A Review on Data Fusion of Multidimensional Medical and Biomedical Data
Abstract
:1. Introduction
2. Overview of Data Types
2.1. Ultrasonography
2.2. Single Photon Emission Computed Tomography (SPECT)
2.3. Positron Emission Tomography (PET)
2.4. Computed Tomography (CT)
2.5. White Light Microscopy
2.6. Macroscopic Imaging
2.7. Mammography
2.8. Magnetic Resonance Imaging (MRI)
2.9. MALDI-IMS
2.10. Fluorescence Lifetime Imaging Microscopy (FLIM)
2.11. Vibrational Spectroscopy
2.12. Biomarkers
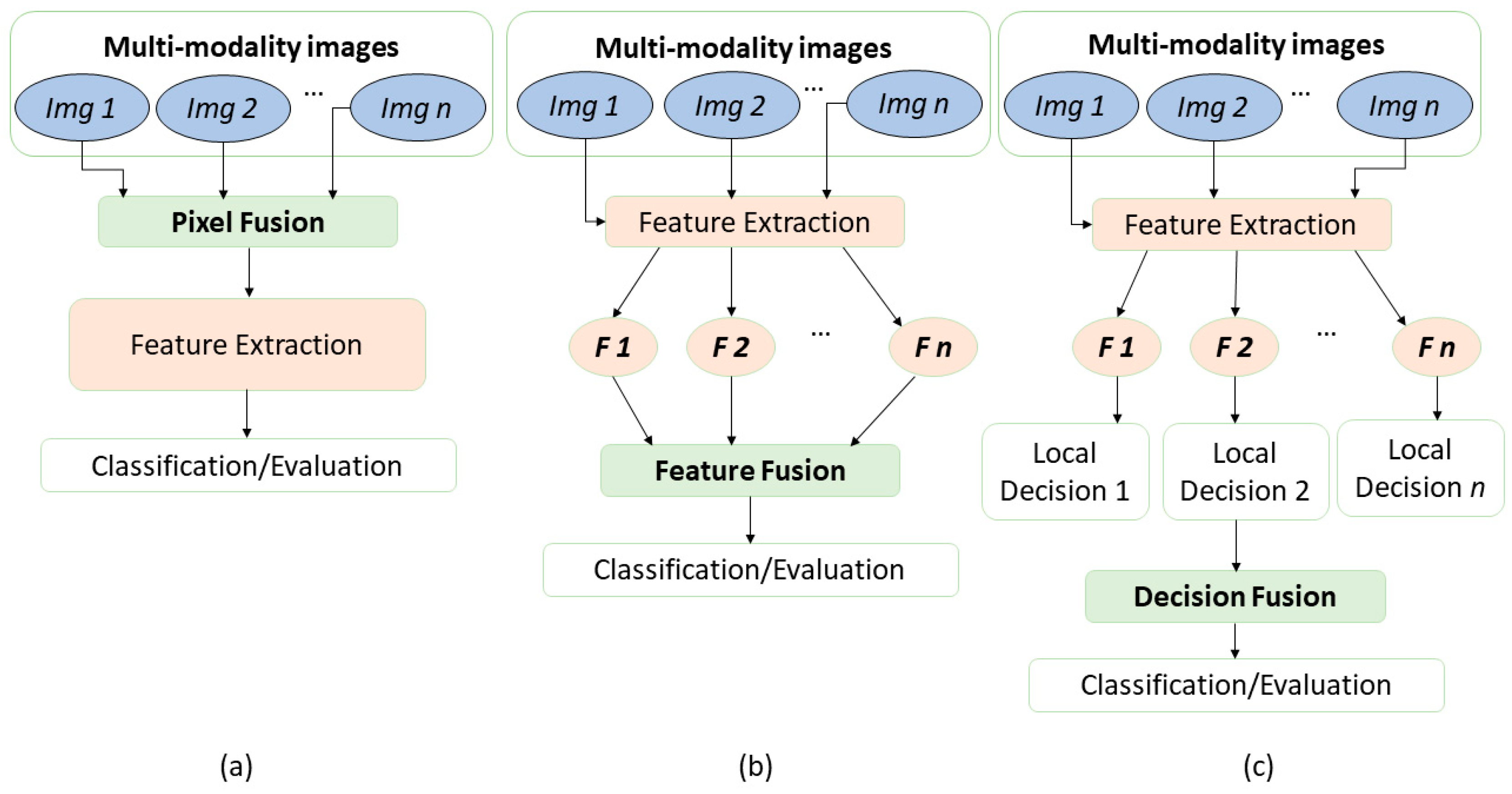
3. Different Data Fusion Techniques
3.1. Data Fusion of Image Data
3.2. Deep Learning in Data Fusion
3.3. Data Fusion of Image and Biomarker Data
3.4. Data Fusion of Spectra Data
3.5. Data Fusion of Spectra and Biomarker Data
4. Summary and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Wang, S.; Celebi, M.E.; Zhang, Y.-D.; Yu, X.; Lu, S.; Yao, X.; Zhou, Q.; Miguel, M.-G.; Tian, Y.; Gorriz, J.M. Advances in data preprocessing for biomedical data fusion: An overview of the methods, challenges, and prospects. Inf. Fusion 2021, 76, 376–421. [Google Scholar] [CrossRef]
- Devanna, H.; Satish Kumar, G.A.E.; Giri Prasad, M.N. A Survey on Multimodal Medical Image Fusion. IOSR J. Comput. Eng. (IOSR-JCE) 2017, 19, 7–14. [Google Scholar] [CrossRef]
- Hermessi, H.; Mourali, O.; Zagrouba, E. Multimodal medical image fusion review: Theoretical background and recent advances. Signal Process. 2021, 183, 108036. [Google Scholar] [CrossRef]
- James, A.P.; Dasarathy, B.V. Medical image fusion: A survey of the state of the art. Inf. Fusion 2014, 19, 4–19. [Google Scholar] [CrossRef] [Green Version]
- Willmann, J.K.; Van Bruggen, N.; Dinkelborg, L.M.; Gambhir, S.S. Molecular imaging in drug development. Nat. Rev. Drug Discov. 2008, 7, 591–607. [Google Scholar] [CrossRef]
- Baker, A.C. Nonlinear effects in ultrasound propagation. In Ultrasound in Medicine; Institute of Physics Publishing: Bristol, UK, 1998; pp. 23–38. [Google Scholar]
- Attlas, N.; Gupta, S. Reduction of speckle noise in ultrasound images using various filtering techniques and discrete wavelet transform: Comparative analysis. Int. J. Res. 2014, 1, 112–117. [Google Scholar]
- Cowan, R.L.; Kessler, R. SPECT Imaging. In Encyclopedia of Psychopharmacology; Stolerman, I.P., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1266–1270. [Google Scholar] [CrossRef]
- Barillot, C.; Lemoine, D.; Le Briquer, L.; Lachmann, F.; Gibaud, B. Data fusion in medical imaging: Merging multimodal and multipatient images, identification of structures and 3D display aspects. Eur. J. Radiol. 1993, 17, 22–27. [Google Scholar] [CrossRef]
- Barra, V.; Boire, J.-Y. A general framework for the fusion of anatomical and functional medical images. NeuroImage 2001, 13, 410–424. [Google Scholar] [CrossRef]
- Wason, J.V.; Nagarajan, A. Image processing techniques for analyzing CT scan images towards the early detection of lung cancer. Bioinformation 2019, 15, 596. [Google Scholar]
- Antony, P.P.M.A.; Trefois, C.; Stojanovic, A.; Baumuratov, A.S.; Kozak, K. Light microscopy applications in systems biology: Opportunities and challenges. Cell Commun. Signal. 2013, 11, 24. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.Q.; Sholl, A.B. Applications of structured light microscopy in clinical pathology. In Proceedings of the Microscopy Histopathology and Analytics, Hollywood, FL, USA, 3–6 April 2018; p. MF1A. 1. [Google Scholar]
- Ramezani, M.; Karimian, A.; Moallem, P. Automatic detection of malignant melanoma using macroscopic images. J. Med. Signals Sens. 2014, 4, 281. [Google Scholar] [PubMed]
- Maxim, L.D.; Niebo, R.; Utell, M.J. Screening tests: A review with examples. Inhal. Toxicol. 2014, 26, 811–828. [Google Scholar] [CrossRef] [PubMed]
- Highnam, R.; Brady, J.; Shepstone, B. A Representation for Mammographic Image Processing; IET: London, UK, 1996. [Google Scholar]
- Heywang-Köbrunner, S.H.; Hacker, A.; Sedlacek, S. Advantages and disadvantages of mammography screening. Breast Care 2011, 6, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Harish, S.; Ahammed, G.A.; Banu, R. An extensive research survey on brain MRI enhancement, segmentation and classification. In Proceedings of the 2017 International Conference on Electrical, Electronics, Communication, Computer, and Optimization Techniques (ICEECCOT), Mysuru, India, 15–16 December 2017; pp. 1–8. [Google Scholar]
- Basu, S.S.; Regan, M.S.; Randall, E.C.; Abdelmoula, W.M.; Clark, A.R.; Gimenez-Cassina Lopez, B.; Cornett, D.S.; Haase, A.; Santagata, S.; Agar, N.Y. Rapid MALDI mass spectrometry imaging for surgical pathology. NPJ Precis. Oncol. 2019, 3, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [Green Version]
- Aichler, M.; Walch, A. MALDI Imaging mass spectrometry: Current frontiers and perspectives in pathology research and practice. Lab. Investig. 2015, 95, 422–431. [Google Scholar] [CrossRef] [Green Version]
- Tomasila, G.; Emanuel, A.W.R. MRI image processing method on brain tumors: A review. AIP Conf. Proc. 2020, 2296, 020023. [Google Scholar]
- Liu, L.; Yang, Q.; Zhang, M.; Wu, Z.; Xue, P. Fluorescence lifetime imaging microscopy and its applications in skin cancer diagnosis. J. Innov. Opt. Health Sci. 2019, 12, 1930004. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zheng, Y.; Zhao, D.; Zhao, Z.; Liu, L.; Pliss, A.; Zhu, F.; Liu, J.; Qu, J.; Luan, P. Applications of fluorescence lifetime imaging in clinical medicine. J. Innov. Opt. Health Sci. 2018, 11, 1830001. [Google Scholar] [CrossRef] [Green Version]
- Datta, R.; Heaster, T.M.; Sharick, J.T.; Gillette, A.A.; Skala, M.C. Fluorescence lifetime imaging microscopy: Fundamentals and advances in instrumentation, analysis, and applications. J. Biomed. Opt. 2020, 25, 071203. [Google Scholar] [CrossRef]
- Guo, S.; Bocklitz, T.; Popp, J. Optimization of Raman-spectrum baseline correction in biological application. Analyst 2016, 141, 2396–2404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramoji, A.; Neugebauer, U.; Bocklitz, T.; Foerster, M.; Kiehntopf, M.; Bauer, M.; Popp, J.R. Toward a spectroscopic hemogram: Raman spectroscopic differentiation of the two most abundant leukocytes from peripheral blood. Anal. Chem. 2012, 84, 5335–5342. [Google Scholar] [CrossRef] [PubMed]
- Mollaoglu, A.D.; Ozyurt, I.; Severcan, F. Applications of infrared spectroscopy and microscopy in diagnosis of obesity. In Infrared Spectroscopy-Principles, Advances, and Applications; IntechOpen: London, UK, 2018. [Google Scholar]
- Franck, P.; Nabet, P.; Dousset, B. Applications of infrared spectroscopy to medical biology. Cell. Mol. Biol. 1998, 44, 273–275. [Google Scholar] [PubMed]
- Matthäus, C.; Bird, B.; Miljković, M.; Chernenko, T.; Romeo, M.; Diem, M. Infrared and Raman microscopy in cell biology. Methods Cell Biol. 2008, 89, 275–308. [Google Scholar]
- Pahlow, S.; Weber, K.; Popp, J.; Bayden, R.W.; Kochan, K.; Rüther, A.; Perez-Guaita, D.; Heraud, P.; Stone, N.; Dudgeon, A. Application of vibrational spectroscopy and imaging to point-of-care medicine: A review. Appl. Spectrosc. 2018, 72, 52–84. [Google Scholar] [CrossRef]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463. [Google Scholar] [CrossRef]
- Blanchet, L.; Smolinska, A.; Attali, A.; Stoop, M.P.; Ampt, K.A.; van Aken, H.; Suidgeest, E.; Tuinstra, T.; Wijmenga, S.S.; Luider, T. Fusion of metabolomics and proteomics data for biomarkers discovery: Case study on the experimental autoimmune encephalomyelitis. BMC Bioinform. 2011, 12, 254. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Cortes, C.; Burges, C.J. The MNIST Database of Handwritten Digits. 1998. Available online: http://yann.lecun.com/exdb/mnist/ (accessed on 20 October 2022).
- Kaur, H.; Koundal, D.; Kadyan, V. Image fusion techniques: A survey. Arch. Comput. Methods Eng. 2021, 28, 4425–4447. [Google Scholar] [CrossRef]
- Yang, L.; Guo, B.; Ni, W. Multimodality medical image fusion based on multiscale geometric analysis of contourlet transform. Neurocomputing 2008, 72, 203–211. [Google Scholar] [CrossRef]
- Kelly, K.M.; Dean, J.; Comulada, W.S.; Lee, S.-J. Breast cancer detection using automated whole breast ultrasound and mammography in radiographically dense breasts. Eur. Radiol. 2010, 20, 734–742. [Google Scholar] [CrossRef] [Green Version]
- Mishra, H.O.S.; Bhatnagar, S. MRI and CT image fusion based on wavelet transform. Int. J. Inf. Comput. Technol. 2014, 4, 47–52. [Google Scholar]
- Muzammil, S.R.; Maqsood, S.; Haider, S.; Damaševičius, R. CSID: A novel multimodal image fusion algorithm for enhanced clinical diagnosis. Diagnostics 2020, 10, 904. [Google Scholar] [CrossRef] [PubMed]
- Tamilselvan, K.S.; Murugesan, G. Survey and analysis of various image fusion techniques for clinical CT and MRI images. Int. J. Imaging Syst. Technol. 2014, 24, 193–202. [Google Scholar] [CrossRef]
- Rajkumar, S.; Kavitha, S. Redundancy discrete wavelet transform and contourlet transform for multimodality medical image fusion with quantitative analysis. In Proceedings of the 2010 3rd International Conference on Emerging Trends in Engineering and Technology, Goa, India, 19–21 November 2010; pp. 134–139. [Google Scholar]
- Agrawal, M.; Tsakalides, P.; Achim, A. Medical image fusion using the convolution of meridian distributions. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina, 31 August–4 September 2010; pp. 3727–3730. [Google Scholar]
- Nandeesh, M.; Meenakshi, M. Image fusion algorithms for medical images-a comparison. Bonfring Int. J. Adv. Image Process. 2015, 5, 23. [Google Scholar] [CrossRef]
- Kavitha, C.; Chellamuthu, C. Multimodal medical image fusion based on integer wavelet transform and neuro-fuzzy. In Proceedings of the 2010 International Conference on Signal and Image Processing, Chennai, India, 15–17 December 2010; pp. 296–300. [Google Scholar]
- Tan, W.; Tiwari, P.; Pandey, H.M.; Moreira, C.; Jaiswal, A.K. Multimodal medical image fusion algorithm in the era of big data. Neural Comput. Appl. 2020, 1–21. [Google Scholar] [CrossRef]
- Singh, R.; Vatsa, M.; Noore, A. Multimodal medical image fusion using redundant discrete wavelet transform. In Proceedings of the 2009 Seventh International Conference on Advances in Pattern Recognition, Kolkata, India, 4–6 February 2009; pp. 232–235. [Google Scholar]
- Adali, T.; Levin-Schwartz, Y.; Calhoun, V.D. Multimodal data fusion using source separation: Application to medical imaging. Proc. IEEE 2015, 103, 1494–1506. [Google Scholar] [CrossRef]
- Bedia, C.; Sierra, À.; Tauler, R. Application of chemometric methods to the analysis of multimodal chemical images of biological tissues. Anal. Bioanal. Chem. 2020, 412, 5179–5190. [Google Scholar] [CrossRef]
- Piqueras, S.; Bedia, C.; Beleites, C.; Krafft, C.; Popp, J.; Maeder, M.; Tauler, R.; de Juan, A. Handling different spatial resolutions in image fusion by multivariate curve resolution-alternating least squares for incomplete image multisets. Anal. Chem. 2018, 90, 6757–6765. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, X.; Ward, R.K.; Wang, Z.J. Medical image fusion via convolutional sparsity based morphological component analysis. IEEE Signal Process. Lett. 2019, 26, 485–489. [Google Scholar] [CrossRef]
- Van de Plas, R.; Yang, J.; Spraggins, J.; Caprioli, R.M. Image fusion of mass spectrometry and microscopy: A multimodality paradigm for molecular tissue mapping. Nat. Methods 2015, 12, 366–372. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.-Q.; Xing, H. Multi-mode medical image fusion algorithm based on principal component analysis. In Proceedings of the 2009 International Symposium on Computer Network and Multimedia Technology, Wuhan, China, 18–20 January 2009; pp. 1–4. [Google Scholar]
- Zhang, Y.; Guo, C.; Zhao, P. Medical Image Fusion Based on Low-Level Features. Comput. Math. Methods Med. 2021, 2021, 8798003. [Google Scholar] [CrossRef] [PubMed]
- Vitor, T.; Martins, K.M.; Ionescu, T.M.; Cunha, M.L.D.; Baroni, R.H.; Garcia, M.R.T.; Wagner, J.; Campos, G.D.C.; Nogueira, S.A.; Guerra, E.G. PET/MRI: A novel hybrid imaging technique. Major clinical indications and preliminary experience in Brazil. Einstein 2017, 15, 115–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandulescu, D.L.; Dumitrescu, D.; Rogoveanu, I.; Saftoiu, A. Hybrid ultrasound imaging techniques (fusion imaging). World J. Gastroenterol. 2011, 17, 49. [Google Scholar] [CrossRef] [PubMed]
- Cal-Gonzalez, J.; Rausch, I.; Shiyam Sundar, L.K.; Lassen, M.L.; Muzik, O.; Moser, E.; Papp, L.; Beyer, T. Hybrid imaging: Instrumentation and data processing. Front. Phys. 2018, 6, 47. [Google Scholar] [CrossRef]
- Gao, J.; Li, P.; Chen, Z.; Zhang, J. A survey on deep learning for multimodal data fusion. Neural Comput. 2020, 32, 829–864. [Google Scholar] [CrossRef]
- Huang, S.-C.; Pareek, A.; Seyyedi, S.; Banerjee, I.; Lungren, M.P. Fusion of medical imaging and electronic health records using deep learning: A systematic review and implementation guidelines. NPJ Digit. Med. 2020, 3, 136. [Google Scholar] [CrossRef]
- Rajalingam, B.; Priya, R. Multimodal medical image fusion based on deep learning neural network for clinical treatment analysis. Int. J. ChemTech Res. 2018, 11, 160–176. [Google Scholar]
- Guo, K.; Li, X.; Hu, X.; Liu, J.; Fan, T. Hahn-PCNN-CNN: An end-to-end multi-modal brain medical image fusion framework useful for clinical diagnosis. BMC Med. Imaging 2021, 21, 111. [Google Scholar] [CrossRef]
- Iqbal, S.; Ghani Khan, M.U.; Saba, T.; Mehmood, Z.; Javaid, N.; Rehman, A.; Abbasi, R. Deep learning model integrating features and novel classifiers fusion for brain tumor segmentation. Microsc. Res. Tech. 2019, 82, 1302–1315. [Google Scholar] [CrossRef]
- Pradhan, P.; Köhler, K.; Guo, S.; Rosin, O.; Popp, J.; Niendorf, A.; Bocklitz, T. Data Fusion of Histological and Immunohistochemical Image Data for Breast Cancer Diagnostics using Transfer Learning. In Proceedings of the ICPRAM, Vienna, Austria, 4–6 February 2021; pp. 495–506. [Google Scholar]
- Zhang, Y.; Liu, H.; Hu, Q. Transfuse: Fusing transformers and cnns for medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; pp. 14–24. [Google Scholar]
- VS, V.; Valanarasu, J.M.J.; Oza, P.; Patel, V.M. Image fusion transformer. arXiv 2021, arXiv:2107.09011. [Google Scholar]
- Wang, T.; Lan, J.; Han, Z.; Hu, Z.; Huang, Y.; Deng, Y.; Zhang, H.; Wang, J.; Chen, M.; Jiang, H. O-Net: A Novel Framework With Deep Fusion of CNN and Transformer for Simultaneous Segmentation and Classification. Front. Neurosci. 2022, 16, 876065. [Google Scholar] [CrossRef]
- Stahlschmidt, S.R.; Ulfenborg, B.; Synnergren, J. Multimodal deep learning for biomedical data fusion: A review. Brief. Bioinform. 2022, 23, bbab569. [Google Scholar] [CrossRef] [PubMed]
- Metsis, V.; Huang, H.; Andronesi, O.C.; Makedon, F.; Tzika, A. Heterogeneous data fusion for brain tumor classification. Oncol. Rep. 2012, 28, 1413–1416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, J.; Liu, J.Y.; Jiang, Y.J.; Sun, X.W.; Zheng, B.; Nie, S.D. Fusion of quantitative imaging features and serum biomarkers to improve performance of computer-aided diagnosis scheme for lung cancer: A preliminary study. Med. Phys. 2018, 45, 5472–5481. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Xue, P.; Li, N.; Zhao, P.; Xu, Z.; Ji, H.; Zhang, Z.; Cui, W.; Dong, E. Fusion of 3D lung CT and serum biomarkers for diagnosis of multiple pathological types on pulmonary nodules. Comput. Methods Programs Biomed. 2021, 210, 106381. [Google Scholar] [CrossRef] [PubMed]
- Neumann, E.K.; Comi, T.J.; Spegazzini, N.; Mitchell, J.W.; Rubakhin, S.S.; Gillette, M.U.; Bhargava, R.; Sweedler, J.V. Multimodal chemical analysis of the brain by high mass resolution mass spectrometry and infrared spectroscopic imaging. Anal. Chem. 2018, 90, 11572–11580. [Google Scholar] [CrossRef] [PubMed]
- Attia, A.S.; Schroeder, K.A.; Seeley, E.H.; Wilson, K.J.; Hammer, N.D.; Colvin, D.C.; Manier, M.L.; Nicklay, J.J.; Rose, K.L.; Gore, J.C. Monitoring the inflammatory response to infection through the integration of MALDI IMS and MRI. Cell Host Microbe 2012, 11, 664–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bocklitz, T.; Bräutigam, K.; Urbanek, A.; Hoffmann, F.; von Eggeling, F.; Ernst, G.; Schmitt, M.; Schubert, U.; Guntinas-Lichius, O.; Popp, J. Novel workflow for combining Raman spectroscopy and MALDI-MSI for tissue based studies. Anal. Bioanal. Chem. 2015, 407, 7865–7873. [Google Scholar] [CrossRef]
- Rabe, J.-H.; Sammour, A.D.; Schulz, S.; Munteanu, B.; Ott, M.; Ochs, K.; Hohenberger, P.; Marx, A.; Platten, M.; Opitz, C.A. Fourier transform infrared microscopy enables guidance of automated mass spectrometry imaging to predefined tissue morphologies. Sci. Rep. 2018, 8, 313. [Google Scholar] [CrossRef] [Green Version]
- Ahlf, D.R.; Masyuko, R.N.; Hummon, A.B.; Bohn, P.W. Correlated mass spectrometry imaging and confocal Raman microscopy for studies of three-dimensional cell culture sections. Analyst 2014, 139, 4578–4585. [Google Scholar] [CrossRef]
- Caspers, P.J.; Lucassen, G.W.; Puppels, G.J. Combined in vivo confocal Raman spectroscopy and confocal microscopy of human skin. Biophys. J. 2003, 85, 572–580. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Du, G.; Tong, D.; Lv, G.; Lv, X.; Si, R.; Tang, J.; Li, H.; Ma, H.; Mo, J. Exploration research on the fusion of multimodal spectrum technology to improve performance of rapid diagnosis scheme for thyroid dysfunction. J. Biophotonics 2020, 13, e201900099. [Google Scholar] [CrossRef] [PubMed]
- Rangaraju, L.P.; Kunapuli, G.; Every, D.; Ayala, O.D.; Ganapathy, P.; Mahadevan-Jansen, A. Classification of burn injury using Raman spectroscopy and optical coherence tomography: An ex-vivo study on porcine skin. Burns 2019, 45, 659–670. [Google Scholar] [CrossRef]
- Placzek, F.; Bautista, E.C.; Kretschmer, S.; Wurster, L.M.; Knorr, F.; González-Cerdas, G.; Erkkilä, M.T.; Stein, P.; Ataman, Ç.; Hermann, G.G. Morpho-molecular ex vivo detection and grading of non-muscle-invasive bladder cancer using forward imaging probe based multimodal optical coherence tomography and Raman spectroscopy. Analyst 2020, 145, 1445–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schie, I.W.; Stiebing, C.; Popp, J. Looking for a perfect match: Multimodal combinations of Raman spectroscopy for biomedical applications. J. Biomed. Opt. 2021, 26, 080601. [Google Scholar] [CrossRef] [PubMed]
- Acar, E.; Bro, R.; Smilde, A.K. Data fusion in metabolomics using coupled matrix and tensor factorizations. Proc. IEEE 2015, 103, 1602–1620. [Google Scholar] [CrossRef]
- Viswanath, S.E.; Tiwari, P.; Lee, G.; Madabhushi, A. Dimensionality reduction-based fusion approaches for imaging and non-imaging biomedical data: Concepts, workflow, and use-cases. BMC Med. Imaging 2017, 17, 2. [Google Scholar] [CrossRef] [Green Version]
- Porta Siegel, T.; Hamm, G.; Bunch, J.; Cappell, J.; Fletcher, J.S.; Schwamborn, K. Mass spectrometry imaging and integration with other imaging modalities for greater molecular understanding of biological tissues. Mol. Imaging Biol. 2018, 20, 888–901. [Google Scholar] [CrossRef] [Green Version]
- Bocklitz, T.; Crecelius, A.; Matthaus, C.; Tarcea, N.; Von Eggeling, F.; Schmitt, M.; Schubert, U.; Popp, J. Deeper understanding of biological tissue: Quantitative correlation of MALDI-TOF and Raman imaging. Anal. Chem. 2013, 85, 10829–10834. [Google Scholar] [CrossRef]
- Ryabchykov, O.; Popp, J.; Bocklitz, T. Fusion of MALDI spectrometric imaging and Raman spectroscopic data for the analysis of biological samples. Front. Chem. 2018, 6, 257. [Google Scholar] [CrossRef] [Green Version]
- Ramoji, A.; Thomas-Rüddel, D.; Ryabchykov, O.; Bauer, M.; Arend, N.; Giamarellos-Bourboulis, E.J.; Eugen-Olsen, J.; Kiehntopf, M.; Bocklitz, T.; Popp, J. Leukocyte Activation Profile Assessed by Raman Spectroscopy Helps Diagnosing Infection and Sepsis. Crit. Care Explor. 2021, 3, e0394. [Google Scholar] [CrossRef] [PubMed]
- Bro, R.; Nielsen, H.J.; Savorani, F.; Kjeldahl, K.; Christensen, I.J.; Brünner, N.; Lawaetz, A.J. Data fusion in metabolomic cancer diagnostics. Metabolomics 2013, 9, 3–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, N.; Girnus, S.; Rösch, P.; Popp, J.R.; Bocklitz, T. Sample-size planning for multivariate data: A Raman-spectroscopy-based example. Anal. Chem. 2018, 90, 12485–12492. [Google Scholar] [CrossRef] [PubMed]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azam, K.S.F.; Ryabchykov, O.; Bocklitz, T. A Review on Data Fusion of Multidimensional Medical and Biomedical Data. Molecules 2022, 27, 7448. https://doi.org/10.3390/molecules27217448
Azam KSF, Ryabchykov O, Bocklitz T. A Review on Data Fusion of Multidimensional Medical and Biomedical Data. Molecules. 2022; 27(21):7448. https://doi.org/10.3390/molecules27217448
Chicago/Turabian StyleAzam, Kazi Sultana Farhana, Oleg Ryabchykov, and Thomas Bocklitz. 2022. "A Review on Data Fusion of Multidimensional Medical and Biomedical Data" Molecules 27, no. 21: 7448. https://doi.org/10.3390/molecules27217448







