Structural Optimization and Improving Antitumor Potential of Moreollic Acid from Gamboge
Abstract
:1. Introduction
2. Results and Discussion
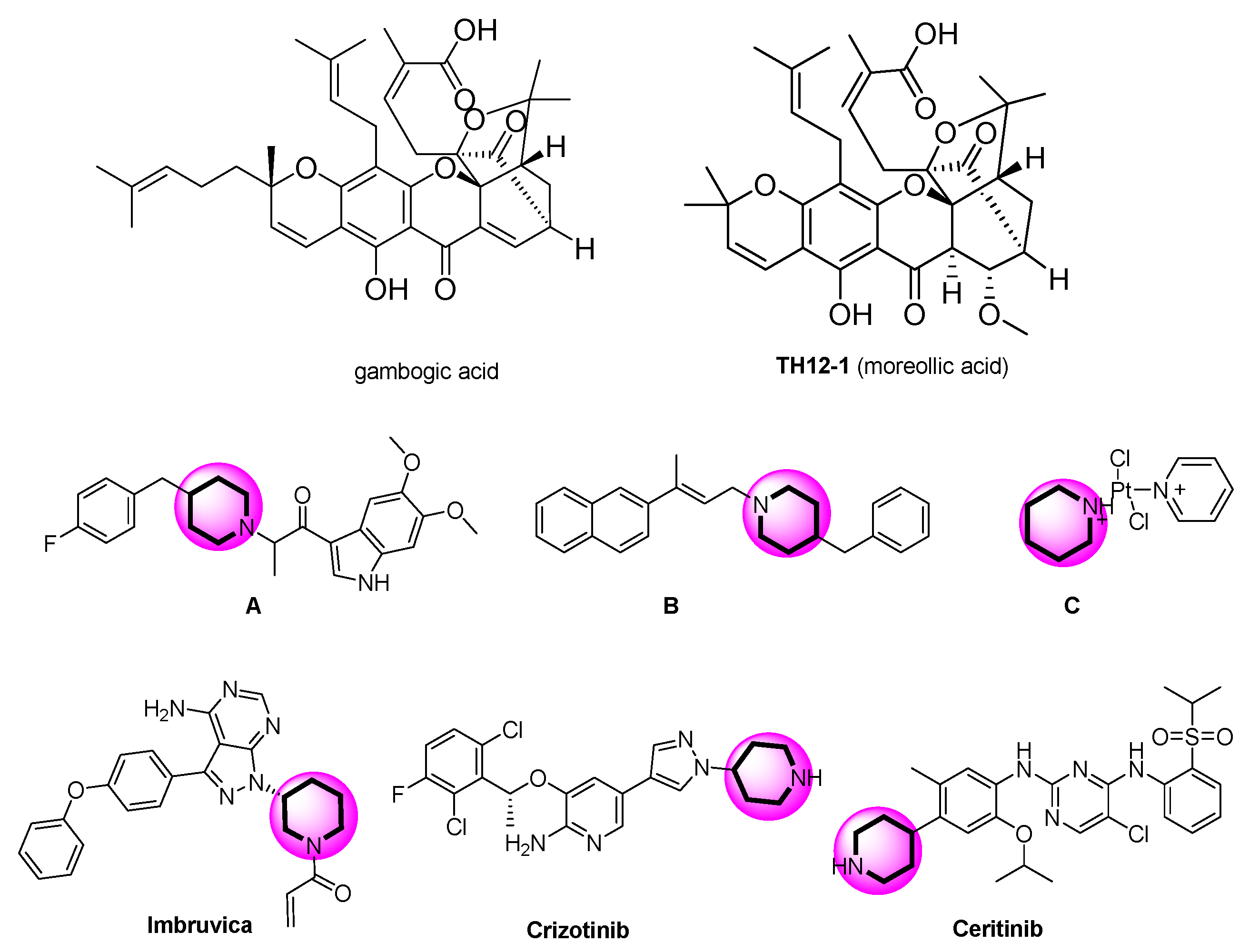
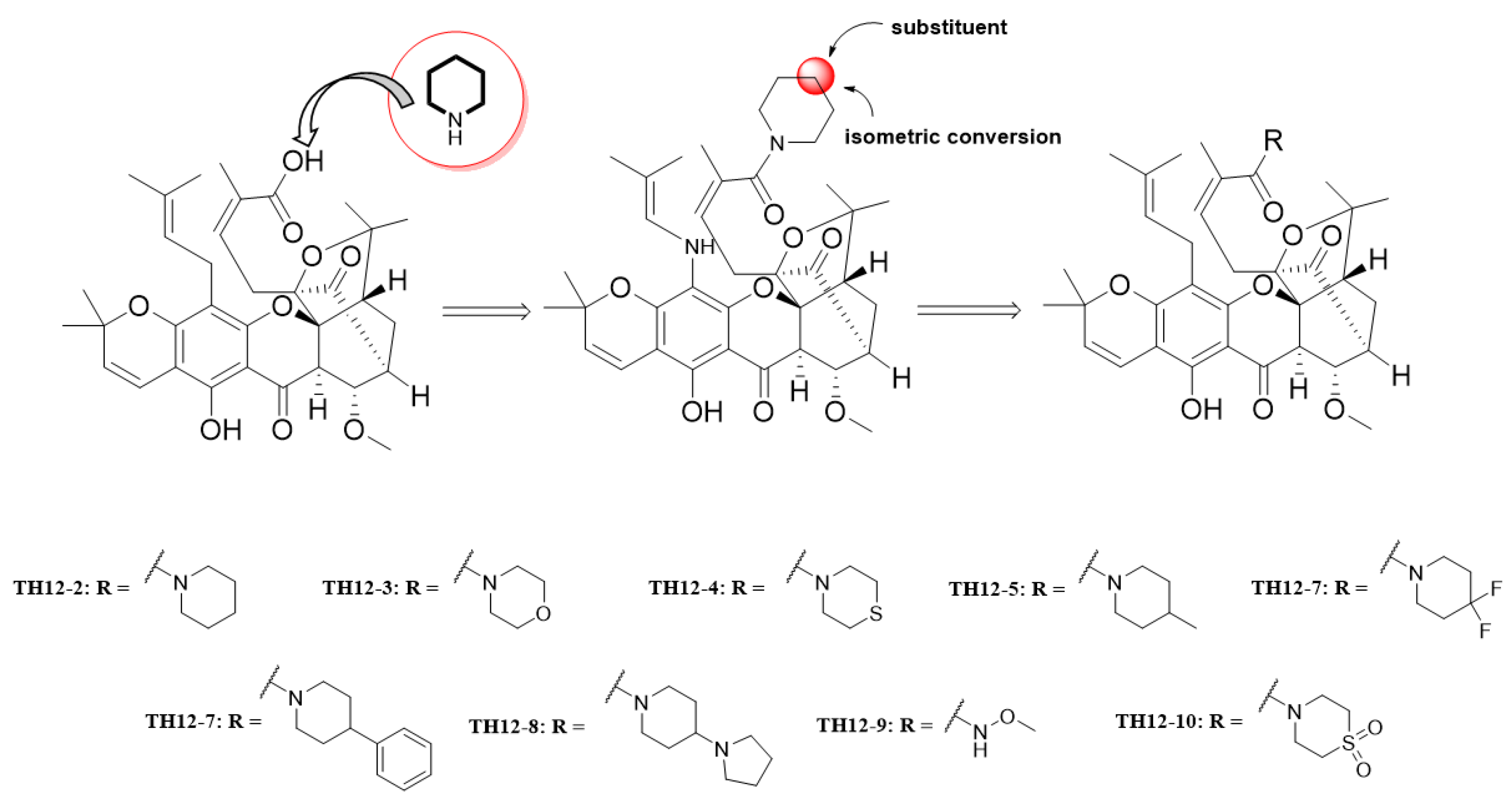
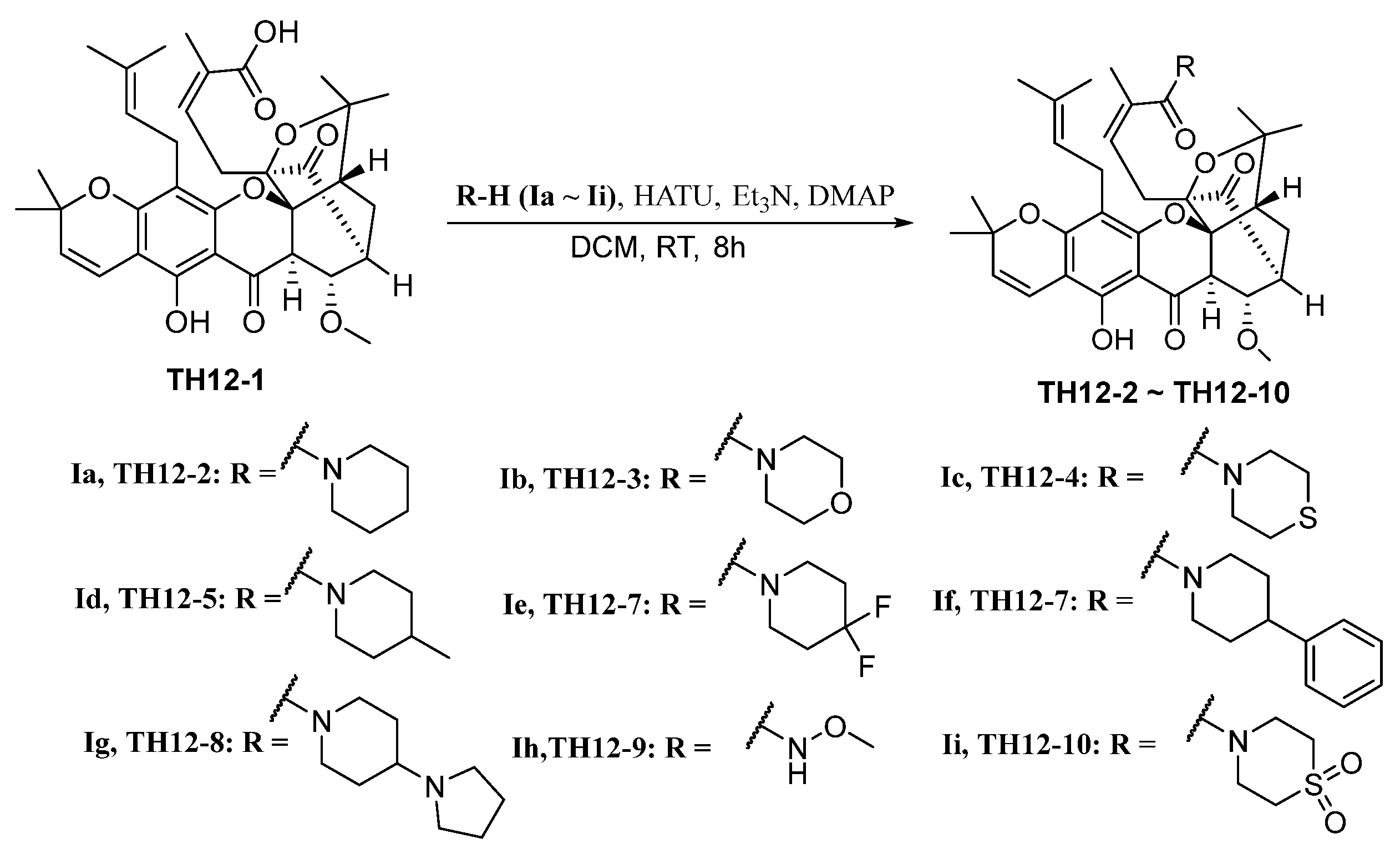
2.1. Chemistry
2.2. Inhibition of Cell Viability in Tumor Cell Lines
2.3. Apparent Structure–Activity Relationship
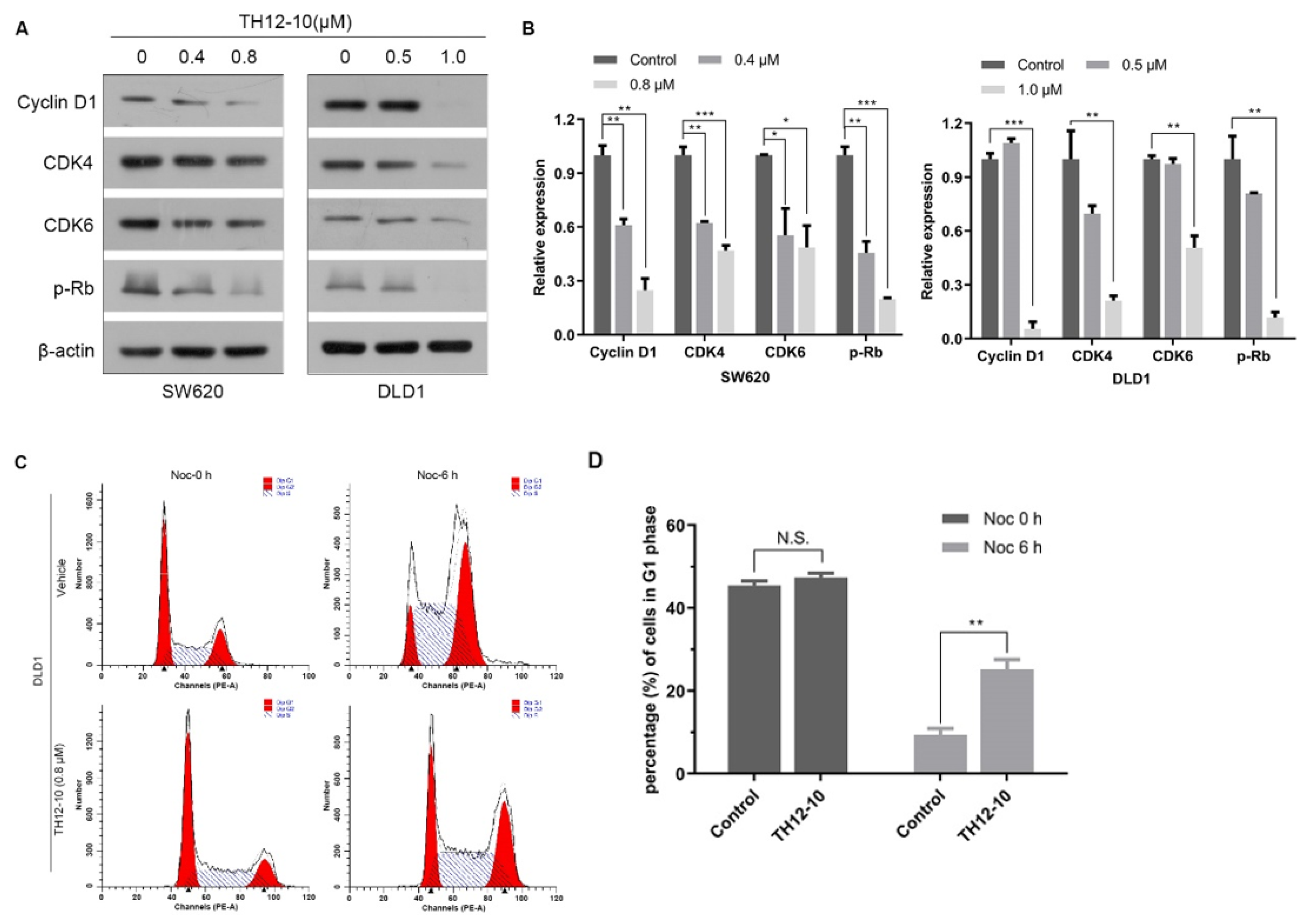
2.4. TH12-10 Effectively Inhibited the Cell Proliferation via Modulation of Cell-Cycle Regulatory Proteins
3. Experimental Section
3.1. General Procedures
3.2. Plant Resins
3.3. Extraction and Isolation of TH12-1
3.4. Synthesis of TH12-2 and Its Analogues (TH12-3~TH12-10)
3.5. Bioactivy Assay
3.5.1. The Cell Culture
3.5.2. Cell Proliferation Assay
3.5.3. Immunoblot
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Sagar, S.; Esau, L.; Moosa, B.; Khashab, N.M.; Bajic, V.B.; Kaur, M. Cytotoxicity and apoptosis induced by a plumbagin derivative in estrogen positive MCF-7 breast cancer cells. Anti-Cancer Agents Med. Chem. 2014, 14, 170–180. [Google Scholar] [CrossRef] [PubMed]
- George, B.P.A.; Abrahamse, H. A Review on Novel Breast Cancer Therapies: Photodynamic Therapy and Plant Derived Agent Induced Cell Death Mechanisms. Anti-Cancer Agents Med. Chem. 2016, 16, 793–801. [Google Scholar] [CrossRef]
- Simon, N.; Odou, P.; Décaudin, B.; Bonnabry, P.; Fleury Souverain, S. Occupational exposure to conventional antineoplastic drugs: Can it be further limited? Eur. J. Hosp. Pharm. 2020, 27, 251–252. [Google Scholar] [CrossRef] [Green Version]
- Mueller-Schoell, A.; Puebla-Osorio, N.; Michelet, R.; Green, M.R.; Künkele, A.; Huisinga, W.; Strati, P.; Chasen, B.; Neelapu, S.S.; Yee, C.; et al. Therapeutic drug monitoring of oral targeted antineoplastic drugs. Eur. J. Clin. Pharmacol. 2021, 77, 441–464. [Google Scholar] [CrossRef]
- Hu, B.Y.; Wang, S.X.; Yan, Y.M.; Liu, J.W.; Qin, D.P.; Cheng, Y.X. Spiromyrrhenes A–D: Unprecedented diterpene–sesquiterpene heterodimers as intermolecular [4 + 2] cycloaddition products from Resina Commiphora that inhibit tumor stemness in esophageal cancer. Org. Chem. Front. 2020, 7, 2710–2718. [Google Scholar] [CrossRef]
- Li, Y.P.; Jiang, X.T.; Qin, F.Y.; Zhang, H.X.; Cheng, Y.X. Gancochlearols E–I, meroterpenoids from Ganoderma cochlear against COX-2 and triple negative breast cancer cells and the absolute configuration assignment of ganomycin K. Bioorg. Chem. 2021, 109, 104706. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Q.; Wang, X.L.; He, D.H.; Cheng, Y.X. Protection against chemotherapy–and radiotherapy–induced side effects: A review based on the mechanisms and therapeutic opportunities of phytochemicals. Phytomedicine 2021, 80, 153402. [Google Scholar] [CrossRef]
- Di, L.; Liu, L.J.; Yan, Y.M.; Fu, R.; Li, Y.; Xu, Y.; Cheng, Y.X.; Wu, Z.Q. Discovery of a natural small–molecule compound that suppresses tumor EMT, stemness and metastasis by inhibiting TGFβ/BMP signaling in triple–negative breast cancer. J. Exp. Clin. Cancer Res. 2019, 38, 134. [Google Scholar] [CrossRef]
- Han, Q.B.; Xu, H.X. Cage Garcinia xanthones: Development since 1937. Curr. Med. Chem. 2009, 16, 3775–3796. [Google Scholar] [CrossRef]
- Chen, Y.; He, S.; Tang, C.; Li, J.; Yang, G. Caged polyprenylated xanthones from the resin of Garcinia hanburyi. Fitoterapia 2016, 109, 106–112. [Google Scholar] [CrossRef]
- Wu, Z.Q.; Guo, Q.L.; You, Q.D.; Zhao, L.; Gu, H.Y. Gambogic acid inhibits proliferation of human lung carcinoma SPC-A1 cells in vivo and in vitro and represses telomerase activity and telomerase reverse transcriptase mRNA expression in the cells. Biol. Pharm. Bull. 2004, 27, 1769–1774. [Google Scholar] [CrossRef] [Green Version]
- Jang, J.H.; Kim, J.Y.; Sung, E.G.; Kim, E.A.; Lee, T.J. Gambogic acid induces apoptosis and sensitizes TRAIL-mediated apoptosis through downregulation of cFLIP(L) in renal carcinoma Caki cells. Int. J. Oncol. 2016, 48, 376–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thida, M.; Kim, D.W.; Tran, T.T.T.; Pham, M.Q.; Lee, H.; Kim, I.; Lee, J.W. Gambogic acid induces apoptotic cell death in T98G glioma cells. Bioorg. Med. Chem. Lett. 2016, 26, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.K.; Sung, B.; Ahn, K.S.; Kunnumakkara, A.B.; Chaturvedi, M.M.; Aggarwal, B.B. Gambogic acid, a novel ligand for transferrin receptor, potentiates TNF-induced apoptosis through modulation of the nuclear factor-kappa B signaling pathway. Blood 2007, 110, 3517–3525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, D.; Zhang, B.; Yao, J.; Liu, Y.; Sun, J.; Ge, C.; Peng, S.; Fang, J. Gambogic acid induces apoptosis in hepatocellular carcinoma SMMC-7721 cells by targeting cytosolic thioredoxin reductase. Free Radic. Biol. Med. 2014, 69, 15–25. [Google Scholar] [CrossRef]
- Pu, P.; Kang, C.; Zhang, Z.; Liu, X.; Jiang, H. Downregulation of PIK3CB by siRNA suppresses malignant glioma cell growth in vitro and in vivo. Technol. Cancer Res. Treat. 2006, 5, 271–280. [Google Scholar] [CrossRef]
- Zhao, L.; Zhen, C.; Wu, Z.; Hu, R.; Zhou, C.; Guo, Q. General pharmacological properties, developmental toxicity, and analgesic activity of gambogic acid, a novel natural anticancer agent. Drug Chem. Toxicol. 2010, 33, 88–96. [Google Scholar] [CrossRef]
- Asano, J.; Chiba, K.; Tada, M.; Yoshii, T. Cytotoxic xanthones from Garcinia hanburyi. Phytochemistry 1996, 41, 815–820. [Google Scholar] [CrossRef]
- Ferro, S.; De Luca, L.; Germano, M.P.; Buemi, M.R.; Ielo, L.; Certo, G.; Kanteev, M.; Fishman, A.; Rapisarda, A.; Gitto, R. Chemical exploration of 4-(4-fluorobenzyl)piperidine fragment for the development of new tyrosinase inhibitors. Eur. J. Med. Chem. 2017, 125, 992–1001. [Google Scholar] [CrossRef]
- Rui, M.; Rossi, D.; Marra, A.; Paolillo, M.; Schinelli, S.; Curti, D.; Tesei, A.; Cortesi, M.; Zamagni, A.; Laurini, E.; et al. Synthesis and biological evaluation of new aryl-alkyl(alkenyl)-4-benzylpiperidines, novel Sigma Receptor (SR) modulators, as potential anticancer-agents. Eur. J. Med. Chem. 2016, 124, 649–665. [Google Scholar] [CrossRef] [Green Version]
- Goel, P.; Alam, O.; Naim, M.J.; Nawaz, F.; Iqbal, M.; Alam, M.I. Recent advancement of piperidine moiety in treatment of cancer- A review. Eur. J. Med. Chem. 2018, 157, 480–502. [Google Scholar] [CrossRef] [PubMed]
- Rubin, S.M.; Sage, J.; Skotheim, J.M. Integrating Old and New Paradigms of G1/S Control. Mol. Cell 2020, 80, 183–192. [Google Scholar] [CrossRef] [PubMed]
| Compound | R | 5 μM | |||
|---|---|---|---|---|---|
| A549 | MDA-MB-231 | KYSE30 | HCT-116 | ||
| TH12-1 | OH | 18.38 ± 0.02 | −4.9 ± 0.04 | 20.76 ± 0.04 | 83.86 ± 1.10 2.07 |
| TH12-2 |  | 2.51 ± 4.32 | 0.14 ± 3.51 | 9.08 ± 2.44 | 86.08 ± 2.83 |
| TH12-3 |  | 11.6 ± 4.25 | 1.71 ± 1.66 | 10.63 ± 2.66 | 87.36 ± 2.49 |
| TH12-4 |  | 11.08 ± 5.41 | 3.13 ± 1.16 | 6.98 ± 1.99 | 16.27 ± 4.56 |
| TH12-5 |  | 14.3 ± 2.84 | 0.69 ± 1.92 | 5.65 ± 3.77 | 85.48 ± 1.45 |
| TH12-6 |  | 12.56 ± 0.71 | 35.5 ± 1.17 | 27.02 ± 3.21 | 87.23 ± 3.07 |
| TH12-7 |  | 4.59 ± 1.27 | 2.99 ± 0.16 | −2.71 ± 0.83 | 87.14 ± 2.37 |
| TH12-8 | 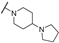 | 4.8 ± 0.63 | 4.32 ± 1.96 | 4.04 ± 1.50 | 87.57 ± 2.76 |
| TH12-9 | 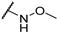 | 8.54 ± 1.11 | −1.76 ± 0.43 | 4.49 ± 1.05 | 83.86 ± 2.07 |
| TH12-10 | 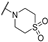 | 6.85 ± 2.16 | 8.09 ± 0.28 | 3.65 ± 2.44 | 88.09 ± 2.78 |
| 5-Fu | 21.14 ± 1.79 | 26.59 ± 2.51 | 27.96 ± 2.27 | 74.45 ± 1.10 | |
| Compound | R | IC50 ± SD (μM) | |||
|---|---|---|---|---|---|
| HCT116 | DLD1 | SW620 | NCM460 | ||
| Th12-1 | OH | 1.71 ± 0.33 | 1.44 ± 0.29 | 1.49 ± 0.37 | 3.16 ± 0.32 |
| Th12-2 | 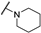 | 1.19 ± 0.25 | 1.39 ± 0.19 | 1.15 ± 0.13 | 4.73 ± 0.04 |
| Th12-3 | 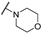 | 1.96 ± 0.20 | 2.308 ± 0.23 | 1.82 ± 0.33 | 4.72 ± 0.03 |
| Th12-4 | 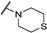 | 7.62 ± 0.93 | 7.89 ± 0.12 | 7.26 ± 0.87 | 19.9 ± 0.84 |
| Th12-5 | 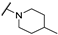 | 2.37 ± 0.63 | 2.49 ± 0.56 | 1.97 ± 0.34 | 5.11 ± 0.21 |
| Th12-6 | 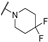 | 1.16 ± 0.23 | 1.22 ± 0.2 | 1.07 ± 0.16 | 2.49 ± 0.16 |
| Th12-7 | 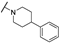 | 1.52 ± 0.14 | 1.45 ± 0.25 | 1.31 ± 0.24 | 2.97 ± 0.08 |
| Th12-8 |  | 1.00 ± 0.13 | 1.05 ± 0.23 | 0.97 ± 0.10 | 2.29 ± 0.01 |
| Th12-9 |  | 1.68 ± 0.26 | 1.67 ± 0.70 | 1.59 ± 0.44 | 1.63 ± 0.02 |
| Th12-10 | 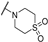 | 0.83 ± 0.10 | 1.10 ± 0.19 | 0.79 ± 0.12 | 4.08 ± 0.11 |
| 5-Fu | 0.62 ± 0.02 | 7.89 ± 0.75 | 2.68 ± 0.01 | 13.59 ± 0.59 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, L.-Z.; Huang, D.-L.; Liao, M.; Li, K.-M.; Wu, Z.-Q.; Cheng, Y.-X. Structural Optimization and Improving Antitumor Potential of Moreollic Acid from Gamboge. Molecules 2022, 27, 482. https://doi.org/10.3390/molecules27020482
Cheng L-Z, Huang D-L, Liao M, Li K-M, Wu Z-Q, Cheng Y-X. Structural Optimization and Improving Antitumor Potential of Moreollic Acid from Gamboge. Molecules. 2022; 27(2):482. https://doi.org/10.3390/molecules27020482
Chicago/Turabian StyleCheng, Li-Zhi, Dan-Ling Huang, Min Liao, Ke-Ming Li, Zhao-Qiu Wu, and Yong-Xian Cheng. 2022. "Structural Optimization and Improving Antitumor Potential of Moreollic Acid from Gamboge" Molecules 27, no. 2: 482. https://doi.org/10.3390/molecules27020482






