Thermostability Improvement of L-Asparaginase from Acinetobacter soli via Consensus-Designed Cysteine Residue Substitution
Abstract
1. Introduction
2. Results
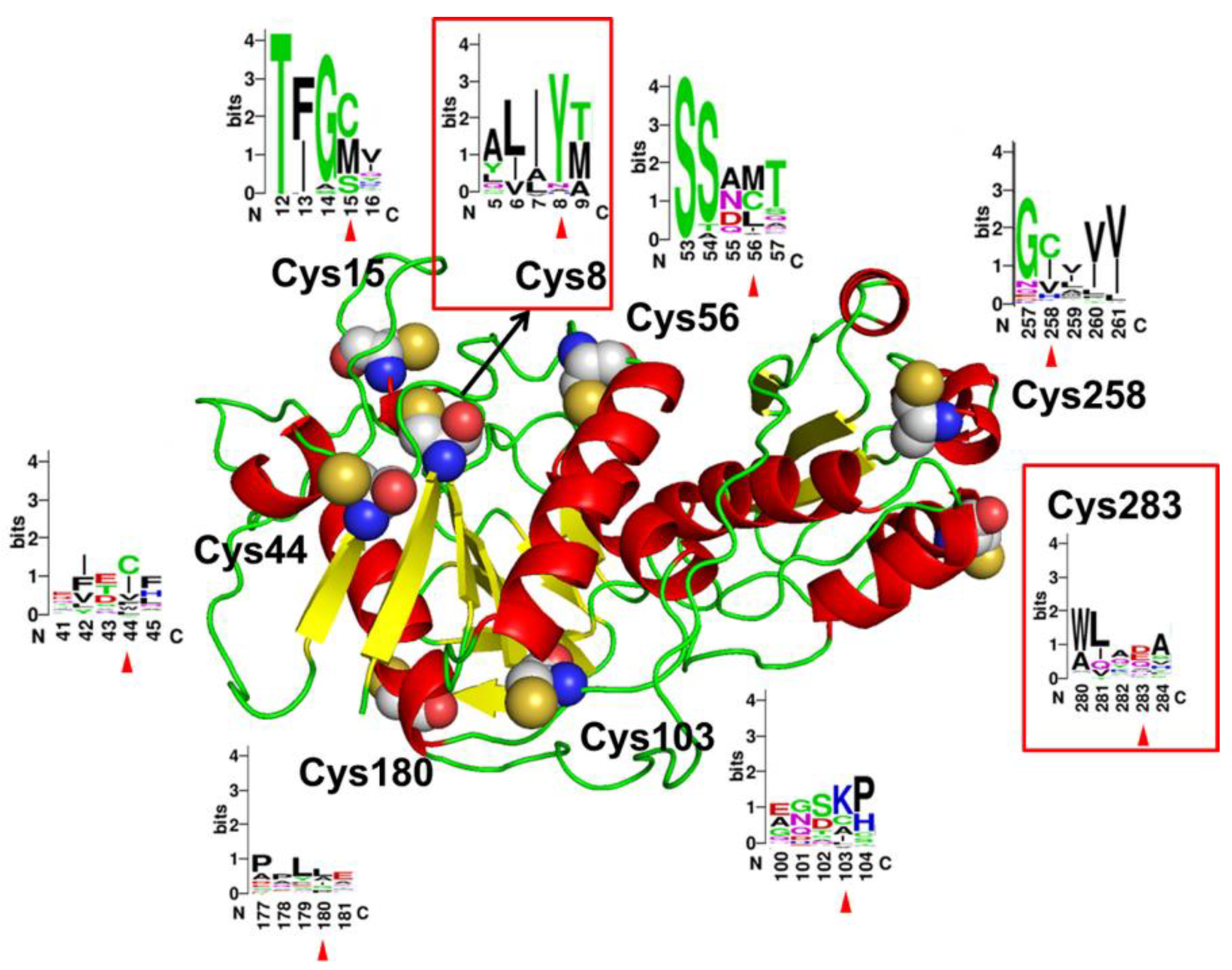
2.1. Cys8 and Cys283 with Low Conservation Selected for Mutagenesis to Enhance the Thermostability of AsA
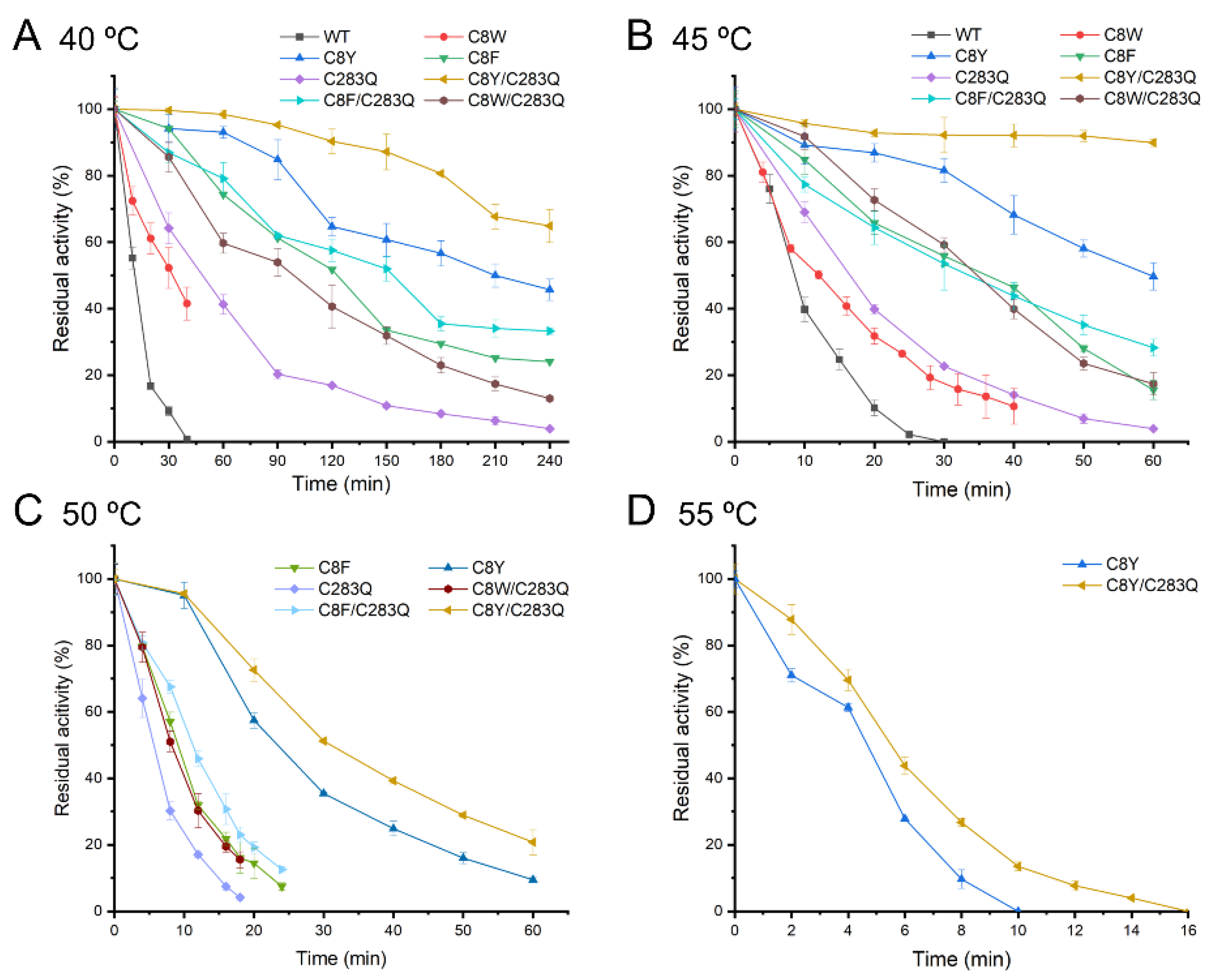
2.2. Thermostability of the AsA Variants
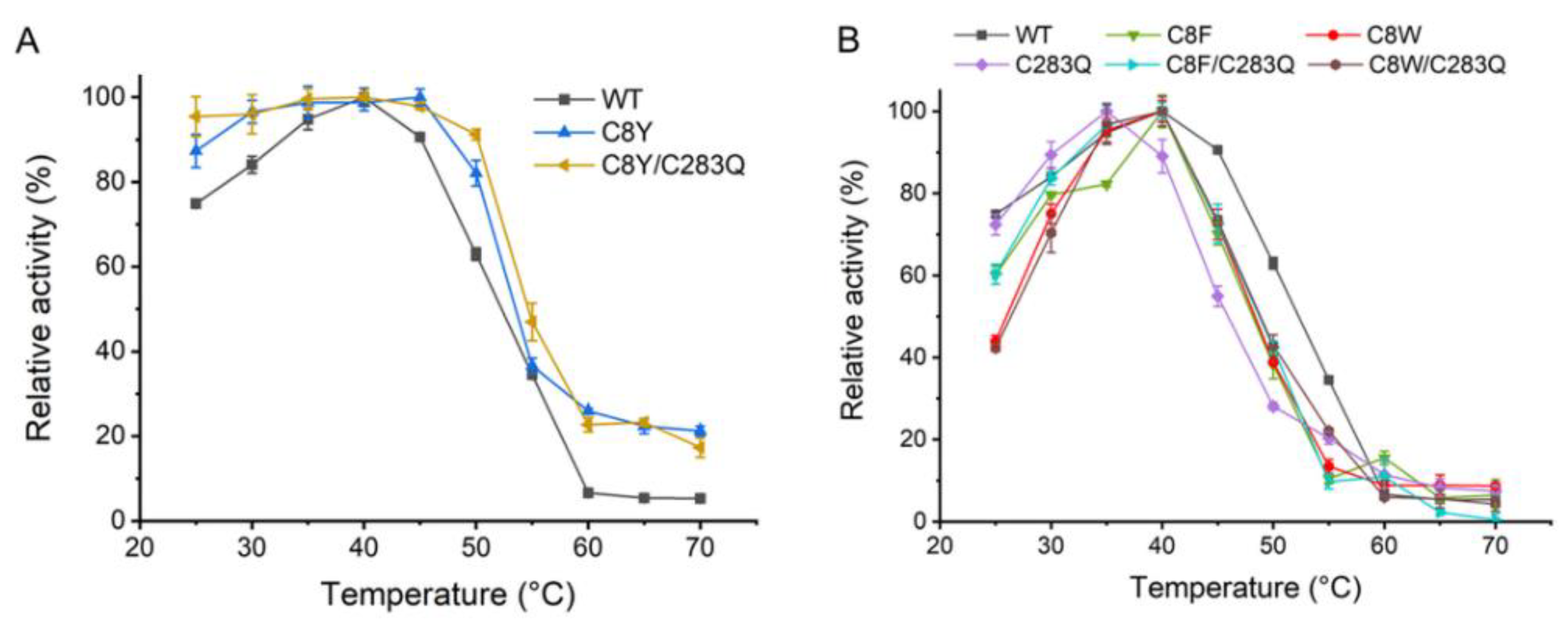
2.3. Optimum Temperature of AsA Variants
2.4. Kinetic Parameters of the AsA Variants
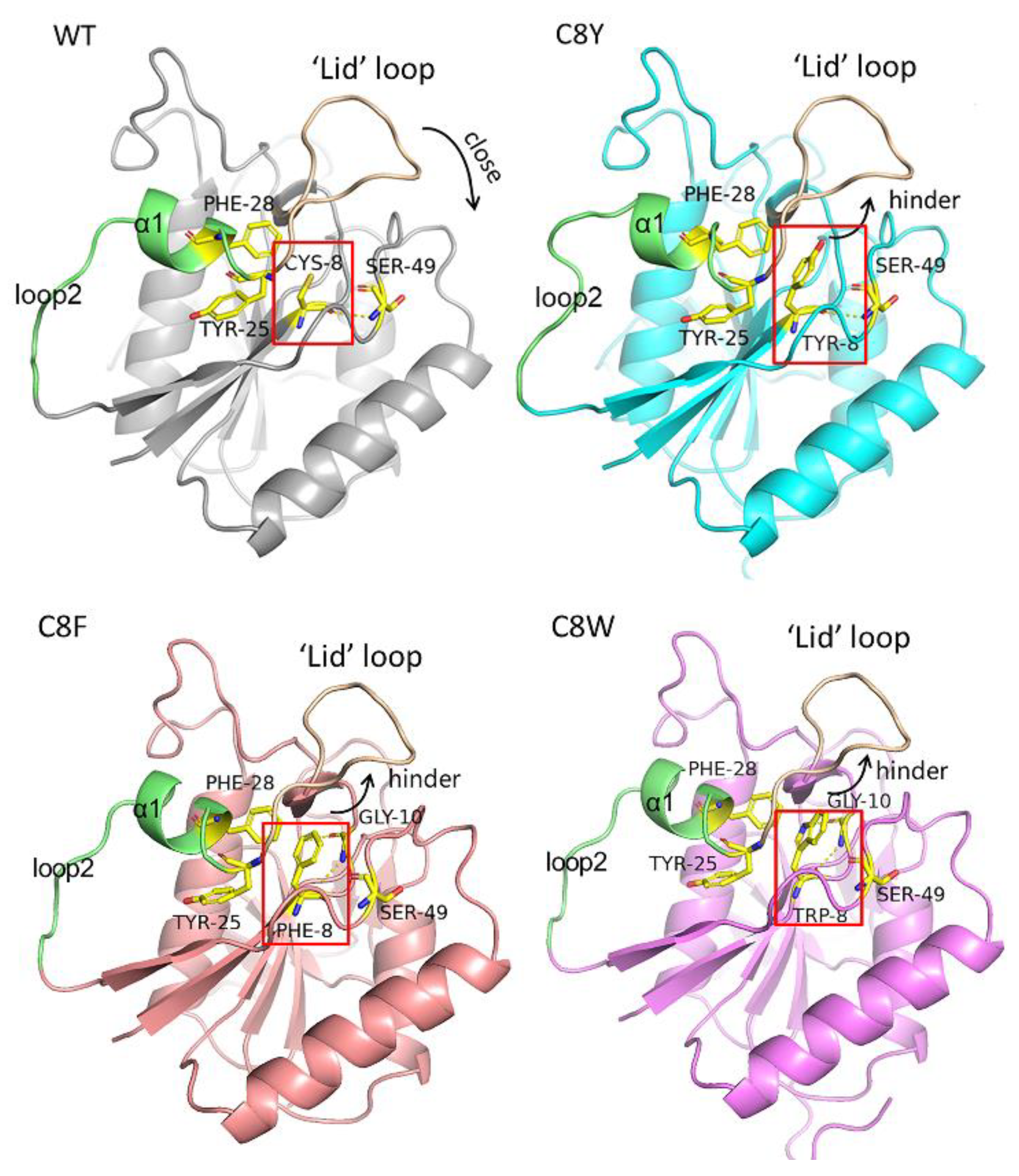
2.5. Model Analysis and MD Simulation of the AsA Variants
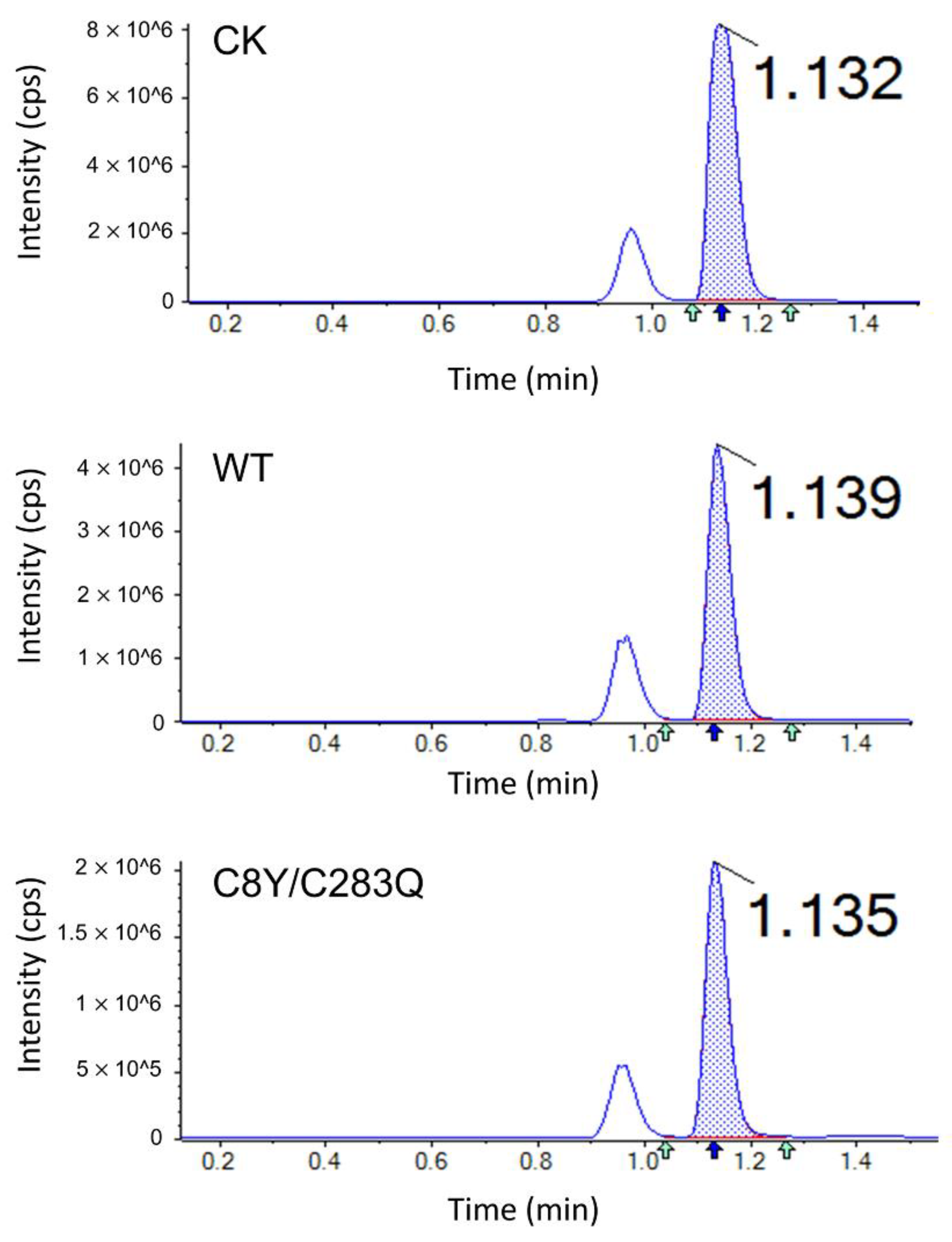
2.6. Acrylamide Inhibition Analysis of AsA Variants
3. Discussion
4. Materials and Methods
4.1. Plasmid, Strains, and Materials
4.2. Site-Directed Saturation Mutagenesis
4.3. Enzyme Expression and Purification
4.4. Activity Assay
4.5. Thermostability Analysis
4.6. Enzymatic Kinetic Analysis
4.7. Consensus Analysis
4.8. Structure Modeling
4.9. MD Simulation
4.10. Acrylamide Inhibition Analysis
4.10.1. Potato Chips Preparation
4.10.2. Acrylamide Extraction and Detection
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nunes, J.C.F.; Cristovao, R.O.; Freire, M.G.; Santos-Ebinuma, V.C.; Faria, J.L.; Silva, C.G.; Tavares, A.P.M. Recent strategies and applications for L-asparaginase confinement. Molecules 2020, 25, 5827. [Google Scholar] [CrossRef]
- Avramis, V.I.; Panosyan, E.H. Pharmacokinetic/pharmacodynamic relationships of asparaginase formulations: The past, the present and recommendations for the future. Clin. Pharmacokinet. 2005, 44, 367–393. [Google Scholar] [CrossRef]
- Marini, B.L.; Perissinotti, A.J.; Bixby, D.L.; Brown, J.; Burke, P.W. Catalyzing improvements in ALL therapy with asparaginase. Blood Rev. 2017, 31, 328–338. [Google Scholar] [CrossRef]
- da Cunha, M.C.; Dos Santos Aguilar, J.G.; de Melo, R.R.; Nagamatsu, S.T.; Ali, F.; de Castro, R.J.S.; Sato, H.H. Fungal L-asparaginase: Strategies for production and food applications. Food Res. Int. 2019, 126, 108658. [Google Scholar] [CrossRef]
- Keramat, J.; Lebail, A.; Prost, C.; Soltanizadeh, N. Acrylamide in foods: Chemistry and analysis. A review. Food Bioprocess Technol. 2011, 4, 340–363. [Google Scholar] [CrossRef]
- Correa, C.L.O.; das Merces Penha, E.; Dos Anjos, M.R.; Pacheco, S.; Freitas-Silva, O.; Luna, A.S.; Gottschalk, L.M.F. Use of asparaginase for acrylamide mitigation in coffee and its influence on the content of caffeine, chlorogenic acid, and caffeic acid. Food Chem. 2021, 338, 128045. [Google Scholar] [CrossRef]
- Sun, Z.; Qin, R.; Li, D.; Ji, K.; Wang, T.; Cui, Z.; Huang, Y. A novel bacterial type II L-asparaginase and evaluation of its enzymatic acrylamide reduction in French fries. Int. J. Biol. Macromol. 2016, 92, 232–239. [Google Scholar] [CrossRef]
- Baskar, G.; Aiswarya, R. Overview on mitigation of acrylamide in starchy fried and baked foods. J. Sci. Food Agric. 2018, 98, 4385–4394. [Google Scholar] [CrossRef]
- Kumari, A.; Bhattacharya, B.; Agarwal, T.; Paul, V.; Chakkaravarthi, S. Integrated approach towards acrylamide reduction in potato-based snacks: A critical review. Food Res. Int. 2022, 156, 111172. [Google Scholar] [CrossRef]
- Hendriksen, H.V.; Kornbrust, B.A.; Ostergaard, P.R.; Stringer, M.A. Evaluating the potential for enzymatic acrylamide mitigation in a range of food products using an asparaginase from Aspergillus oryzae. J. Agric. Food Chem. 2009, 57, 4168–4176. [Google Scholar] [CrossRef]
- Jaeger, K.E.; Eggert, T.; Eipper, A.; Reetz, M.T. Directed evolution and the creation of enantioselective biocatalysts. Appl. Microbiol. Biotechnol. 2001, 55, 519–530. [Google Scholar] [CrossRef]
- Sen, S.; Venkata Dasu, V.; Mandal, B. Developments in directed evolution for improving enzyme functions. Appl. Biochem. Biotechnol. 2007, 143, 212–223. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, Q.; Qu, G.; Feng, Y.; Reetz, M.T. Utility of B-factors in protein science: Interpreting rigidity, flexibility, and internal motion and engineering thermostability. Chem. Rev. 2019, 119, 1626–1665. [Google Scholar] [CrossRef]
- Chen, Q.; Xiao, Y.; Shakhnovich, E.I.; Zhang, W.; Mu, W. Semi-rational design and molecular dynamics simulations study of the thermostability enhancement of cellobiose 2-epimerases. Int. J. Biol. Macromol. 2020, 154, 1356–1365. [Google Scholar] [CrossRef]
- Xia, Y.; Cui, W.; Cheng, Z.; Peplowski, L.; Liu, Z.; Kobayashi, M.; Zhou, Z. Improving thermostability and catalytic efficiency of the subunit-fused nitrile hydratase by semi-rational engineering. Chemcatchem 2017, 10, 1370–1375. [Google Scholar] [CrossRef]
- Modarres, H.P.; Mofrad, M.R.; Sanati-Nezhad, A. Protein thermostability engineering. RSC Adv. 2016, 6, 115252–115270. [Google Scholar] [CrossRef]
- Qian, H.; Zhang, C.; Lu, Z.; Xia, B.; Bie, X.; Zhao, H.; Lu, F.; Yang, G.Y. Consensus design for improved thermostability of lipoxygenase from Anabaena sp. PCC 7120. BMC Biotechnol. 2018, 18, 57. [Google Scholar] [CrossRef]
- Sternke, M.; Tripp, K.W.; Barrick, D. Consensus sequence design as a general strategy to create hyperstable, biologically active proteins. Proc. Natl. Acad. Sci. USA 2019, 116, 11275–11284. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Xu, S.; Xu, M.; Yang, T.; Wang, L.; Zhang, H.; Fang, H.; Osire, T.; Rao, Z. Insight into the thermostability of thermophilic L-asparaginase and non-thermophilic L-asparaginase II through bioinformatics and structural analysis. Appl. Microbiol. Biotechnol. 2019, 103, 7055–7070. [Google Scholar] [CrossRef]
- Jiao, L.; Chi, H.; Lu, Z.; Zhang, C.; Chia, S.R.; Show, P.L.; Tao, Y.; Lu, F. Characterization of a novel type I L-asparaginase from Acinetobacter soli and its ability to inhibit acrylamide formation in potato chips. J. Biosci. Bioeng. 2020, 129, 672–678. [Google Scholar] [CrossRef]
- Austin, C.J.; Kosim-Satyaputra, P.; Smith, J.R.; Willows, R.D.; Jamie, J.F. Mutation of cysteine residues alters the heme-binding pocket of indoleamine 2,3-dioxygenase-1. Biochem. Biophys. Res. Commun. 2013, 436, 595–600. [Google Scholar] [CrossRef]
- Hamza, M.A.; Engel, P.C. Enhancing long-term thermal stability in mesophilic glutamate dehydrogenase from Clostridium symbiosum by eliminating cysteine residues. Enzym. Microb. Technol. 2007, 41, 706–710. [Google Scholar] [CrossRef]
- Yokoyama, K.; Ogaya, D.; Utsumi, H.; Suzuki, M.; Kashiwagi, T.; Suzuki, E.; Taguchi, S. Effect of introducing a disulfide bridge on the thermostability of microbial transglutaminase from Streptomyces mobaraensis. Appl. Microbiol. Biotechnol. 2021, 105, 2737–2745. [Google Scholar] [CrossRef]
- Fremaux, I.; Mazères, S.; Brisson-Lougarre, A.; Arnaud, M.; Ladurantie, C.; Fournier, D. Improvement of Drosophila acetylcholinesterase stability by elimination of a free cysteine. BMC Biochem. 2002, 3, 21. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Tatsumi, M.; Takahashi, K.; Tagami, U.; Sugiki, M.; Kashiwagi, T.; Kameya, M.; Okazaki, S.; Mizukoshi, T.; Asano, Y. Protein engineering for improving the thermostability of tryptophan oxidase and insights from structural analysis. J. Biochem. 2018, 164, 359–367. [Google Scholar] [CrossRef]
- Dehnavi, E.; Moeini, S.; Akbarzadeh, A.; Dabirmanesh, B.; Siadat, S.O.R.; Khajeh, K. Improvement of Selenomonas ruminantium β-xylosidase thermal stability by replacing buried free cysteines via site directed mutagenesis. Int. J. Biol. Macromol. 2019, 136, 352–358. [Google Scholar] [CrossRef]
- Tatko, C.D.; Waters, M.L. Selective aromatic interactions in β-hairpin peptides. J. Am. Chem. Soc. 2002, 124, 9372–9373. [Google Scholar] [CrossRef]
- Georis, J.; Esteves, F.D.L.; Lamotte-Brasseur, J.; Bougnet, V.; Giannotta, F.; Frère, J.M.; Devreese, B.; Granier, B. An additional aromatic interaction improves the thermostability and thermophilicity of a mesophilic family 11 xylanase: Structural basis and molecular study. Protein Sci. 2000, 9, 466–475. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Yang, G.; Xie, Y.; Xu, L.; An, J.; Cui, L.; Feng, Y. Modulation of the thermostability and substrate specificity of Candida rugosa lipase1 by altering the acyl-binding residue Gly414 at the α-helix-connecting bend. Enzym. Microb. Technol. 2016, 82, 34–41. [Google Scholar] [CrossRef]
- Xu, Z.; Cen, Y.-K.; Zou, S.-P.; Xue, Y.-P.; Zheng, Y.-G. Recent advances in the improvement of enzyme thermostability by structure modification. Crit. Rev. Biotechnol. 2020, 40, 83–98. [Google Scholar] [CrossRef]
- Dror, A.; Shemesh, E.; Dayan, N.; Fishman, A. Protein engineering by random mutagenesis and structure-guided consensus of Geobacillus stearothermophilus lipase T6 for enhanced stability in methanol. Appl. Environ. Microbiol. 2014, 80, 1515–1527. [Google Scholar] [CrossRef]
- Kotzia, G.A.; Labrou, N.E. Engineering thermal stability of L-asparaginase by in vitro directed evolution. Febs J. 2009, 276, 1750–1761. [Google Scholar] [CrossRef]
- Sudhir, A.P.; Agarwaal, V.V.; Dave, B.R.; Patel, D.H.; Subramanian, R.B. Enhanced catalysis of L-asparaginase from Bacillus licheniformis by a rational redesign. Enzym. Microb. Technol. 2016, 86, 1–6. [Google Scholar] [CrossRef]
- Chen, Y.; Luo, Q.; Zhou, W.; Xie, Z.; Cai, Y.J.; Liao, X.R.; Guan, Z.B. Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis. Appl. Microbiol. Biotechnol. 2017, 101, 1935–1944. [Google Scholar] [CrossRef]
- Beaufils, C.; Man, H.M.; de Poulpiquet, A.; Mazurenko, I.; Lojou, E. From enzyme stability to enzymatic bioelectrode stabilization processes. Catalysts 2021, 11, 497. [Google Scholar] [CrossRef]
- Miller, S.R. An appraisal of the enzyme stability-activity trade-off. Evolution 2017, 71, 1876–1887. [Google Scholar] [CrossRef]
- Feller, G. Protein stability and enzyme activity at extreme biological temperatures. J. Phys.-Condens. Matter 2010, 22, 323101. [Google Scholar] [CrossRef]
- Nguyen, H.A.; Su, Y.; Lavie, A. Structural insight into substrate selectivity of Erwinia chrysanthemi L-asparaginase. Biochemistry 2016, 55, 1246–1253. [Google Scholar] [CrossRef]
- Ran, T.; Jiao, L.; Wang, W.; Chen, J.; Chi, H.; Lu, Z.; Zhang, C.; Xu, D.; Lu, F. Structures of L-asparaginase from Bacillus licheniformis reveal an essential residue for its substrate stereoselectivity. J. Agric. Food Chem. 2021, 69, 223–231. [Google Scholar] [CrossRef]
- Nguyen, H.A.; Durden, D.L.; Lavie, A. The differential ability of asparagine and glutamine in promoting the closed/active enzyme conformation rationalizes the Wolinella succinogenes L-asparaginase substrate specificity. Sci. Rep. 2017, 7, 41643. [Google Scholar] [CrossRef]
- Lu, X.H.; Chen, J.H.; Jiao, L.S.; Zhong, L.; Lu, Z.X.; Zhang, C.; Lu, F.X. Improvement of the activity of L-asparaginase I improvement of the catalytic activity of L-asparaginase I from Bacillus megaterium H-1 by in vitro directed evolution. J. Biosci. Bioeng. 2019, 128, 683–689. [Google Scholar] [CrossRef]
- Zheng, L.; Baumann, U.; Reymond, J.L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004, 32, e115. [Google Scholar] [CrossRef]
- Abbott, J.A.; Livingston, N.M.; Egri, S.B.; Guth, E.; Francklyn, C.S. Characterization of aminoacyl-tRNA synthetase stability and substrate interaction by differential scanning fluorimetry. Methods 2017, 113, 64–71. [Google Scholar] [CrossRef]
- Jones, B.J.; Kan, C.N.E.; Luo, C.; Kazlauskas, R.J. Consensus Finder web tool to predict stabilizing substitutions in proteins. Methods Enzymol. 2020, 643, 129–148. [Google Scholar]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]


| Mutants | Activities (IU/mL) | Residual Activities after Thermal Treatment (IU/mL) | Half-Life (45 °C, min) |
|---|---|---|---|
| Wild-type | 10.4 ± 0.5 d | 0.0 ± 0.0 g | 4.1 |
| C8V | 22.6 ± 1.4 a | 12.3 ± 1.9 ef | 3.3 |
| C8Y | 20.7 ± 1.3 a | 72.6 ± 0.5 a | 57.3 |
| C8F | 17.7 ± 0.3 b | 34.1 ± 2.1 c | 27.6 |
| C8L | 13.7 ± 0.2 c | 21.8 ± 1.1 d | 3.6 |
| C8W | 16.6 ± 0.7 b | 22.2 ± 0.5 d | 12.3 |
| C283R | 13.8 ± 0.6 c | 16.3 ± 0.9 e | 3.6 |
| C283Q | 11.7 ± 0.4 cd | 33.0 ± 2.7 c | 12.5 |
| C283D | 11.1 ± 0.6 d | 8.6 ± 0.4 f | 2.0 |
| C283L | 12.8 ± 1.1 cd | 15.9 ± 1.4 e | 2.6 |
| C283T | 11.5 ± 1.4 cd | 8.8 ± 0.4 f | 2.9 |
| C283E | 11.0 ± 0.4 d | 44.7 ± 2.8 b | 4.0 |
| C283S | 12.1 ± 0.6 cd | 31.2 ± 1.2 c | 3.7 |
| Mutants | Half-Life (min) | Tm (°C) | |||
|---|---|---|---|---|---|
| 40 °C | 45 °C | 50 °C | 55 °C | ||
| Wild-type | 9.1 | 4.1 | ND | ND | 55.2 ± 0.5 c |
| C8Y | 151.2 | 57.3 | 16.8 | 1.7 | 59.4 ± 0.8 b |
| C8F | 111.1 | 27.6 | 6.5 | ND | 58.2 ± 0.6 b |
| C8W | 25.2 | 12.3 | ND | ND | 57.8 ± 0.5 b |
| C283Q | 52.5 | 12.5 | 4.0 | ND | 59.4 ± 1.2 b |
| C8Y/C283Q | 361.6 | 68.0 | 25.2 | 2.5 | 62.3 ± 0.5 a |
| C8F/C283Q | 145.9 | 33.0 | 7.7 | ND | 59.6 ± 0.3 b |
| C8W/C283Q | 83.5 | 22.9 | 6.5 | ND | 60.8 ± 0.5 ab |
| Mutants | Specific Activity (IU/mg) | Km (mM) | Vmax (μM·min−1·mg−1) | kcat (s−1) | kcat/Km (s−1·mM−1) |
|---|---|---|---|---|---|
| Wild-type | 400.8 ± 8.2 c | 3.8 ± 0.5 c | 327.7 ± 17.3 e | 198.0 | 52.0 |
| C8Y | 625.7 ± 12.1 a | 22.6 ± 5.0 a | 1663.9 ± 65.3 a | 1005.0 | 44.5 |
| C8F | 453.6 ± 41.4 c | 10.1 ± 2.2 bc | 864.7 ± 13.0 c | 522.3 | 51.8 |
| C8W | 144.3 ± 16.7 a | 3.3 ± 0.3 c | 244.3 ± 8.9 e | 147.6 | 44.7 |
| C283Q | 570.2 ± 16.7 b | 14.7 ± 2.8 b | 1165.9 ± 39.7 b | 704.2 | 48.1 |
| C8Y/C283Q | 620.8 ± 20.7 ab | 13.6 ± 2.0 bc | 1150.1 ± 104.0 b | 694.7 | 51.1 |
| C8F/C283Q | 287.5 ± 6.6 d | 7.4 ± 0.9 c | 749.3 ± 42.9 cd | 452.6 | 60.9 |
| C8W/C283Q | 217.5 ± 7.8 e | 6.4 ± 0.9 c | 648.5 ± 43.9 d | 391.7 | 61.0 |
| Mutants | Total Hydrogen Bonds Number inside the Protein | Rg (nm) | SASA (Å2) | |||
|---|---|---|---|---|---|---|
| at 300 K | at 330 K | at 300 K | at 330 K | at 300 K | at 330 K | |
| Wild-type | 457.1 ± 10.6 | 461.8 ± 11.2 | 3.1 ± 0.9 | 2.6 ± 0.2 | 263.4 ± 4.0 | 260.0 ± 3.5 |
| C8Y/C283Q | 489.6 ± 10.0 | 494.8 ± 11.7 | 2.7 ± 0.4 | 2.7 ± 0.5 | 259.5 ± 3.1 | 256.8 ± 3.5 |
| Mutants | Residues | Predicted Hydrogen Bonds Number inside the Protein | Predicted Hydrogen Bonds Number with Other Molecules | ||
|---|---|---|---|---|---|
| at 300 K | at 330 K | at 300 K | at 330 K | ||
| Wild-type | Cys8 | 3.4 | 3.0 | 0.0 | 0.6 |
| Cys283 | 1.8 | 1.6 | 2.0 | 1.8 | |
| C8Y/C283Q | Tyr8 | 5.2 | 5.0 | 0.7 | 1.2 |
| Gln283 | 2.3 | 2.2 | 8.6 | 8.3 | |
| Mutants | Acrylamide Content (mg/kg) | Acrylamide Inhibition Rate (%) |
|---|---|---|
| Control | 1.6559 ± 0.0630 a | - |
| Wild-type | 0.6781 ± 0.0097 b | 59.05 ± 3.63 b |
| C8Y/C283Q | 0.2235 ± 0.0109 c | 86.50 ± 0.66 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, L.; Chi, H.; Xia, B.; Lu, Z.; Bie, X.; Zhao, H.; Lu, F.; Chen, M. Thermostability Improvement of L-Asparaginase from Acinetobacter soli via Consensus-Designed Cysteine Residue Substitution. Molecules 2022, 27, 6670. https://doi.org/10.3390/molecules27196670
Jiao L, Chi H, Xia B, Lu Z, Bie X, Zhao H, Lu F, Chen M. Thermostability Improvement of L-Asparaginase from Acinetobacter soli via Consensus-Designed Cysteine Residue Substitution. Molecules. 2022; 27(19):6670. https://doi.org/10.3390/molecules27196670
Chicago/Turabian StyleJiao, Linshu, Huibing Chi, Bingjie Xia, Zhaoxin Lu, Xiaomei Bie, Haizhen Zhao, Fengxia Lu, and Meirong Chen. 2022. "Thermostability Improvement of L-Asparaginase from Acinetobacter soli via Consensus-Designed Cysteine Residue Substitution" Molecules 27, no. 19: 6670. https://doi.org/10.3390/molecules27196670
APA StyleJiao, L., Chi, H., Xia, B., Lu, Z., Bie, X., Zhao, H., Lu, F., & Chen, M. (2022). Thermostability Improvement of L-Asparaginase from Acinetobacter soli via Consensus-Designed Cysteine Residue Substitution. Molecules, 27(19), 6670. https://doi.org/10.3390/molecules27196670








