
- Guillemin, G.J.; Brew, B.J. Implications of the kynurenine pathway and quinolinic acid in Alzheimer’s disease. Redox Rep. 2002, 7, 199–206. [Google Scholar] [CrossRef] [Green Version]
- Gwanyanya, A.; Godsmark, C.N.; Kelly-Laubscher, R. Ethanolamine: A potential promoiety with additional effects on the brain. CNS Neurol. Disord. Drug Targets 2022, 21, 108–117. [Google Scholar] [CrossRef]
- Schrag, M.; Mueller, C.; Zabel, M.; Crofton, A.; Kirsch, W.; Ghribi, O.; Squitti, R.; Perry, G. Oxidative stress in blood in Alzheimer’s disease and mild cognitive impairment: A meta-analysis. Neurobiol. Dis. 2013, 59, 100–110. [Google Scholar] [CrossRef]
- Shen, H.; Kihara, T.; Hongo, H.; Wu, X.; Kem, W.; Shimohama, S.; Akaike, A.; Niidome, T.; Sugimoto, H. Neuroprotection by donepezil against glutamate excitotoxicity involves stimulation of α7 nicotinic receptors and internalization of NMDA receptors. Br. J. Pharmacol. 2010, 161, 127–139. [Google Scholar] [CrossRef] [Green Version]
- Takada-Takatori, Y.; Kume, T.; Sugimoto, M.; Katsuki, H.; Sugimoto, H.; Akaike, A. Acetylcholinesterase inhibitors used in treatment of Alzheimer’s disease prevent glutamate neurotoxicity via nicotinic acetylcholine receptors and phosphatidylinositol 3-kinase cascade. Neuropharmacology 2006, 51, 474–486. [Google Scholar] [CrossRef]
- Greene, J.G.; Porter, R.H.; Eller, R.V.; Greenamyre, J.T. Inhibition of succinate dehydrogenase by malonic acid produces an “excitotoxic” lesion in rat striatum. J. Neurochem. 1993, 61, 1151–1154. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hong, S.; Shepardson, N.E.; Walsh, D.M.; Shankar, G.M.; Selkoe, D. Soluble oligomers of amyloid β protein facilitate hippocampal long-term depression by disrupting neuronal glutamate uptake. Neuron 2009, 62, 788–801. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Liu, H.; Li, X.; Zhang, A.-H. High-throughput metabolomics analysis discovers salivary biomarkers for predicting mild cognitive impairment and Alzheimer’s disease. RSC Adv. 2016, 6, 75499–75504. [Google Scholar] [CrossRef]
- Guidotti, A.; Badiani, G.; Pepeu, G. Taurine distribution in cat brain. J. Neurochem. 1972, 19, 431–435. [Google Scholar] [CrossRef]
- Santa-María, I.; Hernández, F.; Moreno, F.J.; Avila, J. Taurine, an inducer for tau polymerization and a weak inhibitor for amyloid-β-peptide aggregation. Neurosci. Lett. 2007, 429, 91–94. [Google Scholar] [CrossRef]
- Fang, F.; Liu, G.T. Protective effects of compound FLZ on β-amyloid peptide-(25–35)-induced mouse hippocampal injury and learning and memory impairment. Acta Pharmacol. Sin. 2006, 27, 651–658. [Google Scholar] [CrossRef]
- Dong, H.; Yuede, C.M.; Coughlan, C.A.; Murphy, K.M.; Csernansky, J.G. Effects of donepezil on amyloid-β and synapse density in the Tg2576 mouse model of Alzheimer’s disease. Brain Res. 2009, 1303, 169–178. [Google Scholar] [CrossRef] [Green Version]
- Gu, S.M.; Park, M.H.; Hwang, C.J.; Song, H.S.; Lee, U.S.; Han, S.B.; Oh, K.W.; Ham, Y.W.; Song, M.J.; Son, D.J.; et al. Bee venom ameliorates lipopolysaccharide-induced memory loss by preventing NF-kappaB pathway. J. Neuroinflammation 2015, 12, 124. [Google Scholar] [CrossRef] [Green Version]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, 2584. [Google Scholar] [CrossRef] [Green Version]
- Lommen, A. MetAlign: Interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal. Chem. 2009, 81, 3079–3086. [Google Scholar] [CrossRef]
- Tsugawa, H.; Bamba, T.; Shinohara, M.; Nishiumi, S.; Yoshida, M.; Fukusaki, E. Practical non-targeted gas chromatography/mass spectrometry-based metabolomics platform for metabolic phenotype analysis. J. Biosci. Bioeng. 2011, 112, 292–298. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, 86. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, J.-K.; Lee, G.; Nguyen, C.D.; Park, S.-E.; Kim, E.-J.; Kim, H.-W.; Seo, S.-H.; Cho, K.-M.; Kwon, S.J.; Kim, J.-H.; et al. Effects of Donepezil Treatment on Brain Metabolites, Gut Microbiota, and Gut Metabolites in an Amyloid Beta-Induced Cognitive Impairment Mouse Pilot Model. Molecules 2022, 27, 6591. https://doi.org/10.3390/molecules27196591
Jo J-K, Lee G, Nguyen CD, Park S-E, Kim E-J, Kim H-W, Seo S-H, Cho K-M, Kwon SJ, Kim J-H, et al. Effects of Donepezil Treatment on Brain Metabolites, Gut Microbiota, and Gut Metabolites in an Amyloid Beta-Induced Cognitive Impairment Mouse Pilot Model. Molecules. 2022; 27(19):6591. https://doi.org/10.3390/molecules27196591
Chicago/Turabian StyleJo, Jae-Kwon, Gihyun Lee, Cong Duc Nguyen, Seong-Eun Park, Eun-Ju Kim, Hyun-Woo Kim, Seung-Ho Seo, Kwang-Moon Cho, Sun Jae Kwon, Jae-Hong Kim, and et al. 2022. "Effects of Donepezil Treatment on Brain Metabolites, Gut Microbiota, and Gut Metabolites in an Amyloid Beta-Induced Cognitive Impairment Mouse Pilot Model" Molecules 27, no. 19: 6591. https://doi.org/10.3390/molecules27196591
APA StyleJo, J.-K., Lee, G., Nguyen, C. D., Park, S.-E., Kim, E.-J., Kim, H.-W., Seo, S.-H., Cho, K.-M., Kwon, S. J., Kim, J.-H., & Son, H.-S. (2022). Effects of Donepezil Treatment on Brain Metabolites, Gut Microbiota, and Gut Metabolites in an Amyloid Beta-Induced Cognitive Impairment Mouse Pilot Model. Molecules, 27(19), 6591. https://doi.org/10.3390/molecules27196591







