In Silico Prediction and Validation of CB2 Allosteric Binding Sites to Aid the Design of Allosteric Modulators
Abstract
:1. Introduction
2. Results
2.1. Overview of Seven Predicted Allosteric Binding Sites in CB2-WIN 55,212-2 or CB2-CP55940 Complex
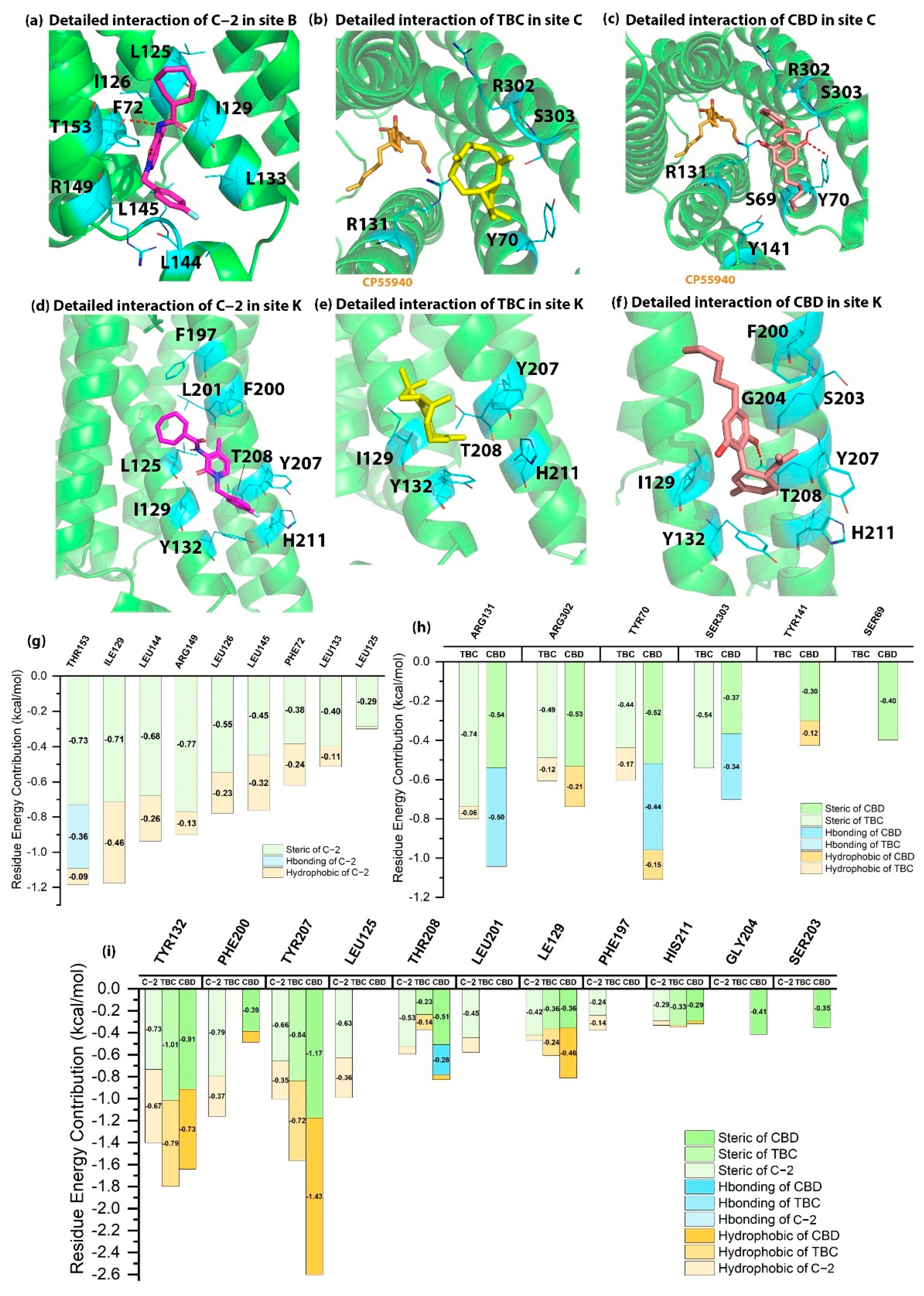
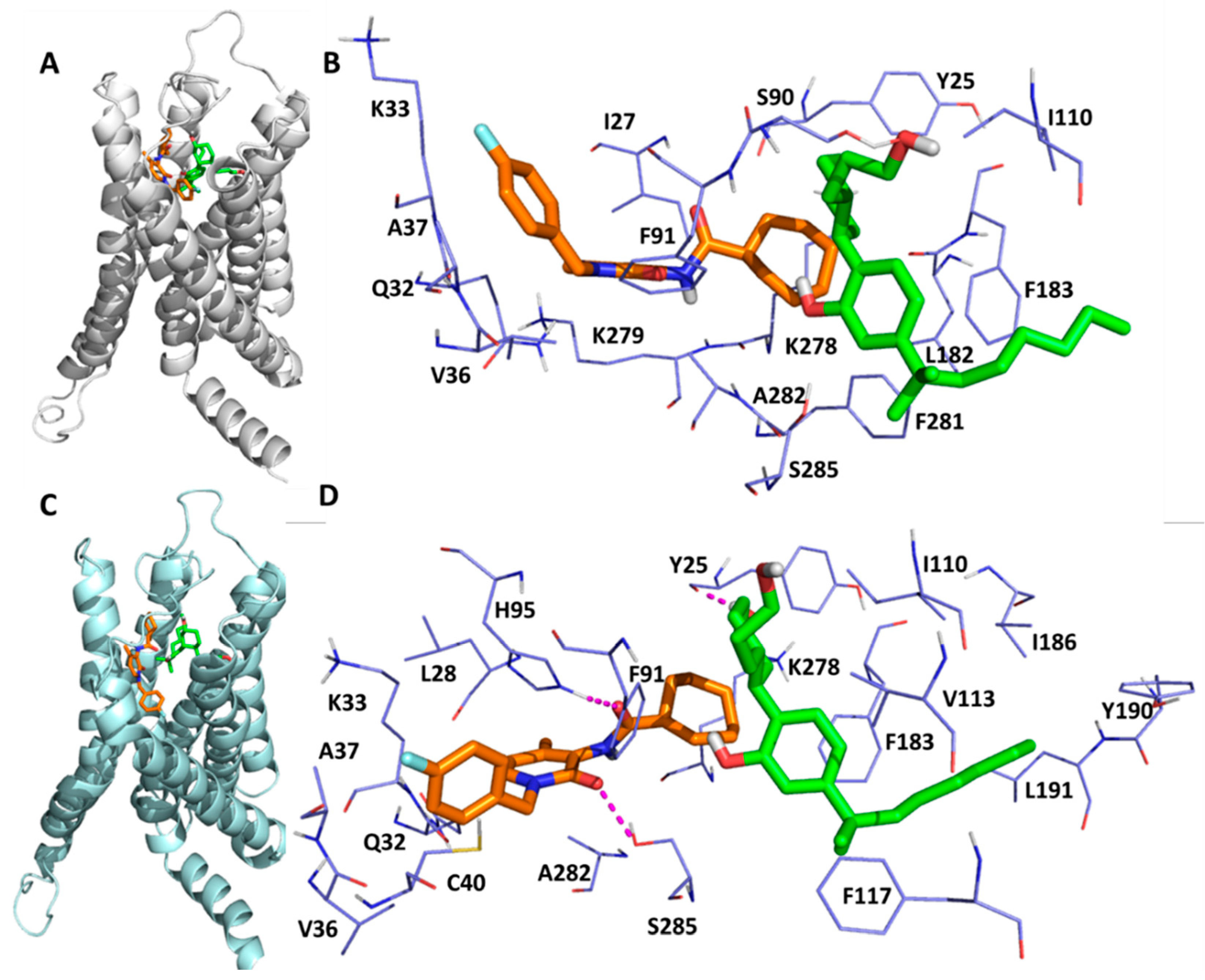
2.2. Detailed Binding Poses and Interactions of C-2, TBC and CBD in Sites B, K and C of CB2
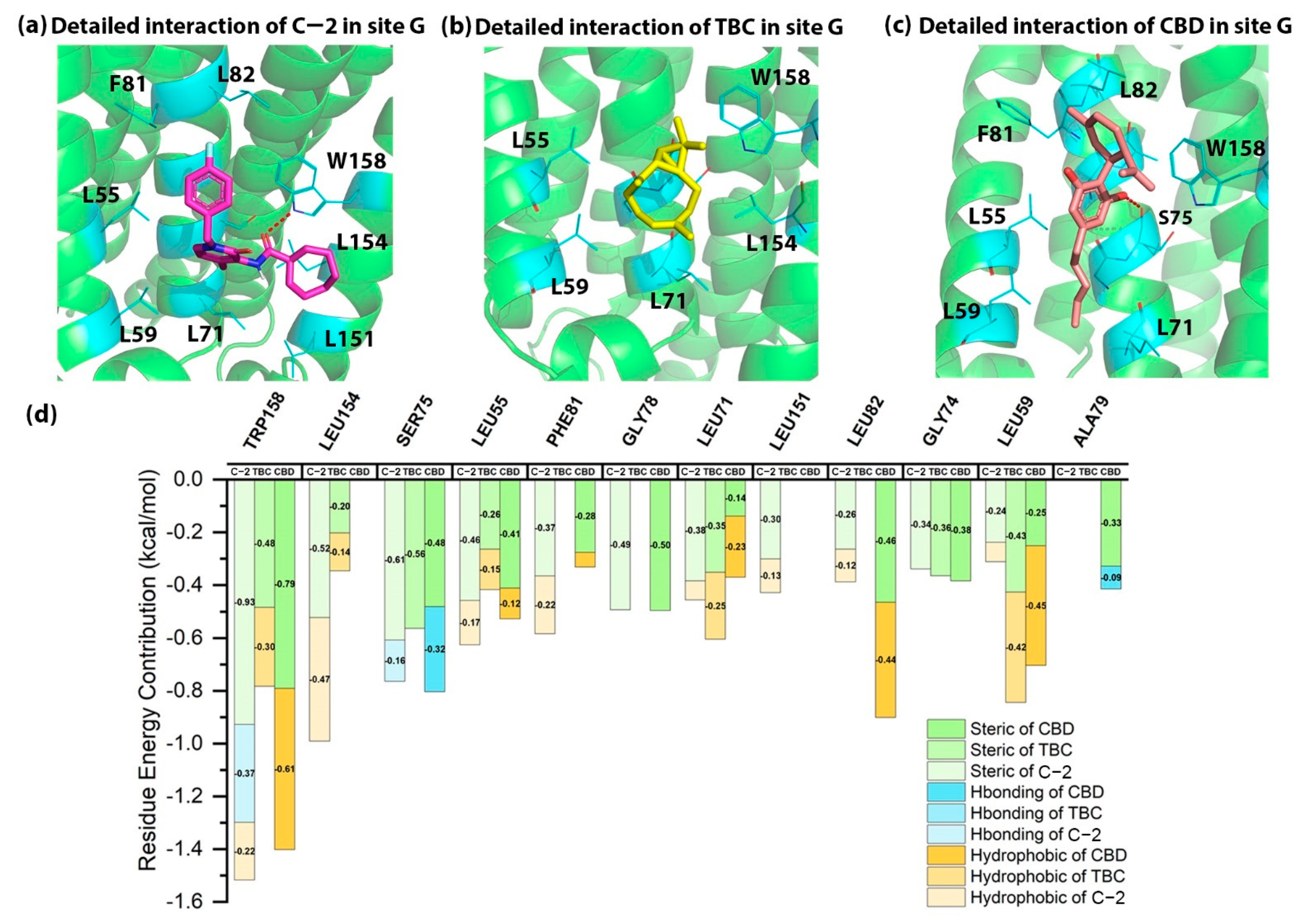
2.3. Detailed Binding Poses and Interactions of C-2, TBC and CBD in Site G of CB2
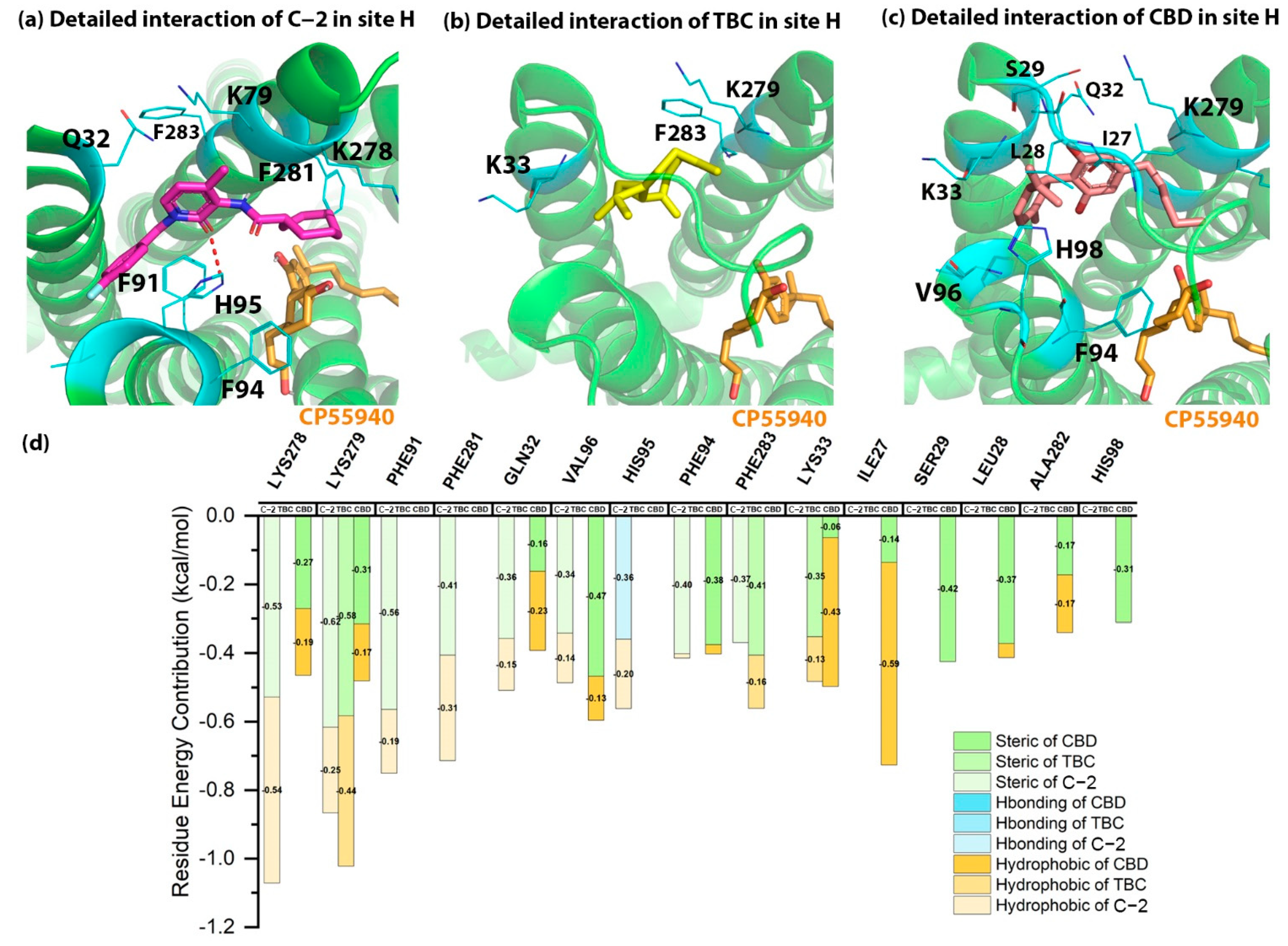
2.4. Detailed Binding Poses and Interactions of C-2, TBC and CBD in Site H of CB2
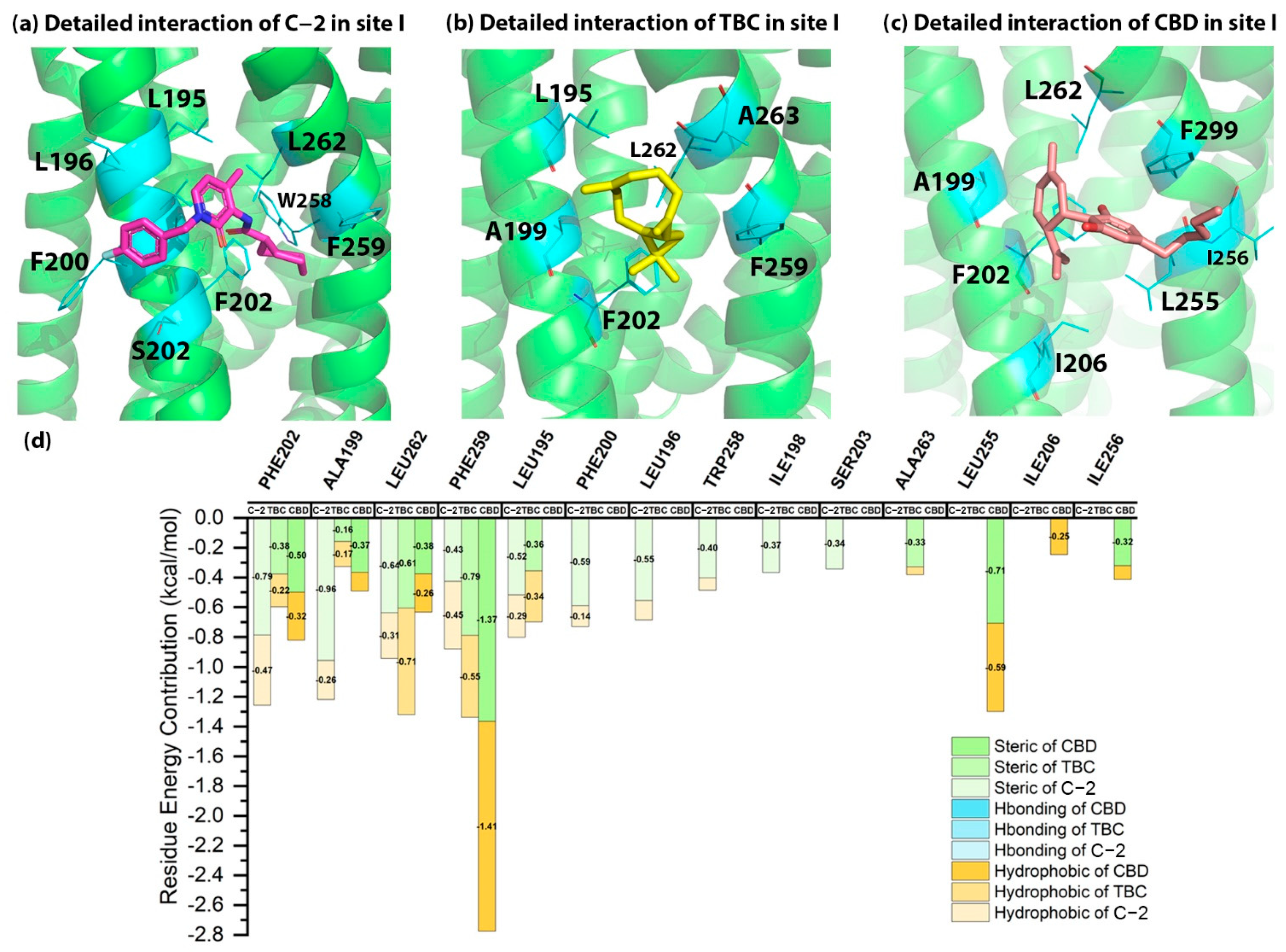
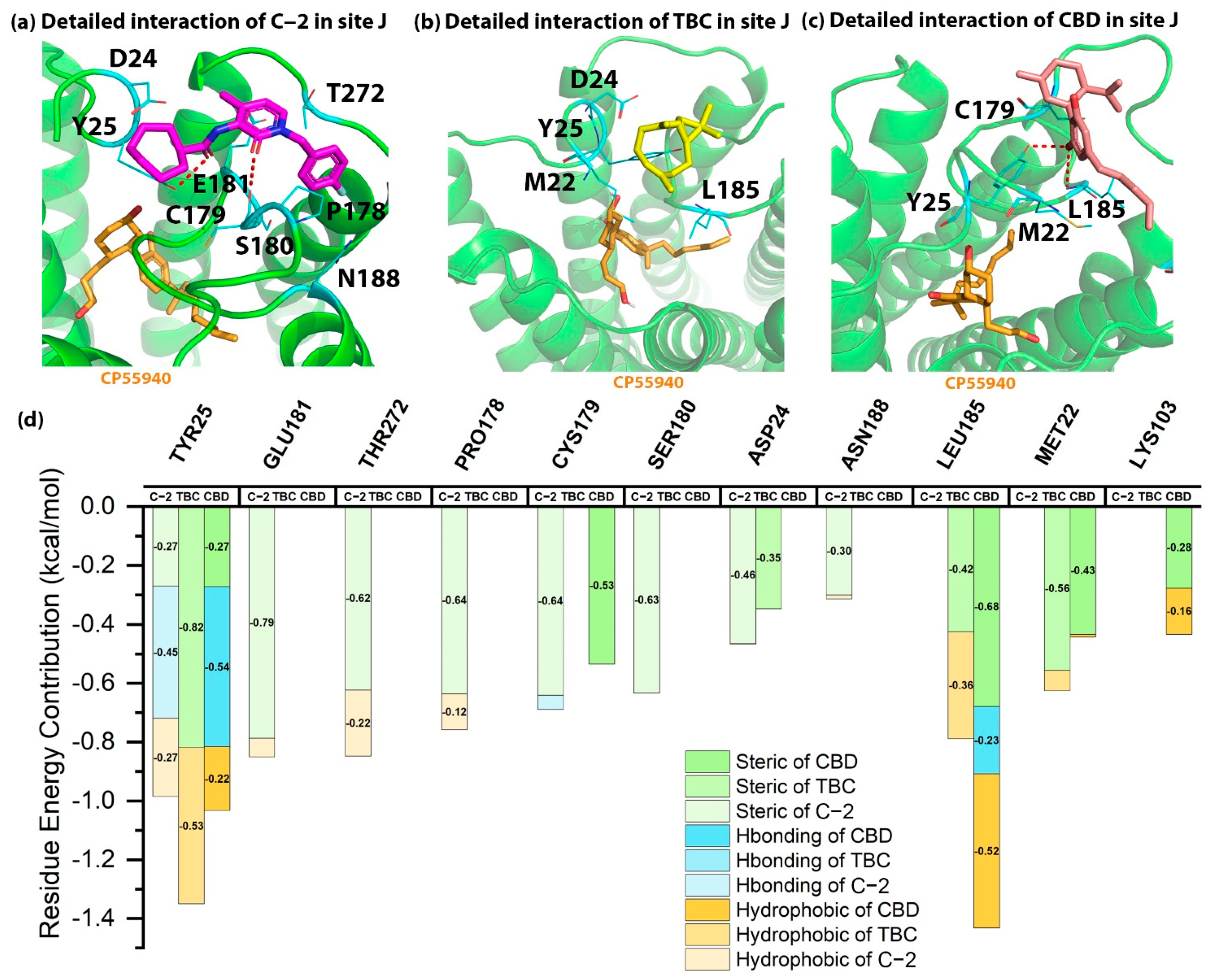
2.5. Detailed Binding Poses and Interactions of C-2, TBC and CBD in Sites I and J of CB2
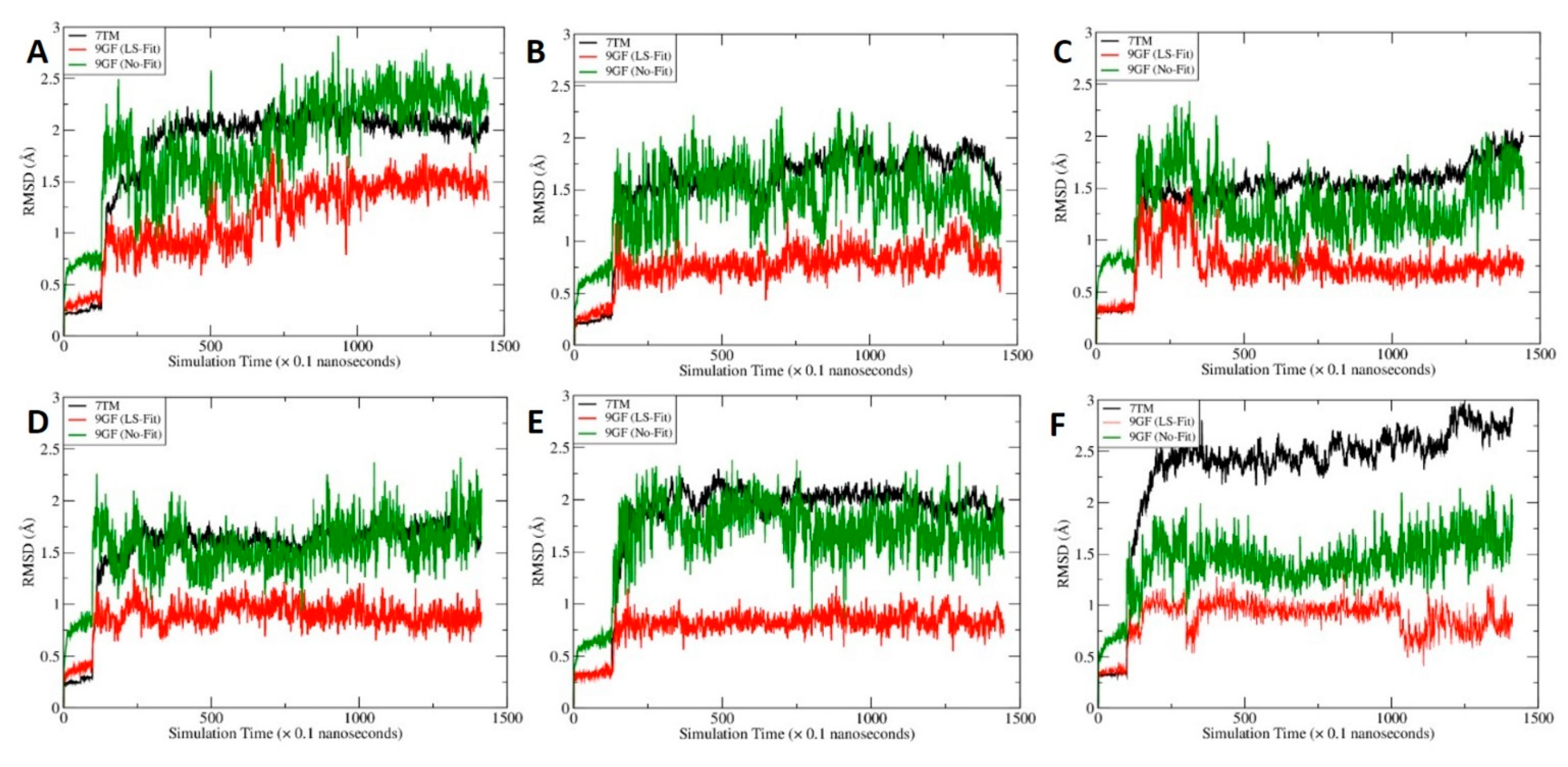
2.6. MD Simulations for C-2 in Five Predicted Allosteric Binding Sites
3. Materials and Methods
3.1. 3D Structures of CB2
3.2. Predictions of Allosteric Binding Sites of CB2
3.3. Molecular Complex Characterizing System (MCCS)
3.4. Molecular Dynamics (MD) Simulation and Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) Calculation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Feng, Z.; Alqarni, M.H.; Yang, P.; Tong, Q.; Chowdhury, A.; Wang, L.; Xie, X.-Q. Modeling, Molecular Dynamics Simulation, and Mutation Validation for Structure of Cannabinoid Receptor 2 Based on Known Crystal Structures of GPCRs. J. Chem. Inf. Model. 2014, 54, 2483–2499. [Google Scholar] [CrossRef] [Green Version]
- Xing, C.; Zhuang, Y.; Xu, T.-H.; Feng, Z.; Zhou, X.E.; Chen, M.; Wang, L.; Meng, X.; Xue, Y.; Wang, J.; et al. Cryo-EM Structure of the Human Cannabinoid Receptor CB2-Gi Signaling Complex. Cell 2020, 180, 645–654.e13. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Feng, Z.; Ma, S.; Zhang, Y.; Tong, Q.; AlQarni, M.H.; Gou, X.; Xie, X.-Q. Difference and Influence of Inactive and Active States of Cannabinoid Receptor Subtype CB2: From Conformation to Drug Discovery. J. Chem. Inf. Model. 2016, 56, 1152–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jordan, C.J.; Feng, Z.-W.; Galaj, E.; Bi, G.-H.; Xue, Y.; Liang, Y.; McGuire, T.; Xie, X.-Q.; Xi, Z.-X. Xie2-64, a novel CB2 receptor inverse agonist, reduces cocaine abuse-related behaviors in rodents. Neuropharmacology 2020, 176, 108241. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Tong, J.; Ding, K.; Zhou, Q.; Zhao, S. GPCR Allosteric Modulator Discovery. Adv. Exp. Med. Biol. 2019, 1163, 225–251. [Google Scholar] [CrossRef] [PubMed]
- Moreira, F.A.; Grieb, M.; Lutz, B. Central side-effects of therapies based on CB1 cannabinoid receptor agonists and antagonists: Focus on anxiety and depression. Best Pract. Res. Clin. Endocrinol. Metab. 2009, 23, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Wenthur, C.J.; Gentry, P.R.; Mathews, T.P.; Lindsley, C.W. Drugs for Allosteric Sites on Receptors. Annu. Rev. Pharmacol. Toxicol. 2014, 54, 165–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Smet, F.; Christopoulos, A.; Carmeliet, P. Allosteric targeting of receptor tyrosine kinases. Nat. Biotechnol. 2014, 32, 1113–1120. [Google Scholar] [CrossRef]
- Jacobson, K.A.; Boeynaems, J.-M. P2Y nucleotide receptors: Promise of therapeutic applications. Drug Discov. Today 2010, 15, 570–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, A.; Armour, D. The Discovery of the CCR5 Receptor Antagonist, UK-427,857, A New Agent for the Treatment of HIV Infection and AIDS. Prog. Med. Chem. 2005, 43, 239–271. [Google Scholar] [CrossRef] [PubMed]
- Wold, E.A.; Chen, J.; Cunningham, K.A.; Zhou, J. Allosteric Modulation of Class A GPCRs: Targets, Agents, and Emerging Concepts. J. Med. Chem. 2018, 62, 88–127. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.E.; Schiedel, A.; Baqi, Y. Allosteric modulators of rhodopsin-like G protein-coupled receptors: Opportunities in drug development. Pharmacol. Ther. 2012, 135, 292–315. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Liang, T.; Wang, S.; Chen, M.; Hou, T.; Zhao, J.; Chen, H.; Zhou, Y.; Xie, X.-Q. Binding Characterization of GPCRs-Modulator by Molecular Complex Characterizing System (MCCS). ACS Chem. Neurosci. 2020, 11, 3333–3345. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Yan, W.; Chapman, K.; Ramesh, K.; Ferrell, A.J.; Yin, J.; Wang, X.; Xu, Q.; Rosenbaum, D.M. Structure of an allosteric modulator bound to the CB1 cannabinoid receptor. Nat. Chem. Biol. 2019, 15, 1199–1205. [Google Scholar] [CrossRef] [PubMed]
- Hua, T.; Li, X.; Wu, L.; Iliopoulos-Tsoutsouvas, C.; Wang, Y.; Wu, M.; Shen, L.; Brust, C.A.; Nikas, S.P.; Song, F.; et al. Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-Gi Complex Structures. Cell 2020, 180, 655–665.e18. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hua, T.; Vemuri, K.; Ho, J.-H.; Wu, Y.; Wu, L.; Popov, P.; Benchama, O.; Zvonok, N.; Locke, K.; et al. Crystal Structure of the Human Cannabinoid Receptor CB2. Cell 2019, 176, 459–467.e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrucci, V.; Chicca, A.; Glasmacher, S.; Paloczi, J.; Cao, Z.; Pacher, P.; Gertsch, J. Pepcan-12 (RVD-hemopressin) is a CB2 receptor positive allosteric modulator constitutively secreted by adrenals and in liver upon tissue damage. Sci. Rep. 2017, 7, 9560. [Google Scholar] [CrossRef]
- Gado, F.; Di Cesare Mannelli, L.; Lucarini, E.; Bertini, S.; Cappelli, E.; Digiacomo, M.; Stevenson, L.A.; Macchia, M.; Tuccinardi, T.; Ghelardini, C.; et al. Identification of the First Synthetic Allosteric Modulator of the CB2 Receptors and Evidence of Its Efficacy for Neuropathic Pain Relief. J. Med. Chem. 2018, 62, 276–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro, G.; Gonzalez, A.; Sánchez-Morales, A.; Casajuana-Martin, N.; Gómez-Ventura, M.; Cordomí, A.; Busqué, F.; Alibés, R.; Pardo, L.; Franco, R. Design of Negative and Positive Allosteric Modulators of the Cannabinoid CB2 Receptor Derived from the Natural Product Cannabidiol. J. Med. Chem. 2021, 64, 9354–9364. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Masoudi, A.; Kahsai, A.W.; Huang, L.-Y.; Pani, B.; Staus, D.P.; Shim, P.J.; Hirata, K.; Simhal, R.K.; Schwalb, A.M.; et al. Mechanism of β2 AR regulation by an intracellular positive allosteric modulator. Science 2019, 364, 1283–1287. [Google Scholar] [CrossRef]
- Lu, J.; Byrne, N.; Wang, J.; Bricogne, G.; Brown, F.K.; Chobanian, H.R.; Colletti, S.L.; Di Salvo, J.; Thomas-Fowlkes, B.; Guo, Y.; et al. Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat. Struct. Mol. Biol. 2017, 24, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Xiao, P.; Yan, W.; Gou, L.; Zhong, Y.-N.; Kong, L.; Wu, C.; Wen, X.; Yuan, Y.; Cao, S.; Qu, C.; et al. Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes. Cell 2021, 184, 943–956.e18. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Qin, L.; Zacarías, N.V.O.; De Vries, H.; Han, G.W.; Gustavsson, M.; Dabros, M.; Zhao, C.; Cherney, R.J.; Carter, P.; et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature 2016, 540, 458–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oswald, C.; Rappas, M.; Kean, J.; Doré, A.S.; Errey, J.C.; Bennett, K.; Deflorian, F.; Christopher, J.A.; Jazayeri, A.; Mason, J.S.; et al. Intracellular allosteric antagonism of the CCR9 receptor. Nature 2016, 540, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ahn, S.; Kahsai, A.W.; Meng, K.-C.; Latorraca, N.R.; Pani, B.; Venkatakrishnan, A.J.; Masoudi, A.; Weis, W.I.; Dror, R.O.; et al. Mechanism of intracellular allosteric β2AR antagonist revealed by X-ray crystal structure. Nat. Cell Biol. 2017, 548, 480–484. [Google Scholar] [CrossRef]
- Liu, X.; Kaindl, J.; Korczynska, M.; Stößel, A.; Dengler, D.; Stanek, M.; Hübner, H.; Clark, M.J.; Mahoney, J.; Matt, R.A.; et al. An allosteric modulator binds to a conformational hub in the β2 adrenergic receptor. Nat. Chem. Biol. 2020, 16, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Robertson, N.; Rappas, M.; Doré, A.S.; Brown, J.; Bottegoni, G.; Koglin, M.; Cansfield, J.; Jazayeri, A.; Cooke, R.M.; Marshall, F.H. Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727. Nature 2018, 553, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Roy, K.K.; Doerksen, R.J. Negative allosteric modulators of cannabinoid receptor 2: Protein modeling, binding site identification and molecular dynamics simulations in the presence of an orthosteric agonist. J. Biomol. Struct. Dyn. 2019, 38, 32–47. [Google Scholar] [CrossRef]
- Chien, E.Y.T.; Liu, W.; Zhao, Q.; Katritch, V.; Han, G.W.; Hanson, M.A.; Shi, L.; Newman, A.H.; Javitch, J.A.; Cherezov, V.; et al. Structure of the Human Dopamine D3 Receptor in Complex with a D2/D3 Selective Antagonist. Science 2010, 330, 1091–1095. [Google Scholar] [CrossRef] [Green Version]
- Lane, J.R.; Chubukov, P.; Liu, W.; Canals, M.; Cherezov, V.; Abagyan, R.; Stevens, R.; Katritch, V. Structure-Based Ligand Discovery Targeting Orthosteric and Allosteric Pockets of Dopamine Receptors. Mol. Pharmacol. 2013, 84, 794–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Hu, G.; Ma, S.; Xie, X.-Q. Computational Advances for the Development of Allosteric Modulators and Bitopic Ligands in G Protein-Coupled Receptors. AAPS J. 2015, 17, 1080–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Goodsell, D.S.; Zardecki, C.; Di Costanzo, L.; Duarte, J.M.; Hudson, B.P.; Persikova, I.; Segura, J.; Shao, C.; Voigt, M.; Westbrook, J.D.; et al. RCSB Protein Data Bank: Enabling biomedical research and drug discovery. Protein Sci. 2019, 29, 52–65. [Google Scholar] [CrossRef] [Green Version]
- Sain, N.M.; Liang, A.; Kane, S.A.; Urban, M.O. Antinociceptive effects of the non-selective cannabinoid receptor agonist CP 55,940 are absent in CB1−/− and not CB2−/− mice in models of acute and persistent pain. Neuropharmacology 2009, 57, 235–241. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, S.; Hu, Q.; Gao, S.; Ma, X.; Zhang, W.; Shen, Y.; Chen, F.; Lai, L.; Pei, J. CavityPlus: A web server for protein cavity detection with pharmacophore modelling, allosteric site identification and covalent ligand binding ability prediction. Nucleic Acids Res. 2018, 46, W374–W379. [Google Scholar] [CrossRef] [PubMed]
- Panjkovich, A.; Daura, X. PARS: A web server for the prediction of Protein Allosteric and Regulatory Sites. Bioinformatics 2014, 30, 1314–1315. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Feng, Z.; Wang, S.; Lin, W.; Xie, X.-Q. MCCS, a novel characterization method for protein–ligand complex. Brief. Bioinform. 2020, 22, bbaa239. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapovalov, M.V.; Dunbrack, R.L. A Smoothed Backbone-Dependent Rotamer Library for Proteins Derived from Adaptive Kernel Density Estimates and Regressions. Structure 2011, 19, 844–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedretti, A.; Villa, L.; Vistoli, G. VEGA—An open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J. Comput. Mol. Des. 2004, 18, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Søndergaard, C.R.; Olsson, M.H.M.; Rostkowski, M.; Jensen, J.H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Leung, K.-S.; Wong, M.-H. idock: A multithreaded virtual screening tool for flexible ligand docking. In Proceedings of the 2012 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), San Diego, CA, USA, 9–12 May 2012; pp. 77–84. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the Performance of the MM/PBSA and MM/GBSA Methods. 1. The Accuracy of Binding Free Energy Calculations Based on Molecular Dynamics Simulations. J. Chem. Inf. Model. 2011, 51, 69–82. [Google Scholar] [CrossRef]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J. Comput. Chem. 2011, 32, 866–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, I.T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K.; et al. AMBER 2018; University of California: San Francisco, CA, USA, 2018; Available online: https://ambermd.org/ (accessed on 1 January 2022).
- Wray, R.; Wang, J.; Iscla, I.; Blount, P. Novel MscL agonists that allow multiple antibiotics cytoplasmic access activate the channel through a common binding site. PLoS ONE 2020, 15, e0228153. [Google Scholar] [CrossRef]
- Wang, J.; Hou, T. Develop and Test a Solvent Accessible Surface Area-Based Model in Conformational Entropy Calculations. J. Chem. Inf. Model. 2012, 52, 1199–1212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Hou, T.; Xu, X. Recent Advances in Free Energy Calculations with a Combination of Molecular Mechanics and Continuum Models. Curr. Comput. Aided-Drug Des. 2006, 2, 287–306. [Google Scholar] [CrossRef] [Green Version]
- Ramesh, K.; Rosenbaum, D.M. Molecular basis for ligand modulation of the cannabinoid CB 1 receptor. Br. J. Pharmacol. 2021. [Google Scholar] [CrossRef]


| Site B | Site C | Site G | Site H | Site I | Site J | Site K | |
|---|---|---|---|---|---|---|---|
| C−2 | −7.26 | N/A * | −6.68 | −7.20 | −7.31 | −5.64 | −6.95 |
| TBC | −6.04 | −4.44 | −5.42 | −2.79 | −6.39 | −4.35 | −5.75 |
| CBD | −6.28 | −4.74 | −5.89 | −4.21 | −5.92 | −4.13 | −5.78 |
| Site | EVDW | Eeel | Eint | Gpol | Gnonpol | TS | GMM-PBSA |
|---|---|---|---|---|---|---|---|
| B | −1213.52 ± 1.02 | −6211.37 ± 5.31 | 7207.92 ± 5.38 | −3699.43 ± 2.93 | 87.94 ± 0.14 | 3406.36 ± 0.68 | −7234.83 ± 8.24 |
| G | −1231.75 ± 0.80 | −6271.31 ± 10.20 | 7216.25 ± 6.29 | −3646.51 ± 16.49 | 89.27 ± 0.10 | 3407.79 ± 0.35 | −7251.85 ± 1.51 |
| H | −1249.43 ± 3.56 | −6356.62 ± 3.06 | 7216.04 ± 2.52 | −3623.11 ± 3.62 | 85.70 ± 0.01 | 3393.90 ± 0.08 | −7321.31 ± 7.11 |
| I | −1257.82 ± 1.33 | −6363.47 ± 9.32 | 7204.73 ± 4.78 | −3520.82 ± 6.20 | 86.77 ± 0.07 | 3395.35 ± 0.13 | −7245.96 ± 11.47 |
| J | −1224.81 ± 1.81 | −6261.38 ± 3.18 | 7224.34 ± 5.16 | −3738.61 ± 7.43 | 87.58 ± 0.04 | 3401.05 ± 0.31 | −7313.93 ± 4.07 |
| Site | ΔEVDW | ΔEeel | ΔGpol | ΔGnonpo | TΔS | ΔGMM-PBSA |
|---|---|---|---|---|---|---|
| B | −27.47 ± 0.49 | 2.42 ± 0.18 | 7.11 ± 0.31 | −2.65 ± 0.03 | −17.80 ± 0.09 | −2.79 ± 0.64 |
| G | −28.83 ± 0.22 | −3.92 ± 0.21 | 12.03 ± 0.43 | −2.30 ± 0.02 | −17.36 ± 0.15 | −5.66 ± 0.68 |
| H | −56.18 ± 0.08 | −24.69 ± 0.50 | 24.41 ± 0.19 | −3.79 ± 0.01 | −26.13 ± 0.08 | −34.12 ± 0.43 |
| I | −32.57 ± 0.08 | −3.01 ± 0.06 | 10.97 ± 0.08 | −2.58 ± 0.02 | −18.47 ± 0.07 | −8.73 ± 0.12 |
| J | −25.14 ± 0.41 | −9.96 ± 0.73 | 23.44 ± 0.55 | −2.15 ± 0.03 | −17.38 ± 0.17 | 3.57 ± 0.44 |
| Site | ΔEVDW | ΔEeel | ΔGpol | ΔGnonpo | TΔS | ΔGMM-PBSA |
|---|---|---|---|---|---|---|
| No Ligand | −53.69 ± 0.21 | −18.76 ± 0.18 | 24.97 ± 0.27 | −4.88 ± 0.01 | −24.96 ± 0.07 | −27.39 ± 0.23 |
| B | −56.87 ± 0.21 | −20.24 ± 0.65 | 25.14 ± 0.27 | −4.96 ± 0.00 | −25.47 ± 0.02 | −31.47 ± 0.18 |
| G | −58.05 ± 0.25 | −9.74 ± 0.36 | 23.54 ± 0.19 | −4.96 ± 0.01 | −25.56 ± 0.09 | −23.66 ± 0.50 |
| H | −56.21 ± 0.27 | −11.57 ± 0.40 | 21.30 ± 0.15 | −4.91 ± 0.01 | −25.31 ± 0.04 | −26.09 ± 0.01 |
| H* | −60.94 ± 0.34 | −12.78 ± 0.38 | 20.00 ± 0.19 | −4.88 ± 0.02 | −26.55 ± 0.05 | −32.04 ± 0.29 |
| I | −57.95 ± 0.21 | −18.73 ± 0.23 | 23.42 ± 0.11 | −4.77 ± 0.01 | −26.20 ± 0.03 | −31.83 ± 0.15 |
| J | −57.69 ± 0.13 | −26.88 ± 0.50 | 24.26 ± 0.16 | −4.80 ± 0.01 | −26.81 ± 0.06 | −38.29 ± 0.40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, J.; Jiang, C.; Wang, J.; Chen, C.-J.; Hao, Y.; Zhao, G.; Feng, Z.; Xie, X.-Q. In Silico Prediction and Validation of CB2 Allosteric Binding Sites to Aid the Design of Allosteric Modulators. Molecules 2022, 27, 453. https://doi.org/10.3390/molecules27020453
Yuan J, Jiang C, Wang J, Chen C-J, Hao Y, Zhao G, Feng Z, Xie X-Q. In Silico Prediction and Validation of CB2 Allosteric Binding Sites to Aid the Design of Allosteric Modulators. Molecules. 2022; 27(2):453. https://doi.org/10.3390/molecules27020453
Chicago/Turabian StyleYuan, Jiayi, Chen Jiang, Junmei Wang, Chih-Jung Chen, Yixuan Hao, Guangyi Zhao, Zhiwei Feng, and Xiang-Qun Xie. 2022. "In Silico Prediction and Validation of CB2 Allosteric Binding Sites to Aid the Design of Allosteric Modulators" Molecules 27, no. 2: 453. https://doi.org/10.3390/molecules27020453
APA StyleYuan, J., Jiang, C., Wang, J., Chen, C.-J., Hao, Y., Zhao, G., Feng, Z., & Xie, X.-Q. (2022). In Silico Prediction and Validation of CB2 Allosteric Binding Sites to Aid the Design of Allosteric Modulators. Molecules, 27(2), 453. https://doi.org/10.3390/molecules27020453







