Investigations on Anticancer Potentials by DNA Binding and Cytotoxicity Studies for Newly Synthesized and Characterized Imidazolidine and Thiazolidine-Based Isatin Derivatives
Abstract
:1. Introduction
2. Results and Discussion
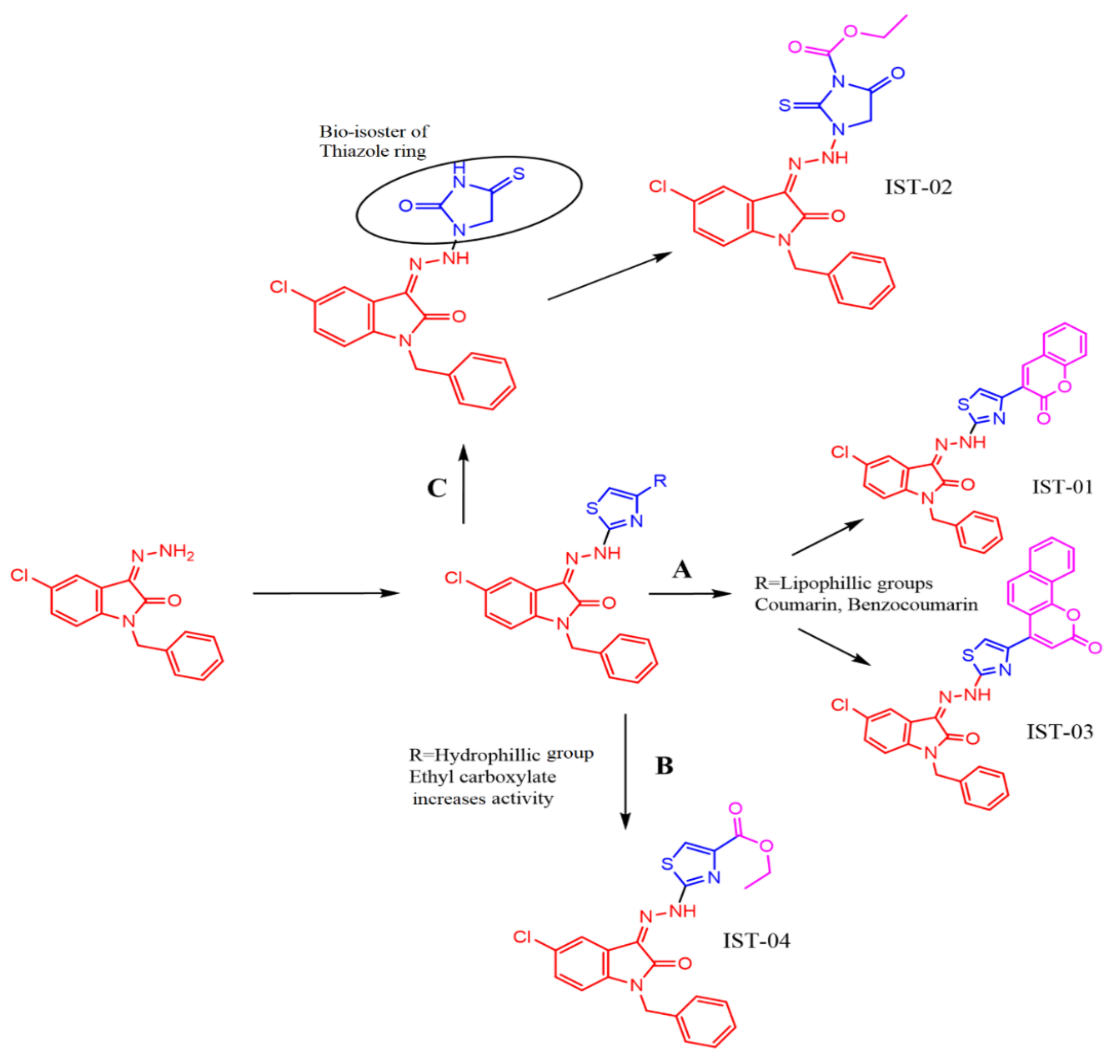
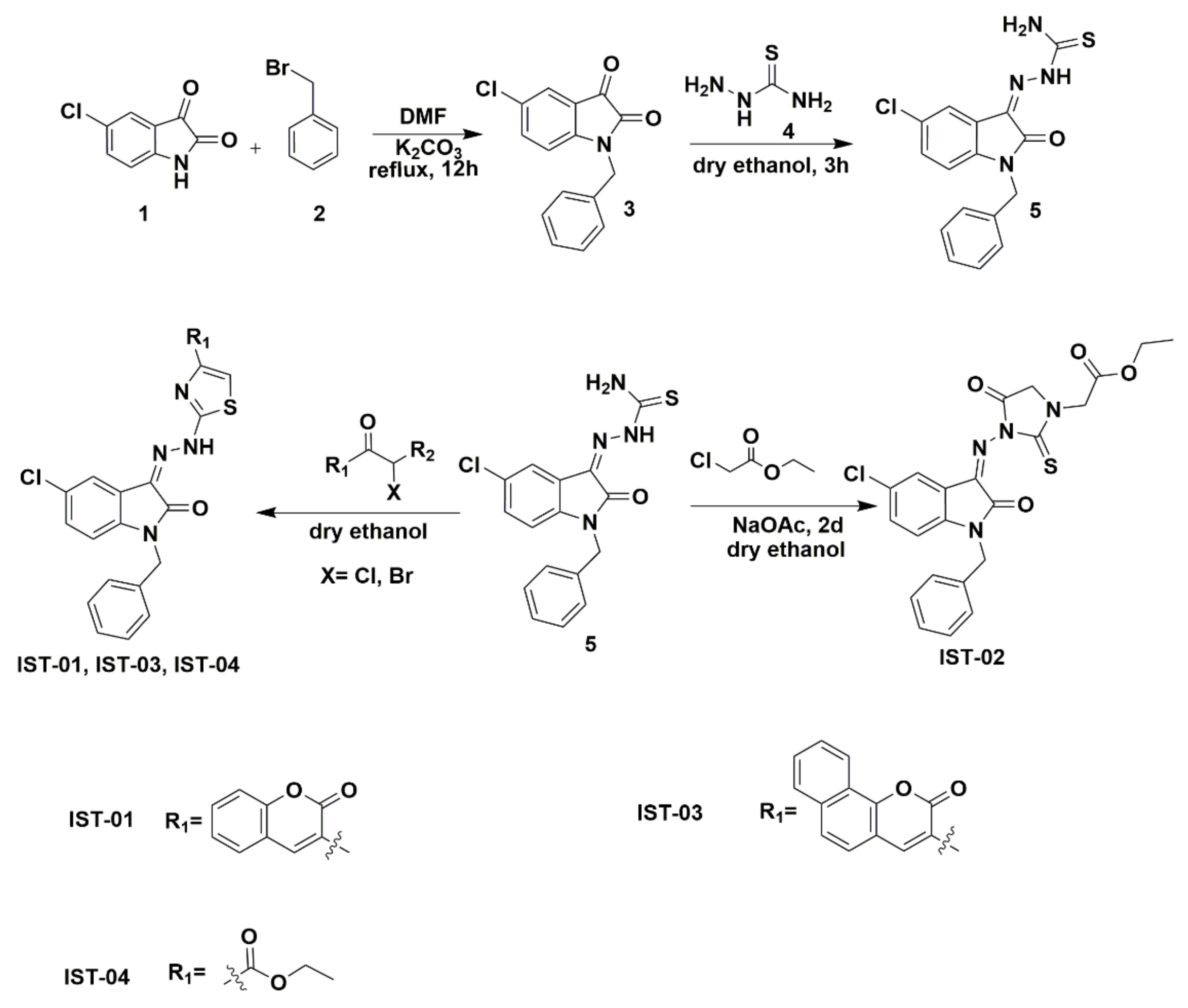
2.1. Chemistry of Synthesized Compounds
2.2. Theoretical Investigations for Compound–DNA Binding
2.2.1. DFT Studies
2.2.2. Molecular Docking Studies
2.3. Experimental Investigations for Compound–DNA Binding
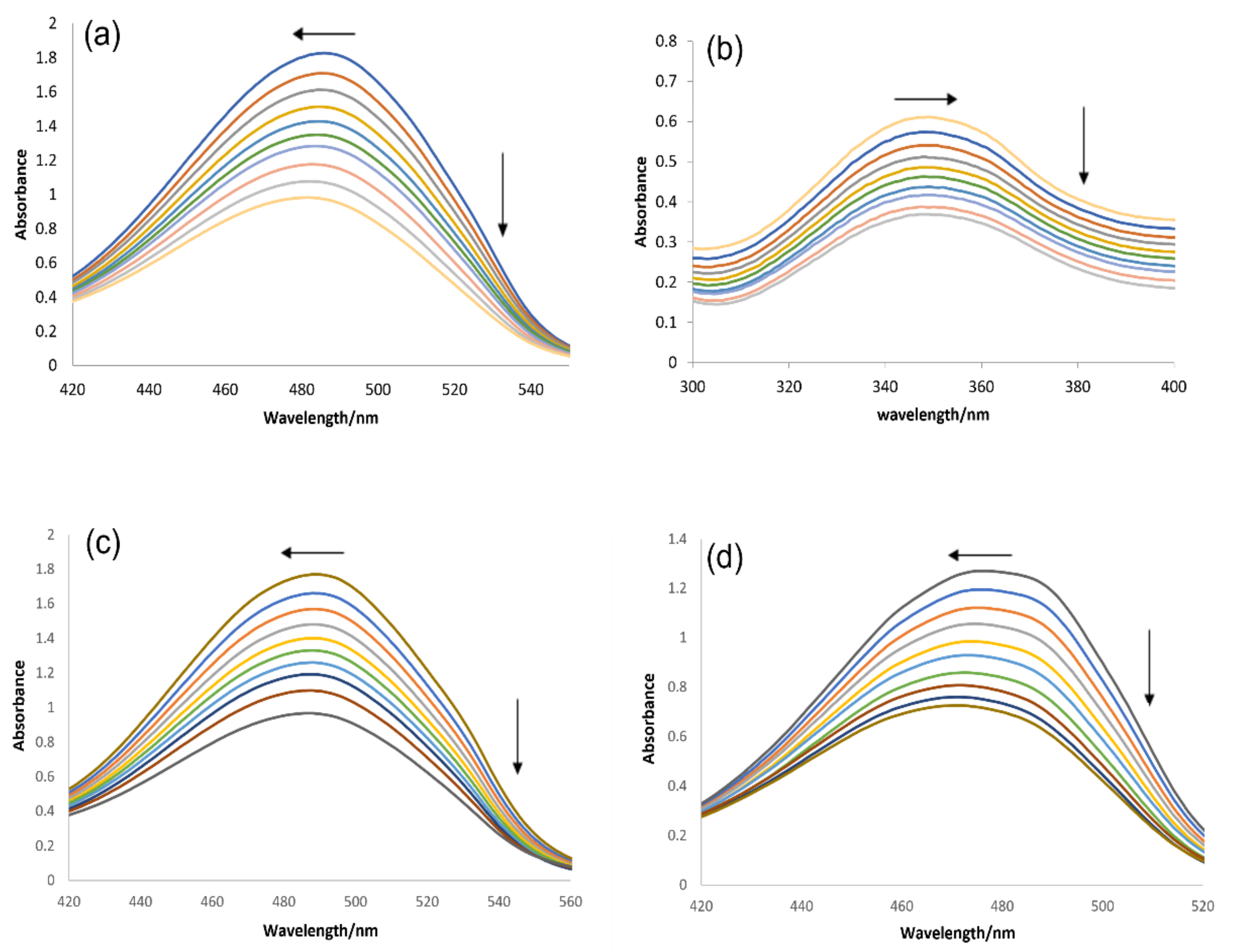
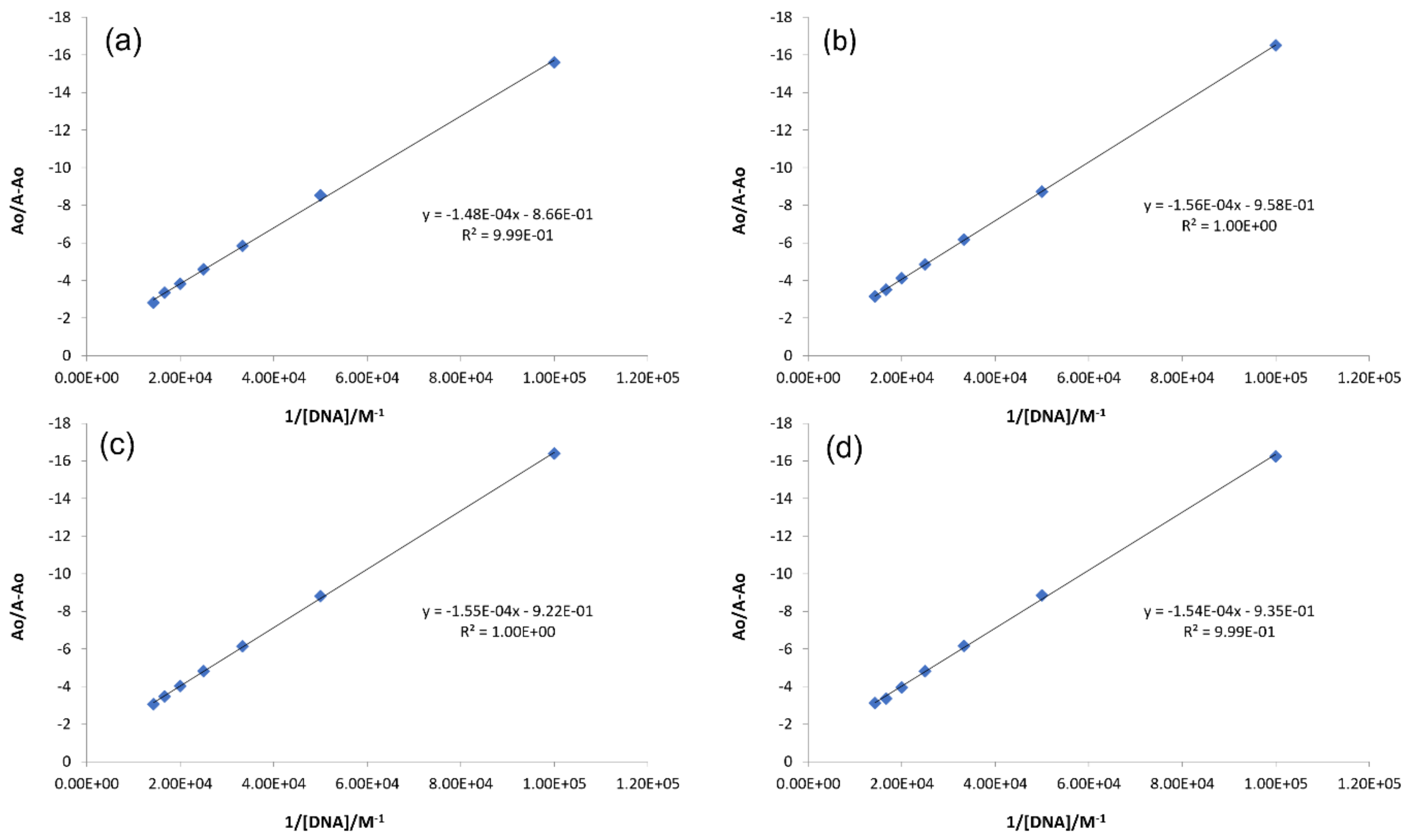
2.3.1. DNA Binding Studies by UV-Visible Spectroscopy
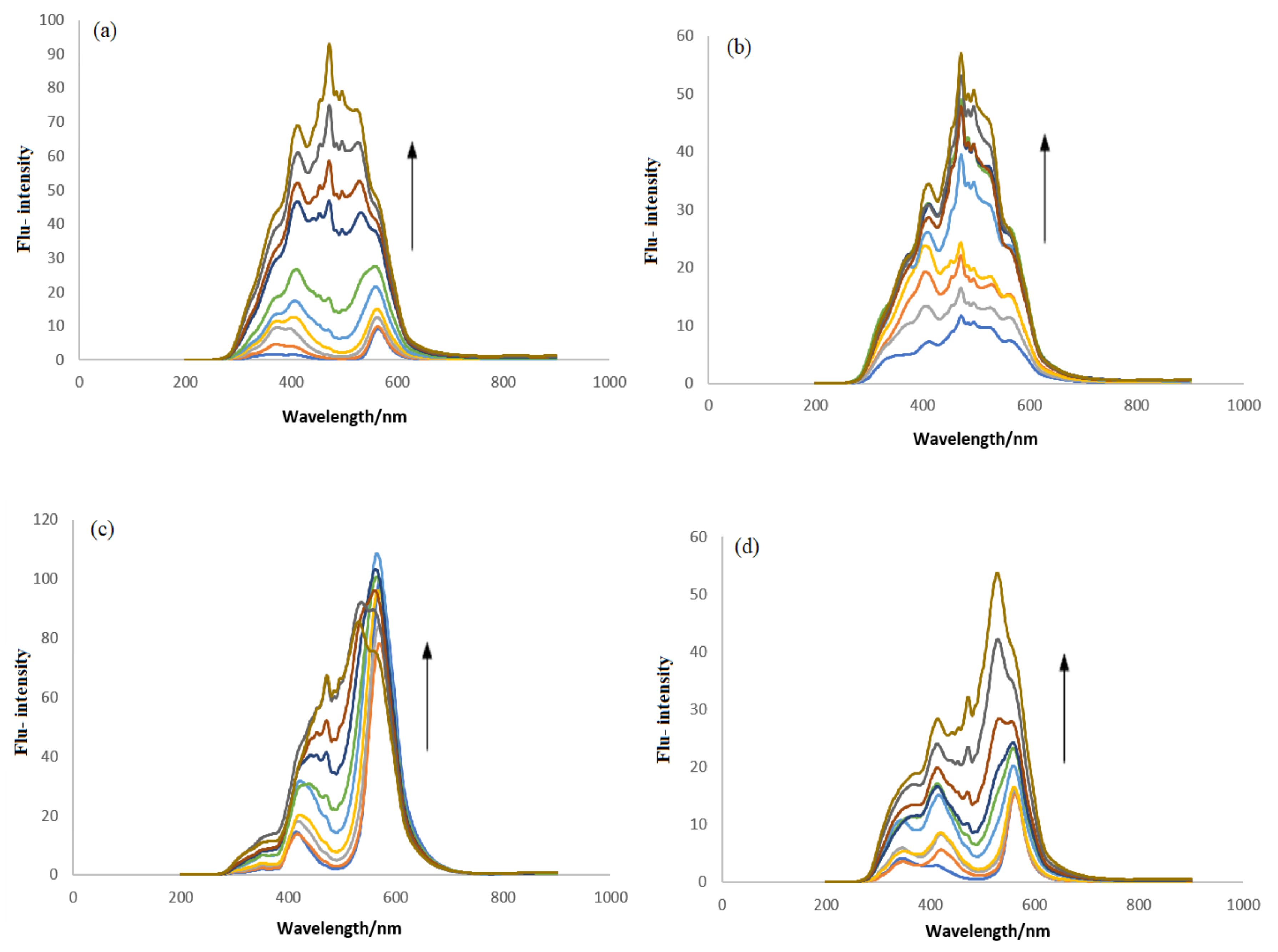
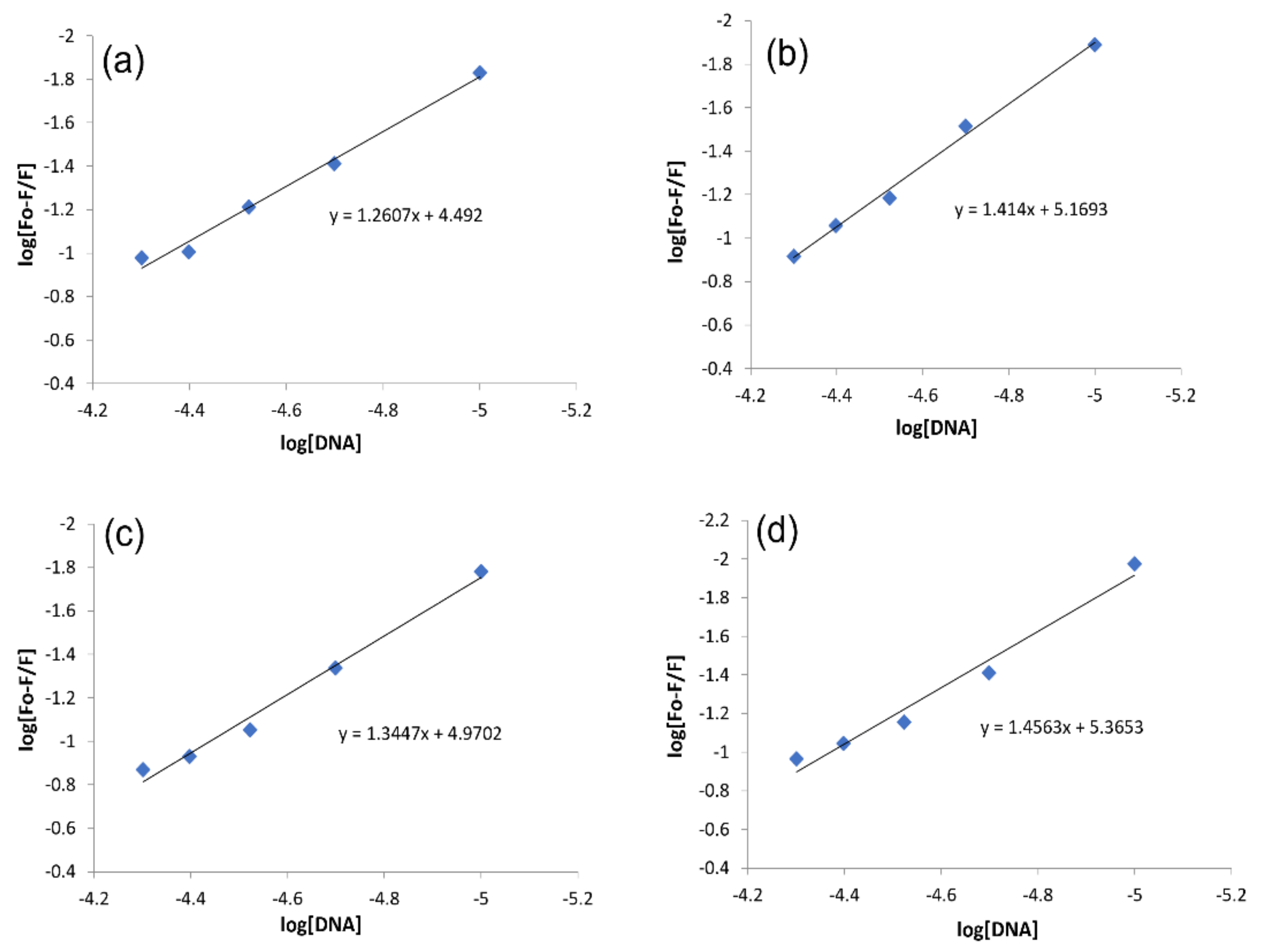
2.3.2. DNA Binding Studies by Fluorescence Spectroscopy
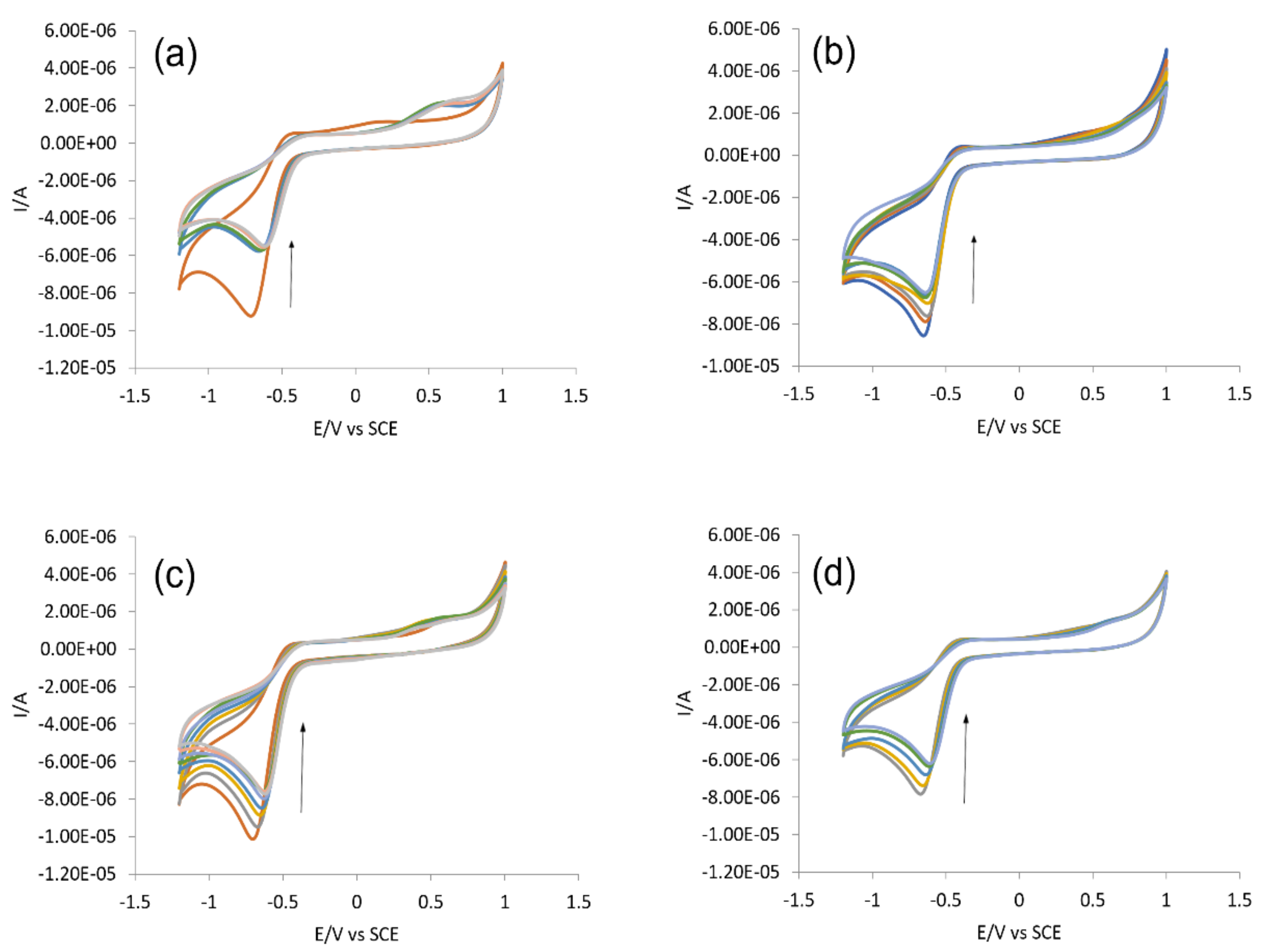
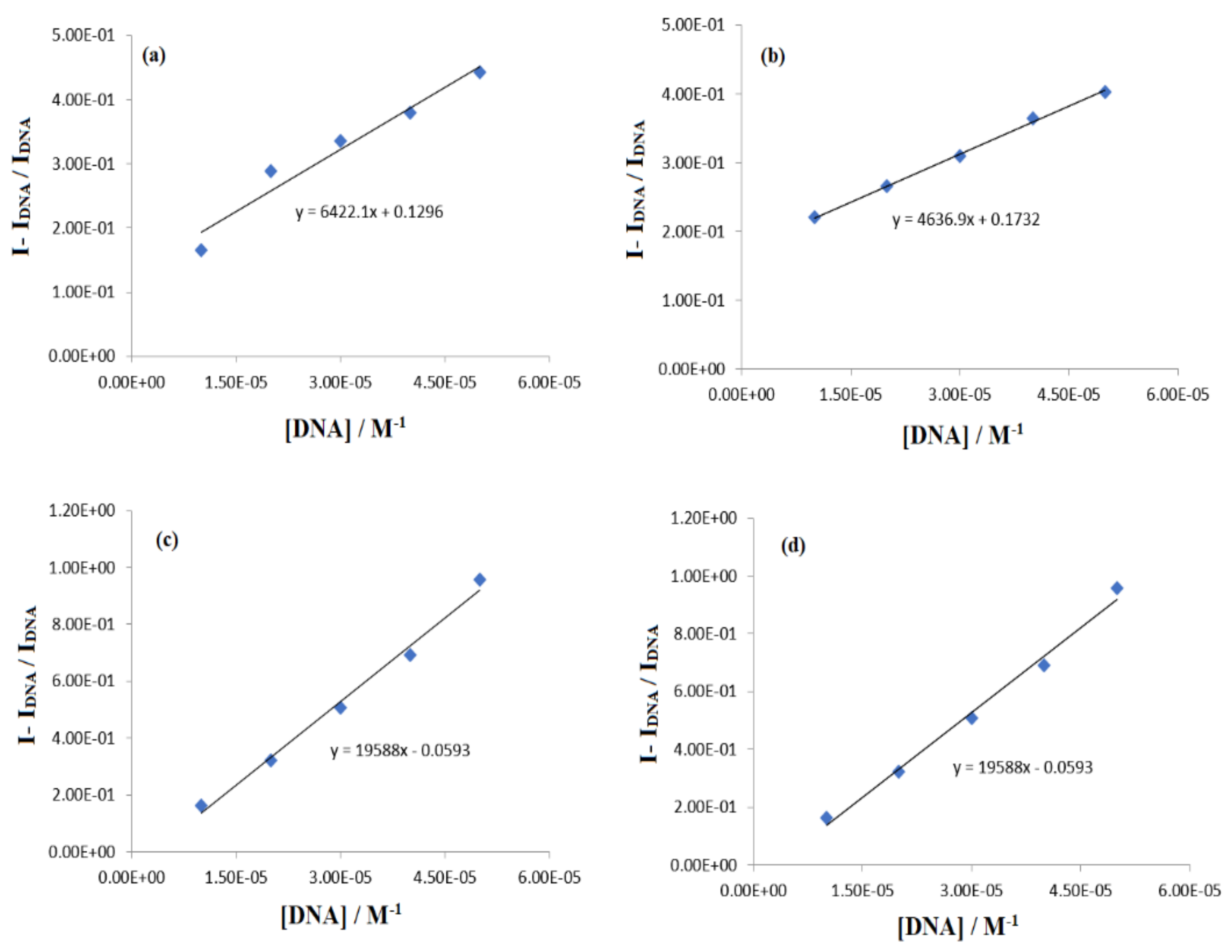
2.3.3. DNA Binding Studies by Cyclic Voltammetry
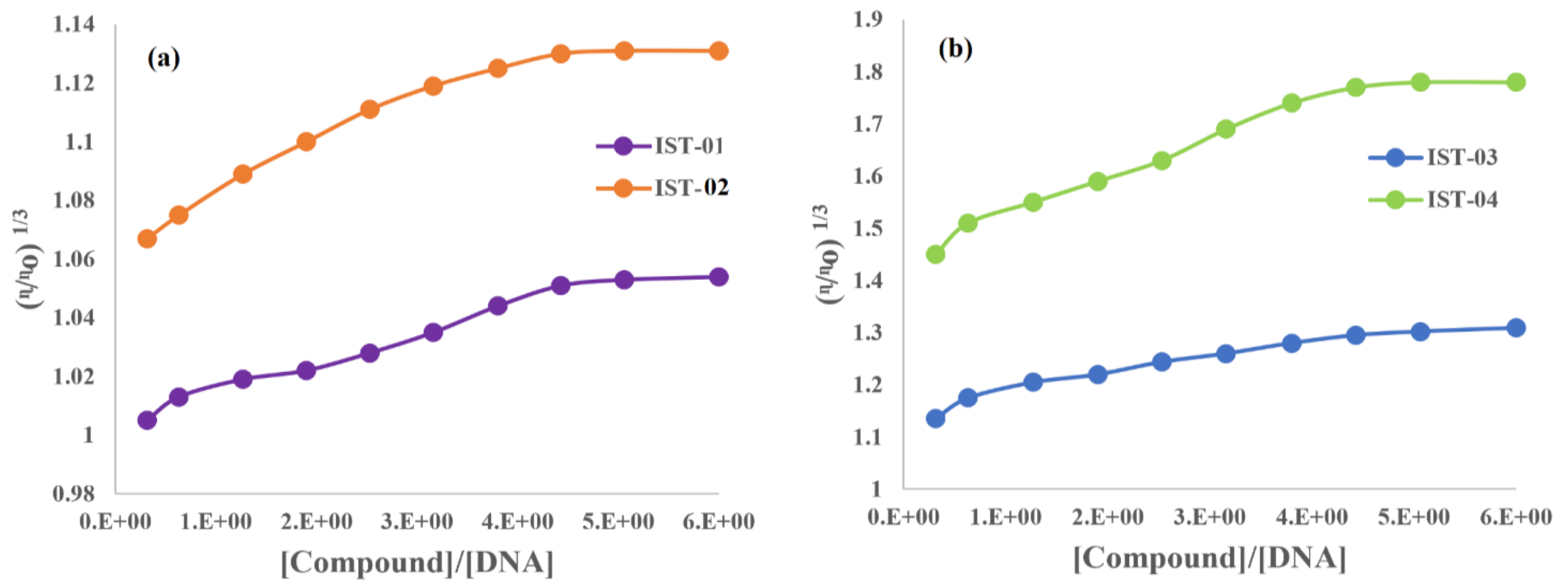
2.3.4. DNA Binding Studies by Viscometry
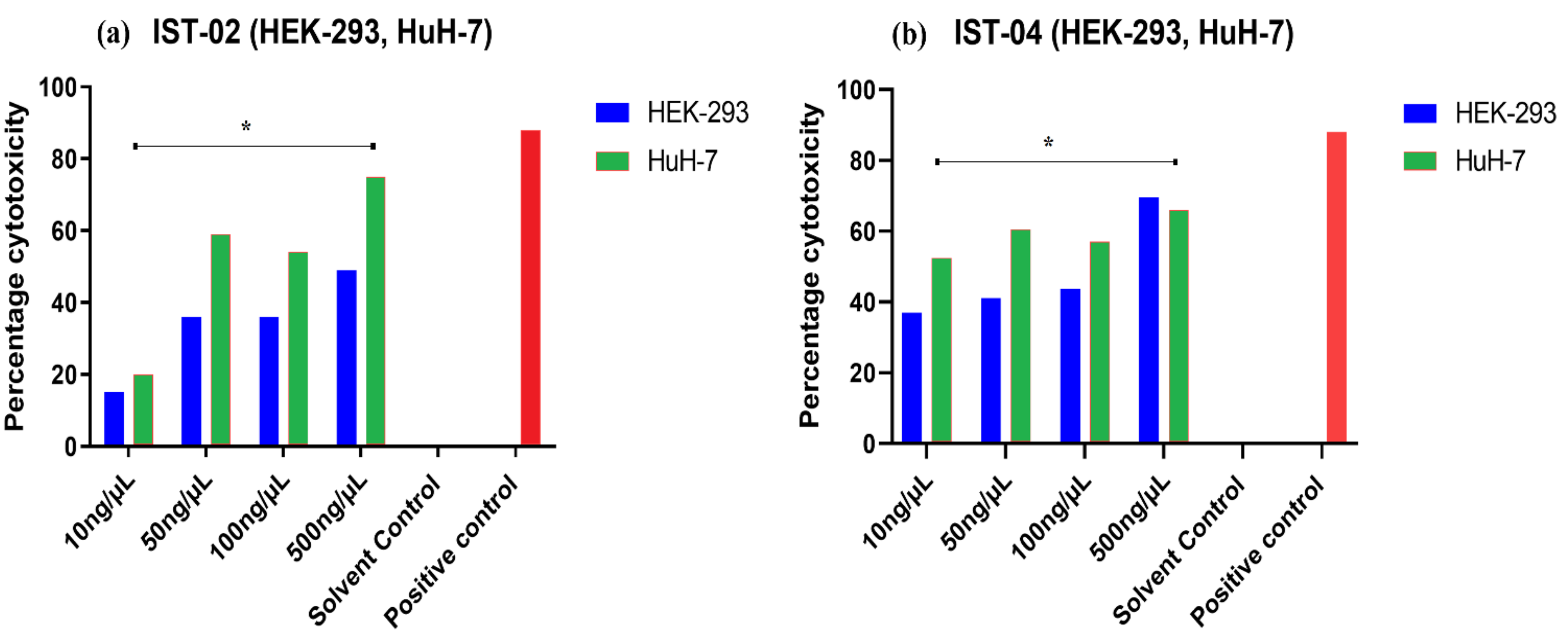
2.4. Cytotoxicity Analysis by Cell Line Studies
3. Experimental
3.1. Materials and Methods
3.2. Instrumentations
3.3. Synthesis of Isatin Derivatives
3.4. Characterization Data
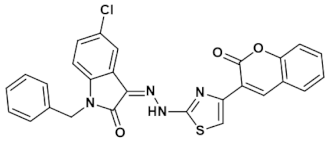
3.4.1. (Z)-1-Benzyl-5-chloro-3-(2-(4-(2-oxo-2H-chromen-3-yl)thiazol-2-yl)hydrazono)indolin-2-one (IST-01)
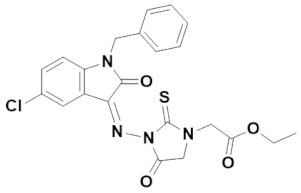
3.4.2. (Z)-Ethyl 2-(3-((1-benzyl-5-chloro-2-oxoindolin-3-ylidene)amino)-4-oxo-2-thioxoimidazolidin-1-yl)acetate (IST-02)
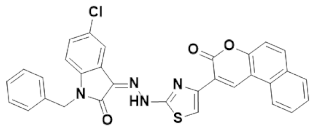
3.4.3. (Z)-1-Benzyl-5-chloro-3-(2-(4-(3-oxo-3H-benzo[f]chromen-2-yl) thiazol-2-yl)hydrazine)indolin-2-one (IST-03)
3.4.4. (Z)-Ethyl 2-(2-(1-benzyl-5-chloro-2-oxoindolin-3-ylidene)hydrazinyl)thiazole-4-carboxylate (IST-04)

3.5. Theoretical Procedures for Compound–DNA Binding
3.5.1. DFT (Density Functional Theory)
3.5.2. Molecular Docking
3.6. Experimental Procedures for Compound–DNA Binding
3.6.1. UV-Visible and Fluorescence Spectrometric Experiments
3.6.2. Cyclic Voltammetric Experiments
3.6.3. Viscosity Experiments
3.7. Cell Line Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Altıntop, M.D.; Ciftci, H.I.; Radwan, M.O.; Sever, B.; Kaplancıklı, Z.A.; Ali, T.F.; Koga, R.; Fujita, M.; Otsuka, M.; Özdemir, A. Design, synthesis, and biological evaluation of novel 1, 3, 4-thiadiazole derivatives as potential antitumor agents against chronic myelogenous leukemia: Striking effect of nitrothiazole moiety. Molecules 2018, 23, 59. [Google Scholar] [CrossRef] [Green Version]
- Strekowski, L.; Wilson, B. Noncovalent interactions with DNA: An overview. Mutat. Res./Fundam. Mol. Mech. Mutagenesis 2007, 623, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Arshad, N.; Abbas, N.; Perveen, F.; Mirza, B.; Almuhaini, A.M.; Alkahtani, S. Molecular docking analysis and spectroscopic investigations of zinc (II), nickel (II) N-phthaloyl-β-alanine complexes for DNA binding: Evaluation of antibacterial and antitumor activities. J. Saudi Chem. Soc. 2021, 25, 101323. [Google Scholar] [CrossRef]
- Arshad, N.; Saeed, A.; Perveen, F.; Ujan, R.; Farooqi, S.I.; Channar, P.A.; Shabir, G.; El-Seedi, H.R.; Javed, A.; Yamin, M. Synthesis, X-ray, Hirshfeld surface analysis, exploration of DNA binding, urease enzyme inhibition and anticancer activities of novel adamantane-naphthyl thiourea conjugate. Bioorganic Chem. 2021, 109, 104707. [Google Scholar] [CrossRef]
- Farooqi, S.I.; Arshad, N.; Channar, P.A.; Perveen, F.; Saeed, A.; Larik, F.A.; Javed, A.; Yamin, M. New aryl Schiff bases of thiadiazole derivative of ibuprofen as DNA binders and potential anticancer drug candidates. J. Biomol. Struct. Dyn. 2021, 39, 3548–3564. [Google Scholar] [CrossRef]
- Lambert, M.; Jambon, S.; Depauw, S.; David-Cordonnier, M.-H. Targeting transcription factors for cancer treatment. Molecules 2018, 23, 1479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baum, H.; Sanders, R.; Straub, G. The occurrence of a diphenyl isatin in California prunes. J. Am. Pharm. Assoc. 1951, 40, 348–349. [Google Scholar] [CrossRef]
- Aboul-Fadl, T.; Bin-Jubair, F.A.; Aboul-Wafa, O. Schiff bases of indoline-2, 3-dione (isatin) derivatives and nalidixic acid carbohydrazide, synthesis, antitubercular activity and pharmacophoric model building. Eur. J. Med. Chem. 2010, 45, 4578–4586. [Google Scholar] [CrossRef]
- Bal, T.R.; Anand, B.; Yogeeswari, P.; Sriram, D. Synthesis and evaluation of anti-HIV activity of isatin β-thiosemicarbazone derivatives. Bioorganic Med. Chem. Lett. 2005, 15, 4451–4455. [Google Scholar] [CrossRef] [PubMed]
- Malawska, B. New anticonvulsant agents. Curr. Top. Med. Chem. 2005, 5, 69–85. [Google Scholar] [CrossRef]
- Singh, U.K.; Pandeya, S.N.; Sunil, J.; Manoj, P.; Srivastava, B.K.; Anamika, S. Synthesis and antimicrobial activity of Schiff’s and Mannich bases of 1H-indole-2, 3-dione derivatives. Pharma Chem. 2010, 2, 392–399. [Google Scholar]
- Khan, K.M.; Khan, M.; Ali, M.; Taha, M.; Rasheed, S.; Perveen, S.; Choudhary, M.I. Synthesis of bis-Schiff bases of isatins and their antiglycation activity. Bioorganic Med. Chem. 2009, 17, 7795–7801. [Google Scholar] [CrossRef] [PubMed]
- Chinnasamy, R.P.; Sundararajan, R.; Govindaraj, S. Synthesis, characterization, and analgesic activity of novel schiff base of isatin derivatives. J. Adv. Pharm. Technol. Res. 2010, 1, 342. [Google Scholar] [CrossRef] [Green Version]
- Raj, R.; Singh, P.; Singh, P.; Gut, J.; Rosenthal, P.J.; Kumar, V. Azide-alkyne cycloaddition en route to 1H-1, 2, 3-triazole-tethered 7-chloroquinoline-isatin chimeras: Synthesis and antimalarial evaluation. Eur. J. Med. Chem. 2013, 62, 590–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kidwai, M.; Jahan, A.; Mishra, N. Isatins: A diversity orientated biological profile. Med. Chem. 2014, 4, 451–468. [Google Scholar] [CrossRef]
- Al-Wabli, R.I.; Zakaria, A.S.; Attia, M.I. Synthesis, spectroscopic characterization and antimicrobial potential of certain new isatin-indole molecular hybrids. Molecules 2017, 22, 1958. [Google Scholar] [CrossRef] [Green Version]
- Eldehna, W.M.; Al-Ansary, G.H.; Bua, S.; Nocentini, A.; Gratteri, P.; Altoukhy, A.; Ghabbour, H.; Ahmed, H.Y.; Supuran, C.T. Novel indolin-2-one-based sulfonamides as carbonic anhydrase inhibitors: Synthesis, in vitro biological evaluation against carbonic anhydrases isoforms I, II, IV and VII and molecular docking studies. Eur. J. Med. Chem. 2017, 127, 521–530. [Google Scholar] [CrossRef]
- Goodman, V.L.; Rock, E.P.; Dagher, R.; Ramchandani, R.P.; Abraham, S.; Gobburu, J.V.; Booth, B.P.; Verbois, S.L.; Morse, D.E.; Liang, C.Y. Approval summary: Sunitinib for the treatment of imatinib refractory or intolerant gastrointestinal stromal tumors and advanced renal cell carcinoma. Clin. Cancer Res. 2007, 13, 1367–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Raghuwanshi, K.; Patel, V.K.; Jain, D.K.; Veerasamy, R.; Dixit, A.; Rajak, H. Assessment of 5-substituted isatin as surface recognition group: Design, synthesis, and antiproliferative evaluation of hydroxamates as novel histone deacetylase inhibitors. Pharm. Chem. J. 2017, 51, 366–374. [Google Scholar] [CrossRef]
- Supuran, C.T.; Winum, J.-Y. Carbonic anhydrase IX inhibitors in cancer therapy: An update. Future Med. Chem. 2015, 7, 1407–1414. [Google Scholar] [CrossRef]
- Maddela, S.; Makula, A. Design, synthesis and docking study of some novel isatin-quinoline hybrids as potential antitubercular agents. Anti-Infect. Agents 2016, 14, 53–62. [Google Scholar] [CrossRef]
- Meleddu, R.; Distinto, S.; Corona, A.; Tramontano, E.; Bianco, G.; Melis, C.; Cottiglia, F.; Maccioni, E. Isatin thiazoline hybrids as dual inhibitors of HIV-1 reverse transcriptase. J. Enzym. Inhib. Med. Chem. 2017, 32, 130–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eldehnaa, W.M.; Al-Wablib, R.I.; Almutairib, M.S.; Adam, B.; Keeton, A.B.; Piazza, G.A.; Abdel-Aziz, H.A.; Attia, M.I. Synthesis and biological evaluation of certain hydrazonoindolin-2-one derivatives as new potent anti-proliferative agents. J. Enzym. Inhib. Med. Chem. 2018, 33, 867–878. [Google Scholar] [CrossRef]
- Ali, A.; Banerjee, S.; Kamaal, S.; Usman, M.; Das, N.; Afzal, M.; Alarifi, A.; Sepay, N.; Roy, P.; Ahmad, M. Ligand substituent effect on the cytotoxicity activity of two new copper (ii) complexes bearing 8-hydroxyquinoline derivatives: Validated by MTT assay and apoptosis in MCF-7 cancer cell line (human breast cancer). RSC Adv. 2021, 11, 14362–14373. [Google Scholar] [CrossRef]
- Teitz, Y.; Ladizensky, E.; Barko, N.; Burstein, E. Selective repression of v-abl-encoded protein by N-methylisatin-beta-4′,4′-diethylthiosemicarbazone and N-allylisatin-beta-4′, 4′-diallylthiosemicarbazone. Antimicrob. Agents Chemother. 1993, 37, 2483–2485. [Google Scholar] [CrossRef] [Green Version]
- Varma, R.S.; Nobles, W.L. Antiviral, antibacterial, and antifungal activities of isatin N-Mannich bases. J. Pharm. Sci. 1975, 64, 881–882. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, S.; Gao, C.; Fan, J.; Zhao, F.; Lv, Z.-S.; Feng, L.-S. Isatin hybrids and their anti-tuberculosis activity. Chin. Chem. Lett. 2017, 28, 159–167. [Google Scholar] [CrossRef]
- Broadbent, T.; Broadbent, H.S. The chemistry and pharmacology of indole-3-carbinol (indole-3-methanol) and 3-(methoxymethyl) indole. [Part II]. Curr. Med. Chem. 1998, 5, 469–491. [Google Scholar] [PubMed]
- Broadbent, T.A.; Broadbent, H.S. The chemistry and pharmacology of indole-3-carbinol (indole-3-methanol) and 3-(methoxymethyl) indole. [Part I]. Curr. Med. Chem. 1998, 5, 337–352. [Google Scholar]
- Eshba, N.; Salama, H. 5-(2-Oxo-3-indolinylidene) thiazolidine-2, 4-dione-1, 3-di-Mannich base derivatives: Synthesis and evaluation for antileukemic activity. Die Pharm. 1985, 40, 320–322. [Google Scholar] [CrossRef] [PubMed]
- Pajouhesh, H.; Parson, R.; Popp, F.D. Potential anticonvulsants VI: Condensation of isatins with cyclohexanone and other cyclic ketones. J. Pharm. Sci. 1983, 72, 318–321. [Google Scholar] [CrossRef]
- Pandeya, S.N.; Smitha, S.; Jyoti, M.; Sridhar, S.K. Biological activities of isatin and its derivatives. Acta Pharm. 2005, 55, 27–46. [Google Scholar]
- Arshad, N.; Rafiq, M.; Ujan, R.; Saeed, A.; Farooqi, S.I.; Perveen, F.; Channar, P.A.; Ashraf, S.; Abbas, Q.; Ahmed, A. Synthesis, X-ray crystal structure elucidation and Hirshfeld surface analysis of N-((4-(1 H-benzo [d] imidazole-2-yl) phenyl) carbamothioyl) benzamide: Investigations for elastase inhibition, antioxidant and DNA binding potentials for biological applications. RSC Adv. 2020, 10, 20837–20851. [Google Scholar]
- Hussain, M.; Arshad, N.; Ujan, R.; Saeed, A.; Channar, P.A.; Perveen, F.; Larik, F.A.; Farooqi, S.I.; Hussain, Z.; Hökelek, T. Synthesis, structure elucidation and surface analysis of a new single crystal N-((2-(benzo [4, 5] imidazo [1, 2-c] quinazolin-6-yl) phenyl) carbamothioyl) heptanamide: Theoretical and experimental DNA binding studies. J. Mol. Struct. 2020, 1205, 127496. [Google Scholar] [CrossRef]
- Tabassum, S.; Ahmad, M.; Afzal, M.; Zaki, M.; Bharadwaj, P.K. Synthesis and structure elucidation of a copper (II) Schiff-base complex: In vitro DNA binding, pBR322 plasmid cleavage and HSA binding studies. J. Photochem. Photobiol. B Biol. 2014, 140, 321–331. [Google Scholar] [CrossRef]
- Farooqi, S.I.; Arshad, N.; Perveen, F.; Channar, P.A.; Saeed, A.; Javed, A. Aroylthiourea derivatives of ciprofloxacin drug as DNA binder: Theoretical, spectroscopic and electrochemical studies along with cytotoxicity assessment. Arch. Biochem. Biophys. 2019, 666, 83–98. [Google Scholar] [CrossRef]
- Farooqi, S.I.; Arshad, N.; Channar, P.A.; Perveen, F.; Saeed, A.; Larik, F.A.; Javeed, A. Synthesis, theoretical, spectroscopic and electrochemical DNA binding investigations of 1, 3, 4-thiadiazole derivatives of ibuprofen and ciprofloxacin: Cancer cell line studies. J. Photochem. Photobiol. B Biol. 2018, 189, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Hajian, R.; Hossaini, P.; Mehrayin, Z.; Woi, P.M.; Shams, N. DNA-binding studies of valrubicin as a chemotherapy drug using spectroscopy and electrochemical techniques. J. Pharm. Anal. 2017, 7, 176–180. [Google Scholar] [CrossRef]
- Shahabadi, N.; Fatahi, A. Multispectroscopic DNA-binding studies of a tris-chelate nickel (II) complex containing 4, 7-diphenyl 1, 10-phenanthroline ligands. J. Mol. Struct. 2010, 970, 90–95. [Google Scholar] [CrossRef]
- Arshad, N.; Abbas, N.; Bhatti, M.H.; Rashid, N.; Tahir, M.N.; Saleem, S.; Mirza, B. Synthesis, crystal structure, DNA binding and in vitro biological studies of Ni (II), Cu (II) and Zn (II) complexes of N-phthaloylglycine. J. Photochem. Photobiol. B Biol. 2012, 117, 228–239. [Google Scholar] [CrossRef]
- Farooqi, S.I.; Arshad, N.; Perveen, F.; Channar, P.A.; Saeed, A.; Javed, A.; Hökelek, T.; Flörke, U. Structure and surface analysis of ibuprofen-organotin conjugate: Potential anti-cancer drug candidacy of the compound is proven by in-vitro DNA binding and cytotoxicity studies. Polyhedron 2020, 192, 114845. [Google Scholar] [CrossRef]
- Arshad, N.; Yunus, U.; Razzque, S.; Khan, M.; Saleem, S.; Mirza, B.; Rashid, N. Electrochemical and spectroscopic investigations of isoniazide and its analogs with ds. DNA at physiological pH: Evaluation of biological activities. Eur. J. Med. Chem. 2012, 47, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Janjua, N.K.; Shaheen, A.; Yaqub, A.; Perveen, F.; Sabahat, S.; Mumtaz, M.; Jacob, C.; Ba, L.A.; Mohammed, H.A. Flavonoid–DNA binding studies and thermodynamic parameters. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2011, 79, 1600–1604. [Google Scholar] [CrossRef]
- Ke, S.; Shi, L.; Yang, Z. Discovery of novel isatin–dehydroepiandrosterone conjugates as potential anticancer agents. Bioorganic Med. Chem. Lett. 2015, 25, 4628–4631. [Google Scholar] [CrossRef]
- Deswal, N.; Shrivastava, A.; Summon Hossain, M.; Gahlyan, P.; Bawa, R.; Gupta, R.D.; Kumar, R. Design, Synthesis, Evaluation and Molecular Docking Studies of Novel Triazole Linked 1, 4-Dihydropyridine-isatin Scaffolds as Potent Anticancer Agents. ChemistrySelect 2021, 6, 717–725. [Google Scholar] [CrossRef]
- Chung, Y.-C.; Tang, F.-Y.; Liao, J.-W.; Chung, C.-H.; Jong, T.-T.; Chen, S.-S.; Tsai, C.-H.; Chiang, E.-P. Isatis indigotica induces hepatocellular cancer cell death via caspase-independent apoptosis-inducing factor translocation apoptotic pathway in vitro and in vivo. Integr. Cancer Ther. 2011, 10, 201–214. [Google Scholar] [CrossRef]
- Wang, X.; Su, H.; Chen, C.; Cao, X. Synthesis and biological evaluation of peptidomimetics containing the tryptamine moiety as a potential antitumor agent. RSC Adv. 2015, 5, 15597–15602. [Google Scholar] [CrossRef]
- Raja, M.; Muralidharan, M.; Arunachalam, M. Comparative analysis of DNA extracted from fish species preserved in formalin in two different periods. Turk. J. Biol. 2011, 35, 331–336. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision D. 01; Gaussian. Inc.: Wallingford, CT, USA, 2009. [Google Scholar]

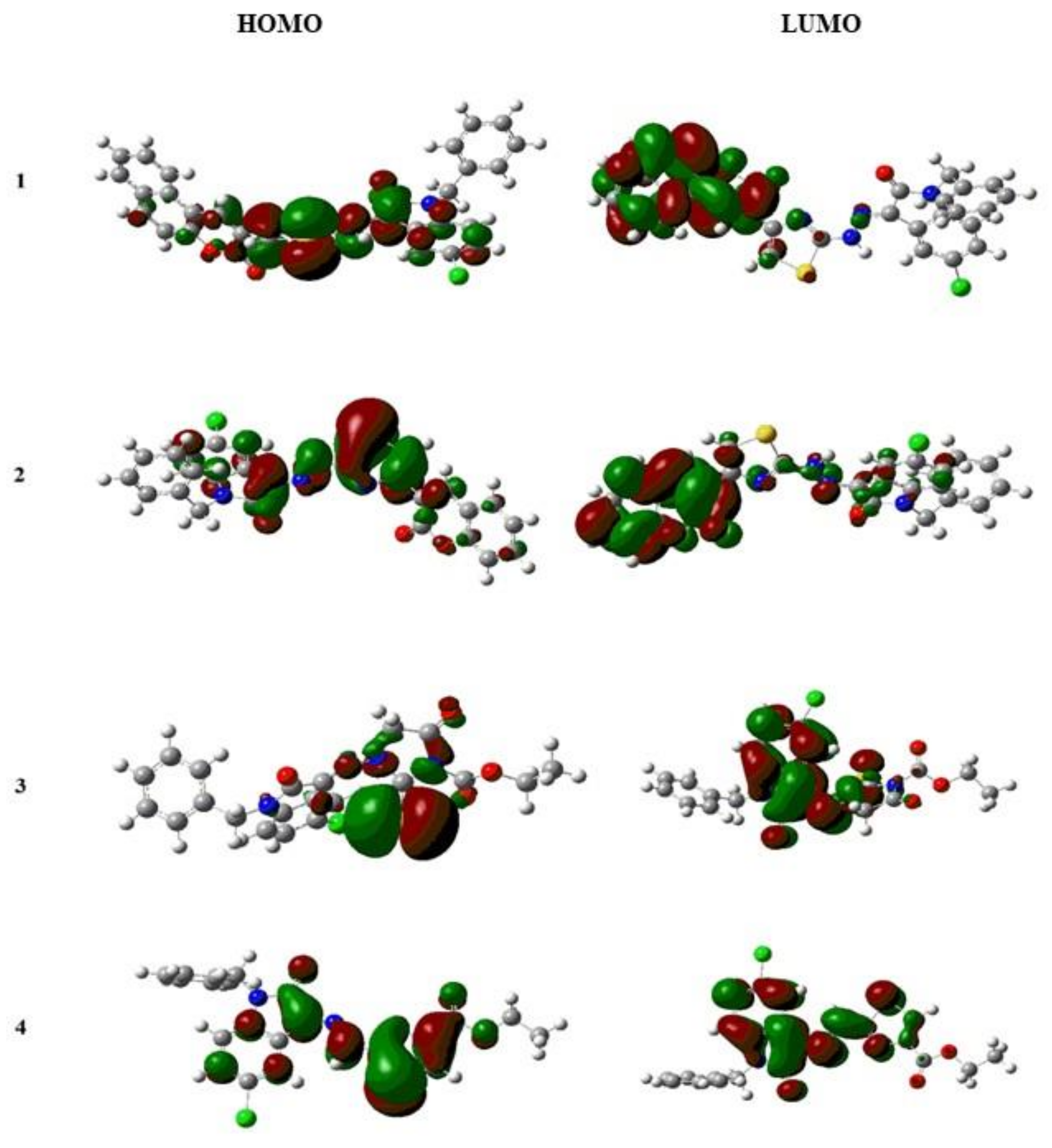



| Compound | HOMO (eV) | LUMO (eV) | Difference (HOMO–LUMO) (eV) |
|---|---|---|---|
| IST-01 | −6.23 | −2.70 | 3.53 |
| IST-02 | −6.56 | −3.18 | 3.38 |
| IST-03 | −6.33 | −2.80 | 3.53 |
| IST-04 | −6.18 | −2.69 | 3.49 |
| Compound | S (kcal.mol−1) | Kb (M−1) |
|---|---|---|
| IST-01 | −4.6204 | 1.81 × 103 |
| IST-02 | −5.4968 | 7.51 × 103 |
| IST-03 | −4.9501 | 3.01 × 103 |
| IST-04 | −5.1164 | 4.01 × 103 |
| Complex | Binding Parameters | UV/Visible | Flu- | CV |
|---|---|---|---|---|
| IST-01–-DNA | Kb/M−1 | 5.85 × 103 | 3.10 × 104 | 6.7 × 104 |
| −∆G/kJmol−1 | −22.36 | −26.65 | −28.52 | |
| n | - | 1.26 | 1.04 | |
| IST-02–ds-DNA | Kb/M−1 | 6.14 × 103 | 1.47 × 105 | 1.43 × 105 |
| −∆G/kJmol−1 | −22.48 | −31.84 | −30.60 | |
| n | - | 1.41 | 3.09 | |
| IST-03–ds-DNA | Kb/M−1 | 5.95 × 103 | 9.34 × 104 | 1.15 × 105 |
| −∆G/kJmol−1 | −22.40 | −29.50 | −30.03 | |
| n | - | 1.34 | 6.89 | |
| IST-04–ds-DNA | Kb/M−1 | 6.07 × 103 | 2.32 × 105 | 1.35 × 105 |
| −∆G/kJmol−1 | −22.45 | −30.66 | −30.44 | |
| n | - | 1.45 | 5.87 |
| Compound | Do (cm2s−1) (without DNA) | Do (cm2s−1) (with DNA) |
|---|---|---|
| IST-01 | 1.92 × 10−5 | 4.52 × 10−6 |
| IST-02 | 5.79 × 10−5 | 8.95 × 10−6 |
| IST-03 | 1.16 × 10−4 | 1.13 × 10−5 |
| IST-04 | 4.47 × 10−5 | 4.36 × 10−6 |
| Conc. (ng/µL) | HuH-7 | HEK-293 | ||
|---|---|---|---|---|
| % Cytotoxicity | IC50 µM | % Cytotoxicity | IC50 µM | |
| IST-02 | ||||
| 0 | 0.0 | 0.0 | ||
| 10 | 20.0 | 15.0 | ||
| 50 | 59.0 | 3.07 ± 9.47 | 36.0 | 488.30 ± 2.44 |
| 100 | 54.0 | 36.0 | ||
| 500 | 75.0 | 49.0 | ||
| IST-04 | ||||
| 0 | 0.0 | 0.0 | ||
| 10 | 52.4 | 37.0 | ||
| 50 | 60.5 | 14.60 ± 2.49 | 41.1 | 197.80 ± 8.75 |
| 100 | 57.0 | 43.7 | ||
| 500 | 66.0 | 69.6 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arshad, N.; Mir, M.I.; Perveen, F.; Javed, A.; Javaid, M.; Saeed, A.; Channar, P.A.; Farooqi, S.I.; Alkahtani, S.; Anwar, J. Investigations on Anticancer Potentials by DNA Binding and Cytotoxicity Studies for Newly Synthesized and Characterized Imidazolidine and Thiazolidine-Based Isatin Derivatives. Molecules 2022, 27, 354. https://doi.org/10.3390/molecules27020354
Arshad N, Mir MI, Perveen F, Javed A, Javaid M, Saeed A, Channar PA, Farooqi SI, Alkahtani S, Anwar J. Investigations on Anticancer Potentials by DNA Binding and Cytotoxicity Studies for Newly Synthesized and Characterized Imidazolidine and Thiazolidine-Based Isatin Derivatives. Molecules. 2022; 27(2):354. https://doi.org/10.3390/molecules27020354
Chicago/Turabian StyleArshad, Nasima, Muhammad Ismail Mir, Fouzia Perveen, Aneela Javed, Memona Javaid, Aamer Saeed, Pervaiz Ali Channar, Shahid Iqbal Farooqi, Saad Alkahtani, and Jamshed Anwar. 2022. "Investigations on Anticancer Potentials by DNA Binding and Cytotoxicity Studies for Newly Synthesized and Characterized Imidazolidine and Thiazolidine-Based Isatin Derivatives" Molecules 27, no. 2: 354. https://doi.org/10.3390/molecules27020354







