Development of a Highly Selective and Sensitive Fluorescent Probe for Imaging RNA Dynamics in Live Cells
Abstract
1. Introduction
2. Results and Discussion
2.1. Molecular Design of Probes QUID−1–QUID−6
2.2. Synthesis of Fluorescent Probes QUID−2–QUID−6
2.3. Screening QUID Fluorescent Probes
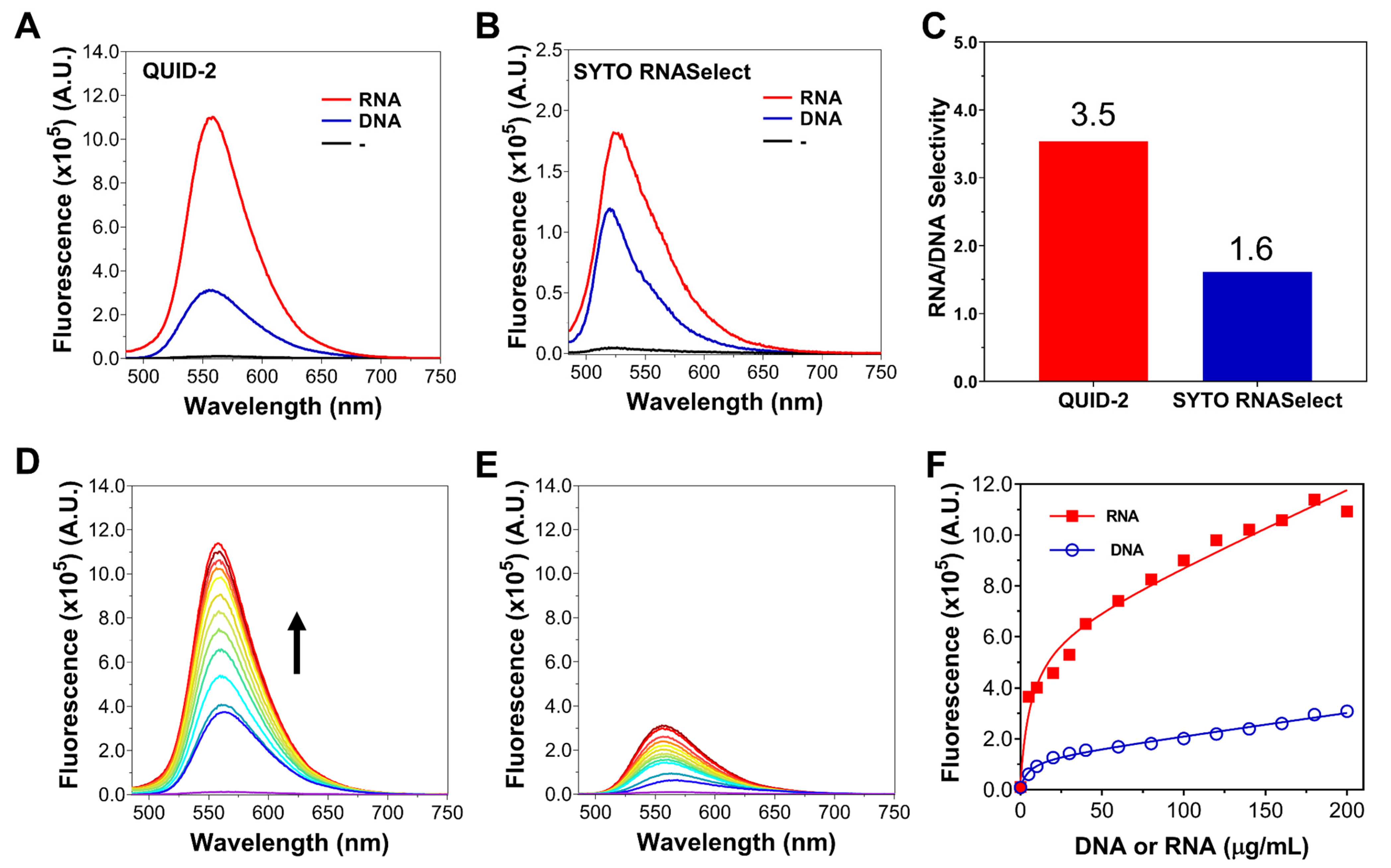
2.4. Specificity Study of QUID−2 towards RNA In Vitro
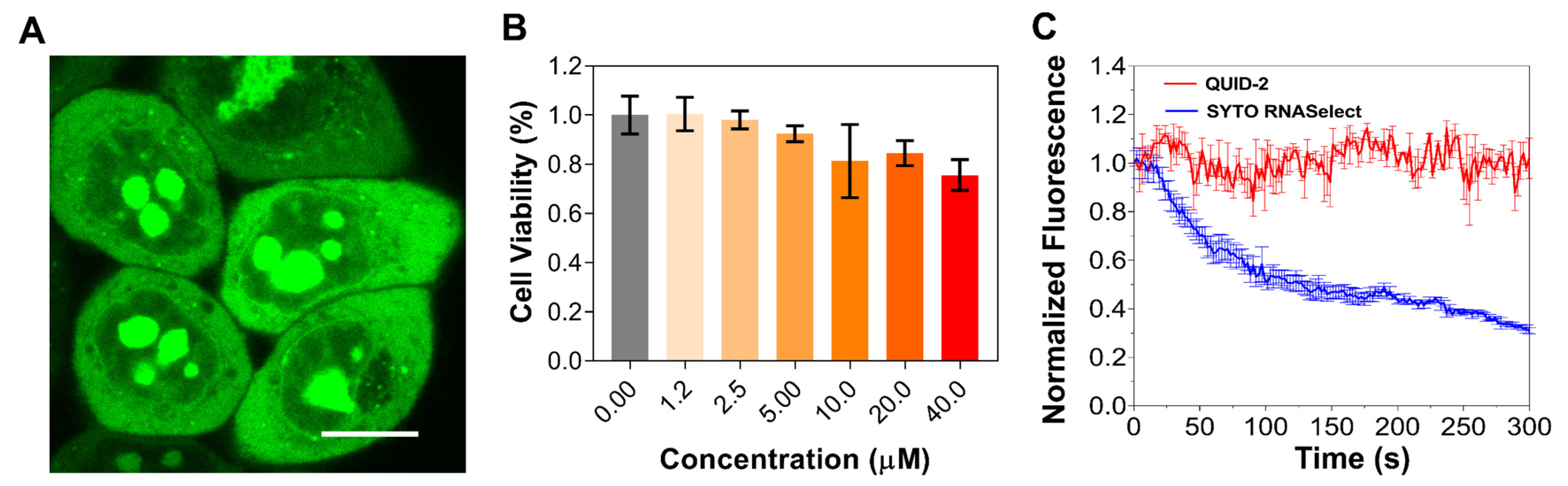
2.5. Specificity Study of QUID−2 towards RNA against Other Nucleic Acids in Cells
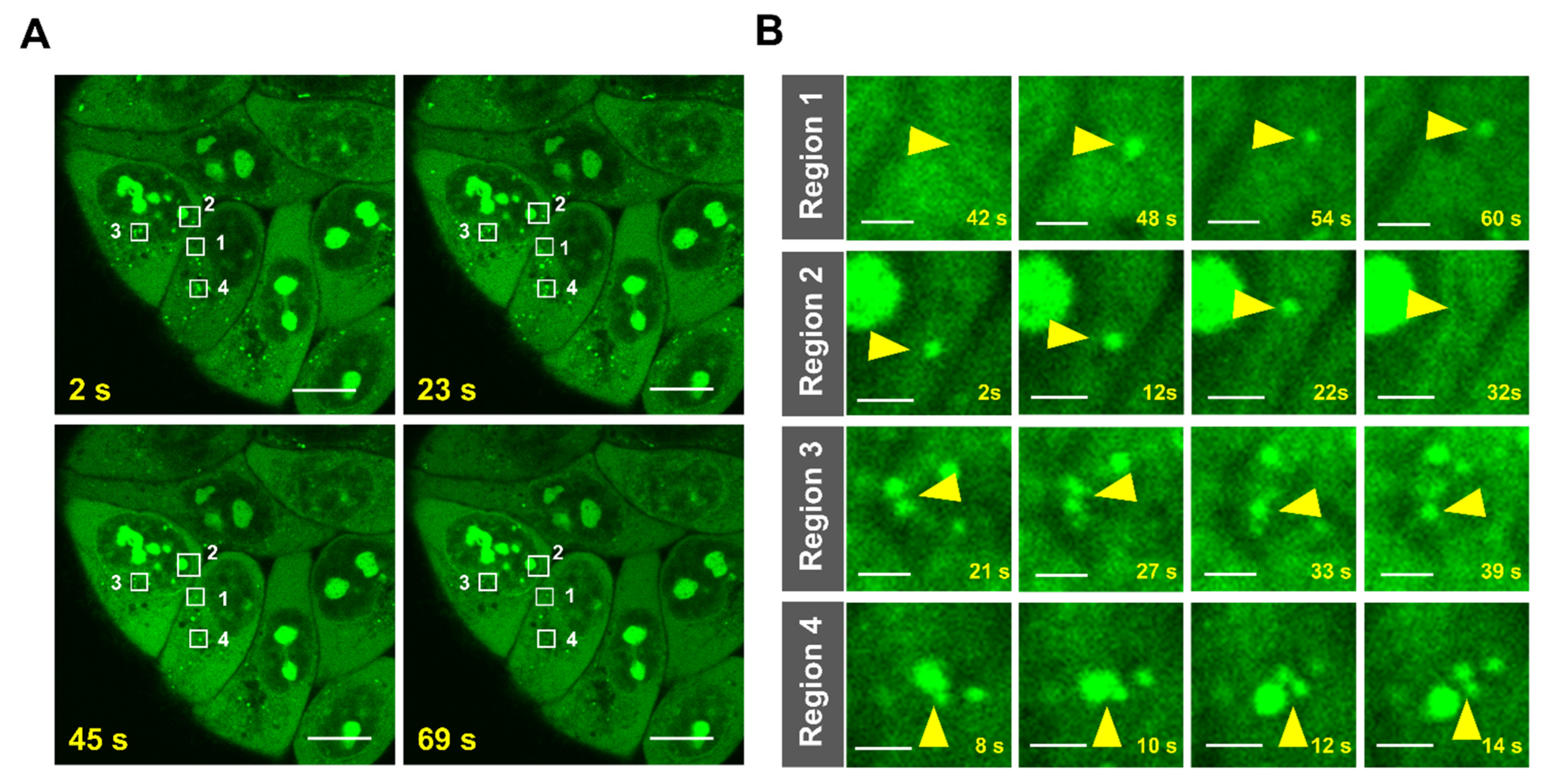
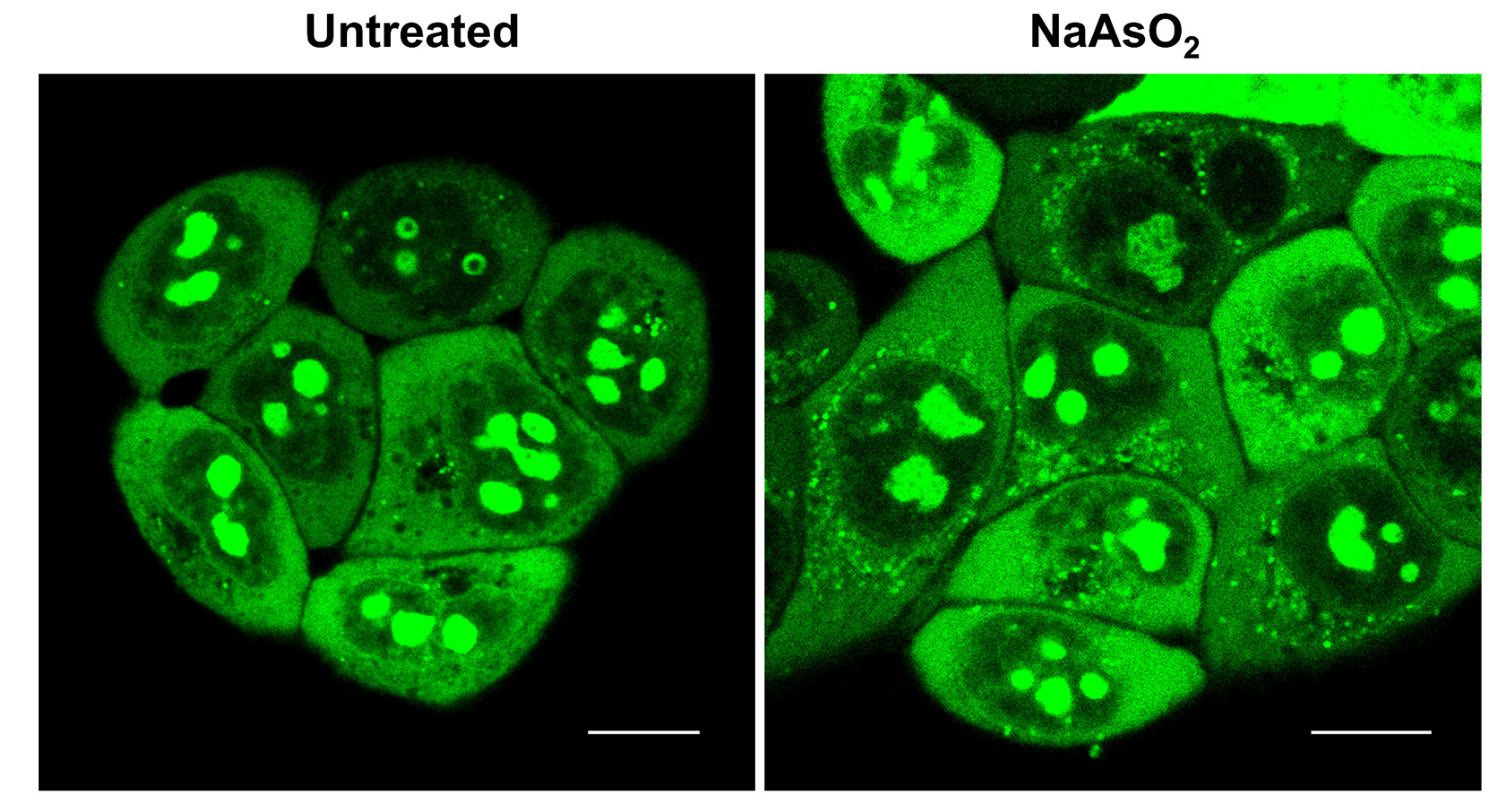
2.6. Imaging of RNA Dynamics in Live Cells
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Synthesis
4.3. pKa Calculations
4.4. UV-Vis Absorption Studies
4.5. Fluorescence Studies
4.6. Fixed Cell Staining Experiments
4.7. Cell Viability Assay
4.8. Plasmid Transfection
4.9. Live Cell Staining Experiments
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Chen, X.; Zhang, D.; Su, N.; Bao, B.; Xie, X.; Zuo, F.; Yang, L.; Wang, H.; Jiang, L.; Lin, Q.; et al. Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nat. Biotechnol. 2019, 37, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Gerstberger, S.; Hafner, M.; Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef] [PubMed]
- Decker, C.J.; Parker, R. P-Bodies and Stress Granules: Possible Roles in the Control of Translation and mRNA Degradation. Cold Spring Harb. Perspect. Biol. 2012, 4, a012286. [Google Scholar] [CrossRef] [PubMed]
- Buchan, J.R.; Parker, R. Eukaryotic Stress Granules: The Ins and Outs of Translation. Mol. Cell 2009, 36, 932–941. [Google Scholar] [CrossRef] [PubMed]
- Rath, A.K.; Rentmeister, A. Genetically encoded tools for RNA imaging in living cells. Curr. Opin. Biotechnol. 2015, 31, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Tomoike, F.; Abe, H. RNA imaging by chemical probes. Adv. Drug Deliv. Rev. 2019, 147, 44–58. [Google Scholar] [CrossRef]
- Bao, G.; Rhee, W.J.; Tsourkas, A. Fluorescent Probes for Live-Cell RNA Detection. Annu. Rev. Biomed. Eng. 2009, 11, 25–47. [Google Scholar] [CrossRef]
- Tian, X.; Murfin, L.C.; Wu, L.; Lewis, S.E.; James, T.D. Fluorescent small organic probes for biosensing. Chem. Sci. 2021, 12, 3406–3426. [Google Scholar] [CrossRef]
- Han, H.-H.; Tian, H.; Zang, Y.; Sedgwick, A.C.; Li, J.; Sessler, J.L.; He, X.-P.; James, T.D. Small-molecule fluorescence-based probes for interrogating major organ diseases. Chem. Soc. Rev. 2021, 50, 9391–9429. [Google Scholar] [CrossRef]
- Wang, L.; Frei, M.S.; Salim, A.; Johnsson, K. Small-Molecule Fluorescent Probes for Live-Cell Super-Resolution Microscopy. J. Am. Chem. Soc. 2019, 141, 2770–2781. [Google Scholar] [CrossRef]
- Cao, C.; Wei, P.; Li, R.; Zhong, Y.; Li, X.; Xue, F.; Shi, Y.; Yi, T. Ribosomal RNA-Selective Light-Up Fluorescent Probe for Rapidly Imaging the Nucleolus in Live Cells. ACS Sensors 2019, 4, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Sun, Y.; Liu, Y.; Wang, X.; Chen, M.; Miao, F.; Zhang, W.; Yu, X.; Jin, J. Low molecular weight fluorescent probes with good photostability for imaging RNA-rich nucleolus and RNA in cytoplasm in living cells. Biomaterials 2014, 35, 2103–2112. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Kim, Y.; Namm, J.; Kulkarni, A.; Rosania, G.R.; Ahn, Y.-H.; Chang, Y.-T. RNA-Selective, Live Cell Imaging Probes for Studying Nuclear Structure and Function. Chem. Biol. 2006, 13, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, S.; Zhang, C.; Dong, J.; Shen, C.; Zhu, J.; Xu, H.; Fu, M.; Yang, G.; Zhang, X. A water-soluble two-photon ratiometric triarylboron probe with nucleolar targeting by preferential RNA binding. Chem. Commun. 2017, 53, 11476–11479. [Google Scholar] [CrossRef]
- Lu, Y.-J.; Deng, Q.; Hu, D.-P.; Wang, Z.-Y.; Huang, B.-H.; Du, Z.-Y.; Fang, Y.-X.; Wong, W.-L.; Zhang, K.; Chow, C.-F. A molecular fluorescent dye for specific staining and imaging of RNA in live cells: A novel ligand integration from classical thiazole orange and styryl compounds. Chem. Commun. 2015, 51, 15241–15244. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Chan, M.S.; Xu, D.; Tam, D.Y.; Bolze, F.; Lo, P.K.; Wong, M.S. Indole-based Cyanine as a Nuclear RNA-Selective Two-Photon Fluorescent Probe for Live Cell Imaging. ACS Chem. Biol. 2015, 10, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, K.; Sato, Y.; Togashi, N.; Suzuki, M.; Yoshino, Y.; Nishizawa, S. Bright and Light-Up Sensing of Benzo[c,d]indole-oxazolopyridine Cyanine Dye for RNA and Its Application to Highly Sensitive Imaging of Nucleolar RNA in Living Cells. ACS Omega 2022, 7, 23744–23748. [Google Scholar] [CrossRef]
- Yoshino, Y.; Sato, Y.; Nishizawa, S. Deep-Red Light-up Signaling of Benzo[c,d]indole-Quinoline Monomethine Cyanine for Imaging of Nucleolar RNA in Living Cells and for Sequence-Selective RNA Analysis. Anal. Chem. 2019, 91, 14254–14260. [Google Scholar] [CrossRef]
- Li, Z.; Sun, S.; Yang, Z.; Zhang, S.; Zhang, H.; Hu, M.; Cao, J.; Wang, J.; Liu, F.; Song, F.; et al. The use of a near-infrared RNA fluorescent probe with a large Stokes shift for imaging living cells assisted by the macrocyclic molecule CB7. Biomaterials 2013, 34, 6473–6481. [Google Scholar] [CrossRef]
- Zhou, B.; Liu, W.; Zhang, H.; Wu, J.; Liu, S.; Xu, H.; Wang, P. Imaging of nucleolar RNA in living cells using a highly photostable deep-red fluorescent probe. Biosens. Bioelectron. 2015, 68, 189–196. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Wang, W.; Zhao, C.; Lin, W. A single small molecule fluorescent probe for imaging RNA distribution and detecting endogenous SO2 through distinct fluorescence channels. New J. Chem. 2021, 45, 19812–19817. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Wang, W.; Zhao, C.; Lin, W. A novel fluorescent probe with high photostability for imaging distribution of RNA in living cells and tissues. New J. Chem. 2021, 45, 2614–2619. [Google Scholar] [CrossRef]
- Feng, R.; Li, L.; Li, B.; Li, J.; Peng, D.; Yu, Y.; Mu, Q.; Zhao, N.; Yu, X.; Wang, Z. Turn-on fluorescent probes that can light up endogenous RNA in nucleoli and cytoplasm of living cells under a two-photon microscope. RSC Adv. 2017, 7, 16730–16736. [Google Scholar] [CrossRef]
- Wang, L.; Xia, Q.; Liu, R.; Qu, J. A red fluorescent probe for ribonucleic acid (RNA) detection, cancer cell tracing and tumor growth monitoring. Sens. Actuators B Chem. 2018, 273, 935–943. [Google Scholar] [CrossRef]
- Wang, C.; Lu, Y.-J.; Cai, S.-Y.; Long, W.; Zheng, Y.-Y.; Lin, J.-W.; Yan, Y.; Huang, X.-H.; Wong, W.-L.; Zhang, K.; et al. Advancing small ligands targeting RNA for better binding affinity and specificity: A study of structural influence through molecular design approach. Sens. Actuators B Chem. 2018, 262, 386–394. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, W.; Sun, Y.; Song, G.; Miao, F.; Guo, F.; Tian, M.; Yu, X.; Sun, J.Z. Two-photon fluorescence imaging of RNA in nucleoli and cytoplasm in living cells based on low molecular weight probes. Dye. Pigments 2014, 103, 191–201. [Google Scholar] [CrossRef]
- Chen, X.; Chen, S.; Dai, J.; Yuan, J.; Ou, T.; Huang, Z.; Tan, J. Tracking the Dynamic Folding and Unfolding of RNA G-Quadruplexes in Live Cells. Angew. Chem. Int. Ed. 2018, 57, 4702–4706. [Google Scholar] [CrossRef] [PubMed]
- Fürstenberg, A.; Deligeorgiev, T.G.; Gadjev, N.I.; Vasilev, A.A.; Vauthey, E. Structure–Fluorescence Contrast Relationship in Cyanine DNA Intercalators: Toward Rational Dye Design. Chem.—A Eur. J. 2007, 13, 8600–8609. [Google Scholar] [CrossRef]
- Means, J.A.; Hines, J.V. Fluorescence resonance energy transfer studies of aminoglycoside binding to a T box antiterminator RNA. Bioorganic Med. Chem. Lett. 2005, 15, 2169–2172. [Google Scholar] [CrossRef]
- Wong, C.-H.; Hendrix, M.; Priestley, E.S.; Greenberg, W.A. Specificity of aminoglycoside antibiotics for the A-site of the decoding region of ribosomal RNA. Chem. Biol. 1998, 5, 397–406. [Google Scholar] [CrossRef][Green Version]
- Aristova, D.; Kosach, V.; Chernii, S.; Slominsky, Y.; Balanda, A.; Filonenko, V.; Yarmoluk, S.; Rotaru, A.; Özkan, H.G.; Mokhir, A.; et al. Monomethine cyanine probes for visualization of cellular RNA by fluorescence microscopy. Methods Appl. Fluoresc. 2021, 9, 045002. [Google Scholar] [CrossRef]
- Nygren, J.; Svanvik, N.; Kubista, M. The interactions between the fluorescent dye thiazole orange and DNA. Biopolymers 1998, 46, 39–51. [Google Scholar] [CrossRef]
- Granzhan, A.; Ihmels, H.; Viola, G. 9-Donor-Substituted Acridizinium Salts: Versatile Environment-Sensitive Fluorophores for the Detection of Biomacromolecules. J. Am. Chem. Soc. 2007, 129, 1254–1267. [Google Scholar] [CrossRef]
- Bouhedda, F.; Fam, K.T.; Collot, M.; Autour, A.; Marzi, S.; Klymchenko, A.; Ryckelynck, M. A dimerization-based fluorogenic dye-aptamer module for RNA imaging in live cells. Nat. Chem. Biol. 2020, 16, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Braselmann, E.; Wierzba, A.J.; Polaski, J.T.; Chromiński, M.; Holmes, Z.E.; Hung, S.-T.; Batan, D.; Wheeler, J.R.; Parker, R.; Jimenez, R.; et al. A multicolor riboswitch-based platform for imaging of RNA in live mammalian cells. Nat. Chem. Biol. 2018, 14, 964–971. [Google Scholar] [CrossRef]
- Shao, W.; Zeng, S.-T.; Yu, Z.-Y.; Tang, G.-X.; Chen, S.-B.; Huang, Z.-S.; Chen, X.-C.; Tan, J.-H. Tracking Stress Granule Dynamics in Live Cells and In Vivo with a Small Molecule. Anal. Chem. 2021, 93, 16297–16301. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Kedersha, N. RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 2009, 10, 430–436. [Google Scholar] [CrossRef]
- Kedersha, N.; Stoecklin, G.; Ayodele, M.; Yacono, P.; Lykke-Andersen, J.; Fritzler, M.J.; Scheuner, D.; Kaufman, R.J.; Golan, D.E.; Anderson, P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005, 169, 871–884. [Google Scholar] [CrossRef]




| Compound | λabs (nm) | λem (nm) | LOD (ng/mL) | ΦFa (%) | ||
|---|---|---|---|---|---|---|
| Buffer | DNA | RNA | ||||
| QUID−1 | 478 | 560 | 26.5 | 0.45 | 6.58 | 16.55 |
| QUID−2 | 475 | 555 | 1.8 | 0.61 | 8.64 | 36.37 |
| QUID−3 | 475 | 556 | 24.8 | 0.90 | 10.83 | 18.61 |
| QUID−4 | 475 | 558 | 44.6 | 0.17 | 2.51 | 15.80 |
| QUID−5 | 474 | 556 | 65.0 | 0.63 | 5.25 | 15.31 |
| QUID−6 | 470 | 549 | 10.7 | 1.71 | 19.33 | 36.63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, L.; Shao, W.; Zeng, S.-T.; Tang, G.-X.; Yan, J.-T.; Chen, S.-B.; Huang, Z.-S.; Tan, J.-H.; Chen, X.-C. Development of a Highly Selective and Sensitive Fluorescent Probe for Imaging RNA Dynamics in Live Cells. Molecules 2022, 27, 6927. https://doi.org/10.3390/molecules27206927
Fang L, Shao W, Zeng S-T, Tang G-X, Yan J-T, Chen S-B, Huang Z-S, Tan J-H, Chen X-C. Development of a Highly Selective and Sensitive Fluorescent Probe for Imaging RNA Dynamics in Live Cells. Molecules. 2022; 27(20):6927. https://doi.org/10.3390/molecules27206927
Chicago/Turabian StyleFang, Lan, Wen Shao, Shu-Tang Zeng, Gui-Xue Tang, Jia-Tong Yan, Shuo-Bin Chen, Zhi-Shu Huang, Jia-Heng Tan, and Xiu-Cai Chen. 2022. "Development of a Highly Selective and Sensitive Fluorescent Probe for Imaging RNA Dynamics in Live Cells" Molecules 27, no. 20: 6927. https://doi.org/10.3390/molecules27206927
APA StyleFang, L., Shao, W., Zeng, S.-T., Tang, G.-X., Yan, J.-T., Chen, S.-B., Huang, Z.-S., Tan, J.-H., & Chen, X.-C. (2022). Development of a Highly Selective and Sensitive Fluorescent Probe for Imaging RNA Dynamics in Live Cells. Molecules, 27(20), 6927. https://doi.org/10.3390/molecules27206927







