Small Heterocyclic Ligands as Anticancer Agents: QSAR with a Model G-Quadruplex
Abstract
1. Introduction
2. Results
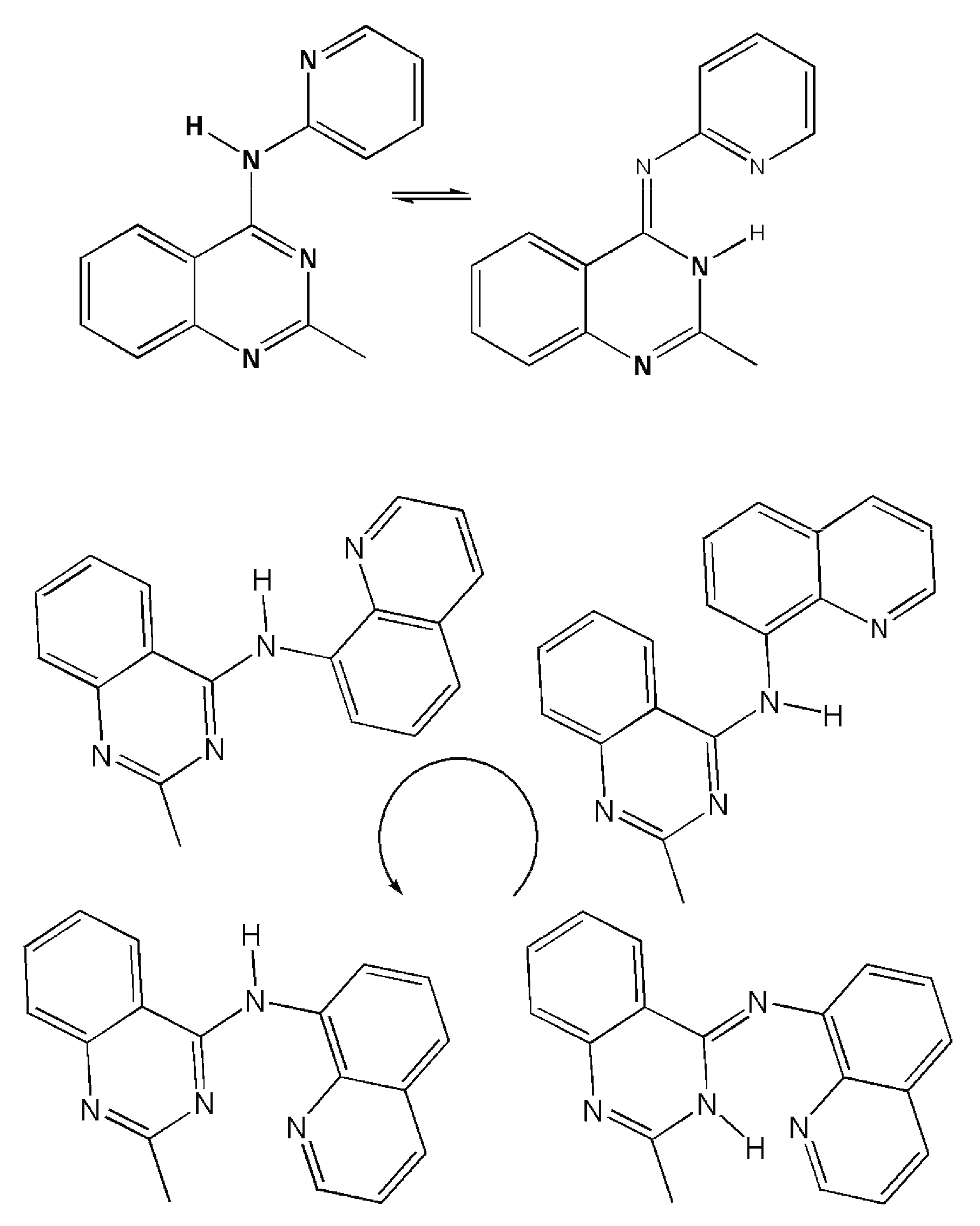
2.1. Synthesis
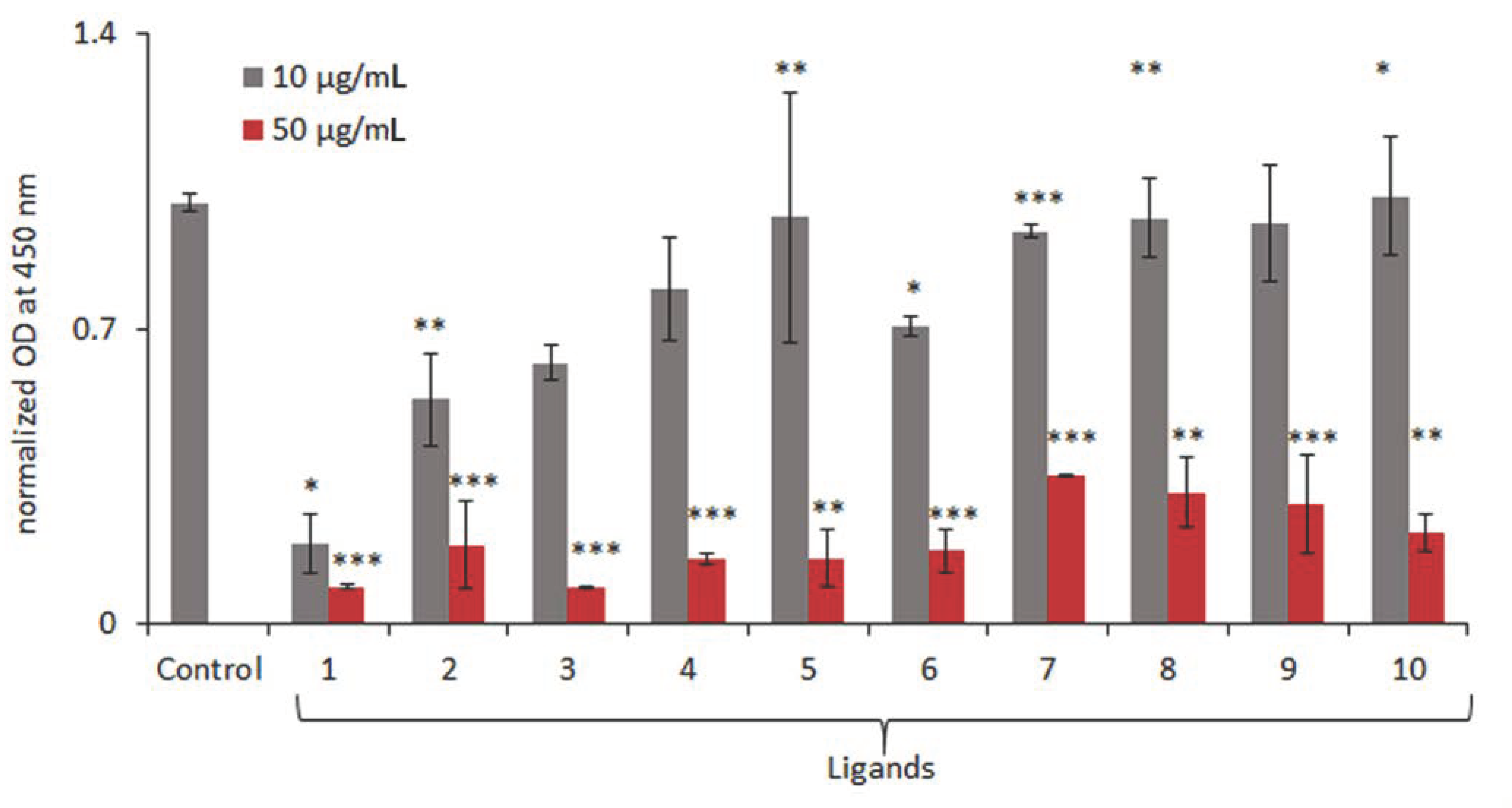
2.2. Biological Evaluation of the Effect of the Studied Heterocyclic Ligands
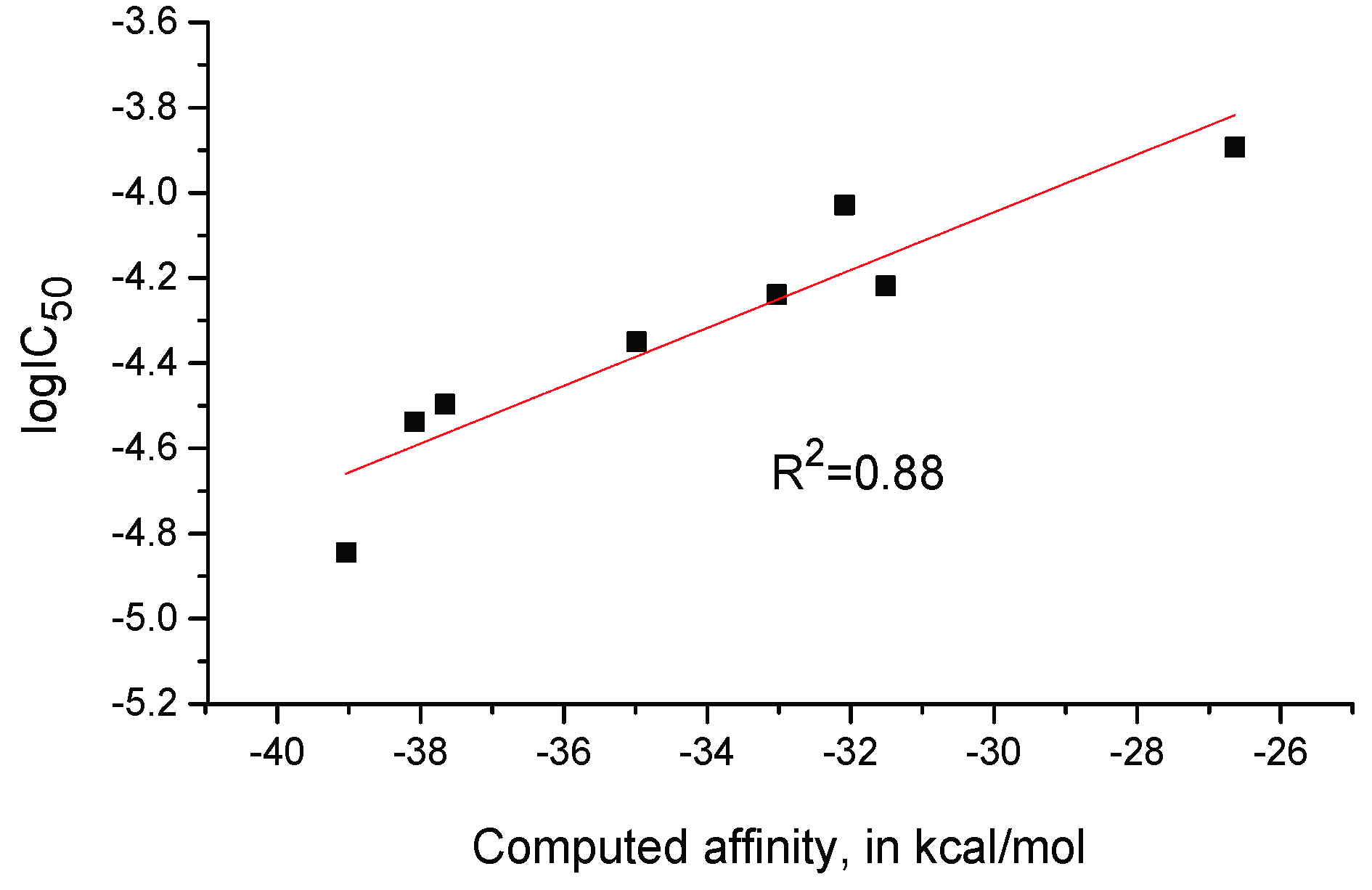
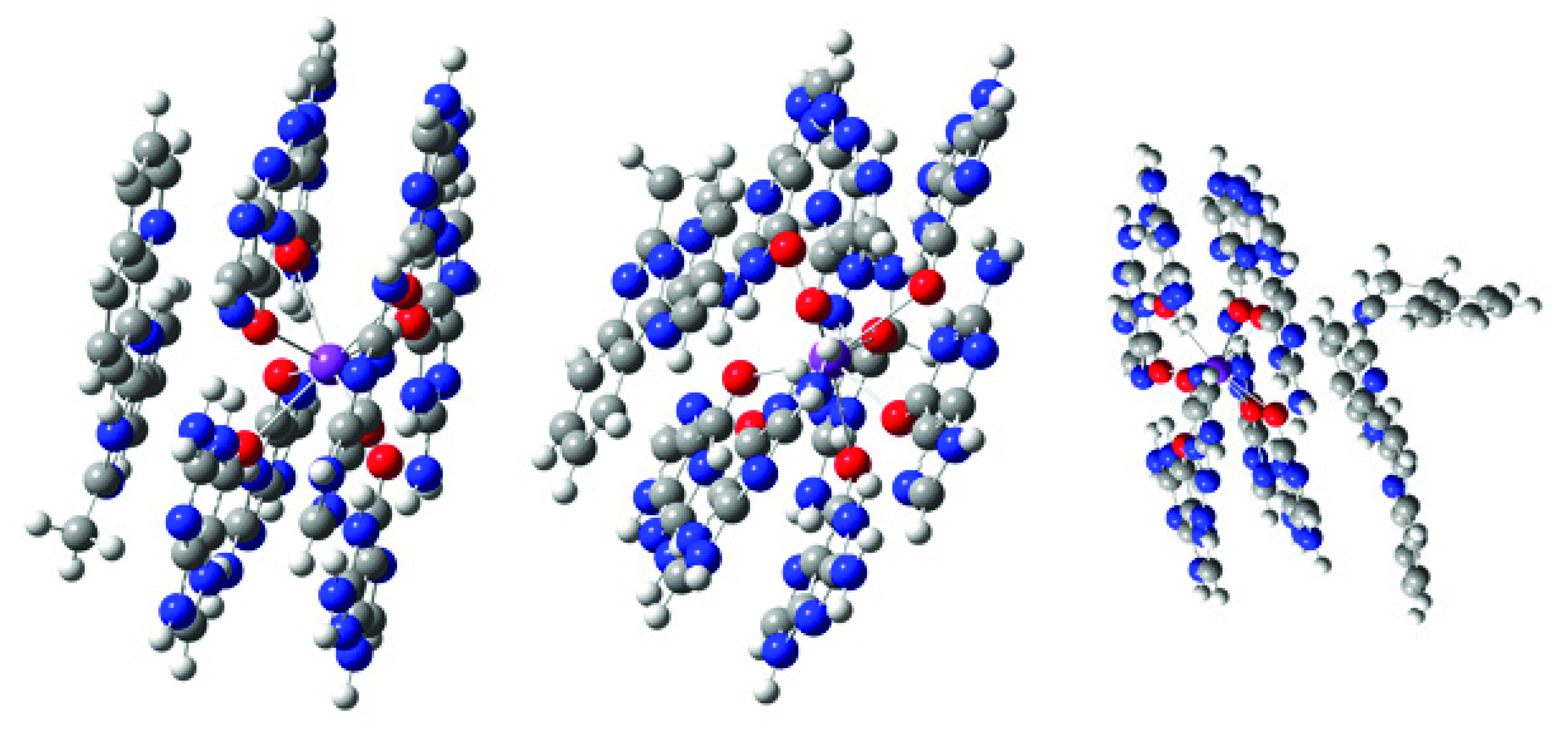
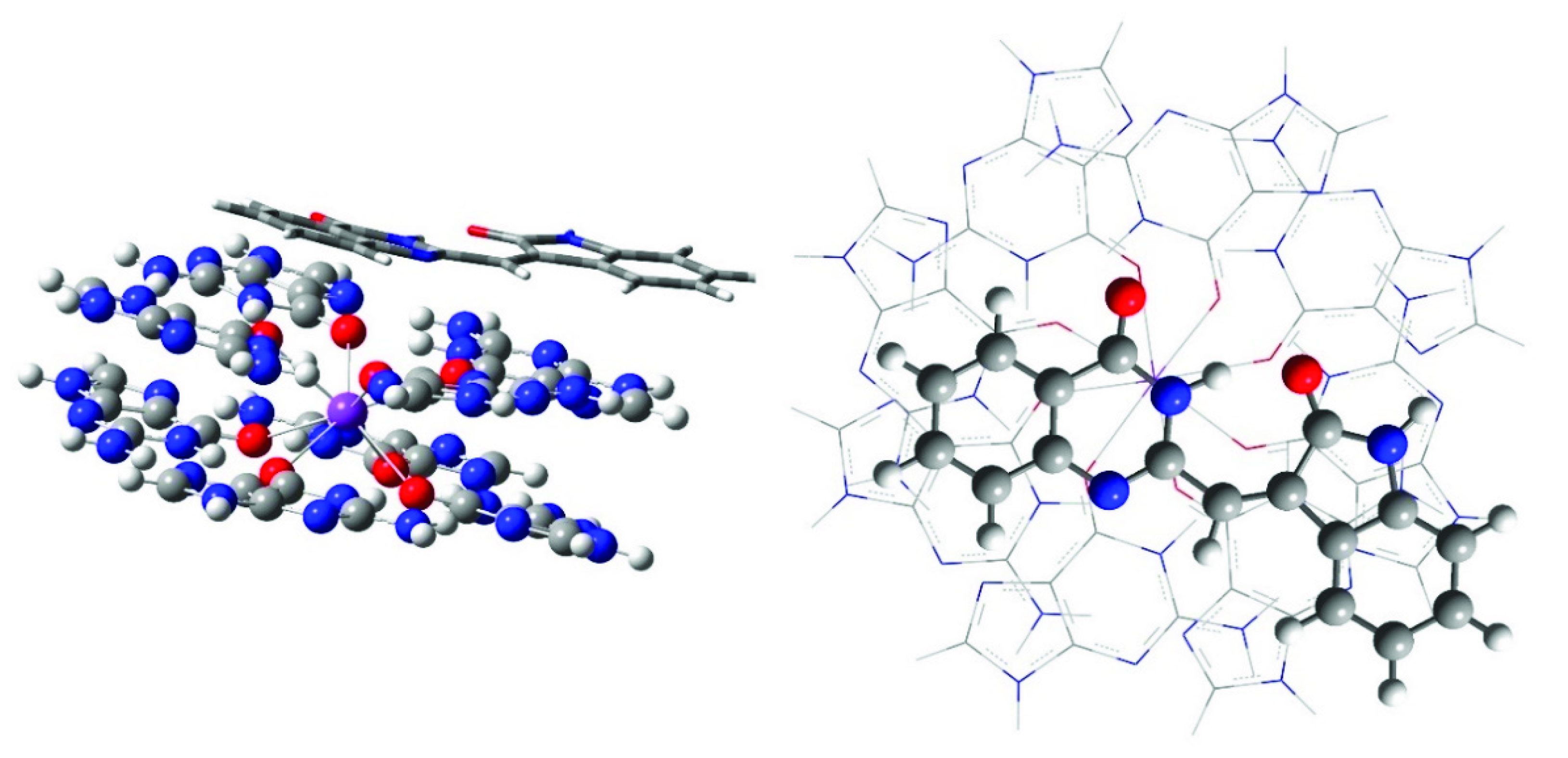
2.3. Computational Modeling
3. Discussion
4. Materials and Methods
4.1. Synthesis
4.1.1. Synthesis of 4-Chloroquinazolines
4.1.2. Synthesis of 4-Aminoquinazolines
4.1.3. Synthesis of Perimidines
4.2. Biological Experiments
4.2.1. Cells and Cell Culture
4.2.2. WST-1 Cell Proliferation Assay
4.3. Computational Details
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Che, T.; Wang, Y.-Q.; Huang, Z.-L.; Tan, J.-H.; Huang, Z.-S.; Chen, S.-B. Natural alkaloids and heterocycles as G-quadruplex ligands and potential anticancer agents. Molecules 2018, 23, 493. [Google Scholar] [CrossRef] [PubMed]
- Chadeneau, C.; Hay, K.; Hirte, H.W.; Gallinger, S.; Bacchetti, S. Telomerase activity associated with acquisition of malignancy in human colorectal cancer. Cancer Res. 1995, 55, 2533–2536. [Google Scholar]
- Phatak, P.; Cookson, J.C.; Dai, F.; Smith, V.; Gartenhaus, R.B.; Stevens, M.F.; Burger, A.M. Telomere uncapping by the G-quadruplex ligand RHPS4 inhibits clonogenic tumour cell growth in vitro and in vivo consistent with a cancer stem cell targeting mechanism. Brit. J. Cancer 2007, 96, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.N.; Lee, M.P.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef]
- Hu, M.-H.; Chen, S.-B.; Wang, B.; Ou, T.-M.; Gu, L.-Q.; Tan, J.-H.; Huang, Z.-S. Specific targeting of telomeric multimeric G-quadruplexes by a new triaryl-substituted imidazole. Nucleic Acids Res. 2016, 45, 1606–1618. [Google Scholar] [CrossRef]
- Yang, Y.O.; Wang, C.; Zhao, B.; Xiong, H.; Xiao, Z.; Zhang, B.; Zheng, P.; Hu, J.; Gao, Y.; Zhang, M.; et al. Design, synthesis, antiproliferative activity and docking studies of quinazoline derivatives bearing oxazole or imidazole as potential EGFR inhibitors. New J. Chem. 2018, 42, 17203–17215. [Google Scholar] [CrossRef]
- Wang, K.-B.; Elsayed, M.; Wu, G.; Deng, N.; Cushman, M.; Yang, D. Indenoisoquinoline topoisomerase inhibitors strongly bind and stabilize the MYC promoter G-quadruplex and downregulate MYC. J. Am. Chem. Soc. 2019, 141, 11059–11070. [Google Scholar] [CrossRef]
- Neidle, S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009, 19, 239–250. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef]
- Neidle, S. Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 2016, 59, 5987–6011. [Google Scholar] [CrossRef]
- Kosiol, N.; Juranek, S.; Brossart, P.; Heine, A.; Paeschke, K. G-quadruplexes: A promising target for cancer therapy. Mol. Cancer 2021, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Alessandrini, I.; Recagni, M.; Zaffaroni, N.; Folini, M. On the road to fight cancer: The potential of G-quadruplex ligands as novel therapeutic agents. Int. J. Mol. Sci. 2021, 22, 5947. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.; Sun, X.; Li, W.; Hou, G.; Gao, F. 1,2,3-Triazole-containing compounds as anti-lung cancer agents: Current developments, mechanisms of action and structure-activity relationship. Front. Pharmacol. 2021, 12, 661173. [Google Scholar] [CrossRef] [PubMed]
- Andréasson, M.; Bhuma, N.; Pemberton, N.; Chorell, E. Using macrocyclic G-quadruplex ligands to decipher the interactions between small molecules and G-quadruplex DNA. Chem. Eur. J. 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Lazareno, S.; Birdsall, N.J. Estimation of competitive antagonist affinity from functional inhibition curves using the Gaddum, Schild and Cheng-Prusoff equations. Brit. J. Pharmacol. 1993, 109, 1110–1119. [Google Scholar] [CrossRef]
- Kaneti, J.; Georgieva, M.; Rangelov, M.; Philipova, I.; Vasileva, B.; Angelov, I.; Staneva, D.; Miloshev, G.; Bakalova, S. Biological activity of quinazoline analogues and molecular modeling of their interactions with G-quadruplexes. Biochim. Biophys. Acta-Gen. Subj. 2021, 1865, 129773. [Google Scholar] [CrossRef]
- Islam, B.; Stadlbauer, P.; Neidle, S.; Haider, S.; Sponer, J. Can we execute reliable MM-PBSA free energy computations of relative stabilities of different guanine quadruplex folds? J. Phys. Chem. B 2016, 120, 2899–2912. [Google Scholar] [CrossRef]
- Sponer, J.; Mladek, A.; Spackova, N.; Cang, X.; Cheatham III, T.E.; Grimme, S. Relative stability of different DNA guanine quadruplex stem topologies derived using large-scale quantum-chemical computations. J. Am. Chem. Soc. 2013, 135, 9785–9796. [Google Scholar] [CrossRef]
- Hohenberg, P.; Kohn, W. Inhomogeneous electron gas. Phys. Rev. 1964, 136, B864–B871. [Google Scholar] [CrossRef]
- Kohn, W.; Sham, L.J. Self-consistent equations including exchange and correlation effects. Phys. Rev. 1965, 140, A1133–A1138. [Google Scholar] [CrossRef]
- Hehre, W.; Radom, L.; Schleyer, P.V.R.; Pople, J.A. Ab Initio Molecular Orbital Theory; John Wiley and Sons: Hoboken, NY, USA, 1986. [Google Scholar]
- Scarborough, H.C.; Lawers, B.C.; Mikielli, J.L.; Compton, J.L. Pyrrolidines. VI. Synthesis of 4(1-Substituted 3-Pyrrolidinylmethy1amino)- and 4-(1-Substituted 3-Pyrrolidinylmethoxy)Quinazolines. J. Org. Chem. 1962, 27, 957–961. [Google Scholar] [CrossRef]
- Giani, A.M.; Lamperti, M.; Maspero, A.; Cimino, A.; Negri, R.; Giovenzana, G.B.; Palmisano, G.; Nardo, L. Fluorescence Studies on 2-(Het)Aryl Perimidine Derivatives. J. Lumines. 2016, 179, 384–392. [Google Scholar] [CrossRef]
- Che, T.; Chen, S.-B.; Tu, J.-L.; Wang, B.; Wang, Y.-Q.; Zhang, Y.; Wang, J.; Wang, Z.-Q.; Zhang, Z.-P.; Ou, T.-M.; et al. Discovery of novel schizocommunin derivatives as telomeric G-quadruplex ligands that trigger telomere dysfunction and the deoxyribonucleic acid (DNA) damage response. J. Med. Chem. 2018, 61, 3436–3453. [Google Scholar] [CrossRef] [PubMed]
- Lech, C.J.; Heddi, B.; Phan, A.T. Guanine base stacking in G-quadruplex nucleic acids. Nucleic Acids Res. 2013, 41, 2034–2046. [Google Scholar] [CrossRef] [PubMed]
- Machireddy, B.; Sullivan, H.-J.; Wu, C. Binding of BRACO19 to a telomeric G-quadruplex DNA probed by all-atom molecular dynamics simulations with explicit solvent. Molecules 2019, 24, 1010. [Google Scholar] [CrossRef] [PubMed]
- Moshkina, T.N.; Nosova, E.V.; Permyakova, J.V.; Lipunova, G.N.; Valova, M.S.; Slepukhin, P.A.; Sadieva, L.K.; Charushin, V.N. Synthesis and photophysical properties of 2-aryl-4-(morpholin-4-yl)quinazoline chromophores: The effect of π-linker moiety. Dyes Pigm. 2022, 206, 110592. [Google Scholar] [CrossRef]
- Lockman, J.W.; Klimova, Y.; Anderson, M.B.; Willardsen, J.A. Synthesis of substituted quinazolines: Application to the synthesis of verubulin. Synth. Commun. 2012, 42, 1715–1723. [Google Scholar] [CrossRef]
- Kim, S.-H.; Kim, J.-H.; Cui, J.-Z.; Gal, Y.-S.; Jin, S.-H.; Koh, K. Absorption spectra, aggregation and photofading behaviour of near-infrared absorbing squarylium dyes containing perimidine moiety. Dyes Pigm. 2002, 55, 1–7. [Google Scholar] [CrossRef]
- Georgieva, M.; Vasileva, B.; Speranza, G.; Wang, D.; Stoyanov, K.; Draganova-Filipova, M.; Zagorchev, P.; Sarafian, V.; Miloshev, G.; Krasteva, N. Amination of graphene oxide leads to increased cytotoxicity in hepatocellular carcinoma cells. Int. J. Mol. Sci. 2020, 21, 2427. [Google Scholar] [CrossRef]
- Krasteva, N.; Staneva, D.; Vasileva, B.; Miloshev, G.; Georgieva, M. Bioactivity of PEGylated graphene oxide nanoparticles combined with near-infrared laser irradiation studied in colorectal carcinoma cells. Nanomaterials 2021, 11, 3061. [Google Scholar] [CrossRef]
- Grimme, S. Improved second-order Møller–Plesset perturbation theory by separate scaling of parallel- and antiparallel-spin pair correlation energies. J. Chem. Phys. 2003, 118, 9095–9102. [Google Scholar] [CrossRef]
- Chai, J.-D.; Head-Gordon, M. Long-range corrected hybrid density functionals with damped atom–atom dispersion corrections. Phys. Chem. Chem. Phys. 2008, 10, 6615–6620. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2019. [Google Scholar]
- Schmidt, M.W.; Baldridge, K.K.; Boatz, J.A.; Elbert, S.T.; Gordon, M.S.; Jensen, J.H.; Koseki, S.; Matsunaga, N.; Nguyen, K.A.; Su, S.J.; et al. General atomic and molecular electronic structure system. J. Comput. Chem. 1993, 14, 1347–1363. [Google Scholar] [CrossRef]
- Feyereisen, M.; Fitzgerald, G.; Komornicki, A. Use of approximate integrals in ab initio theory. An application in MP2 energy calculations. Chem. Phys. Lett. 1993, 208, 359–363. [Google Scholar] [CrossRef]
- Drygin, D.; Siddiqui-Jain, A.; O’Brien, S.; Schwaebe, M.; Lin, A.; Bliesath, J.; Ho, C.B.; Proffitt, C.; Trent, K.; Whitten, J.P.; et al. Anticancer activity of CX-3543: A direct inhibitor of rRNA biogenesis. Cancer Res. 2009, 69, 7653–7661. [Google Scholar] [CrossRef]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent progress of targeted G-quadruplex-preferred ligands toward cancer therapy. Molecules 2019, 24, 429. [Google Scholar] [CrossRef]
| No | Formula | IC50 | Affinity (DFT) | Affinity (RI-MP2) |
|---|---|---|---|---|
| 1 | 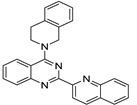 | 1.43 × 10−5 | −39.04 | −28.76 |
| 2 | 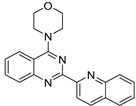 | 2.90 × 10−5 | −38.08 | −41.79 |
| 3 | 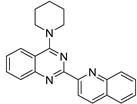 | 3.19 × 10−5 | −37.63 | −44.42 |
| 4 | 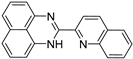 | 4.47 × 10−5 | −35.02 | −41.04 |
| 5 | 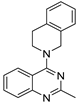 | 5.77 × 10−5 | −33.03 | −33.44 |
| 6 | 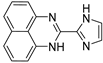 | 6.05 × 10−5 | −31.51 | −35.71 |
| 7 | 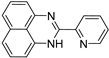 | 9.37 × 10−5 | −32.08 | −38.74 |
| 8 | 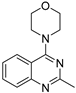 | 1.28 × 10−4 | −26.64 | −26.76 |
| 9 | 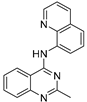 | 9.78 × 10−5 | −35.23 | −45.02 |
| 10 | 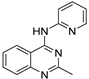 | 1.61 × 10−4 | −31.29 | −37.96 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaneti, J.; Kurteva, V.; Georgieva, M.; Krasteva, N.; Miloshev, G.; Tabakova, N.; Petkova, Z.; Bakalova, S.M. Small Heterocyclic Ligands as Anticancer Agents: QSAR with a Model G-Quadruplex. Molecules 2022, 27, 7577. https://doi.org/10.3390/molecules27217577
Kaneti J, Kurteva V, Georgieva M, Krasteva N, Miloshev G, Tabakova N, Petkova Z, Bakalova SM. Small Heterocyclic Ligands as Anticancer Agents: QSAR with a Model G-Quadruplex. Molecules. 2022; 27(21):7577. https://doi.org/10.3390/molecules27217577
Chicago/Turabian StyleKaneti, Jose, Vanya Kurteva, Milena Georgieva, Natalia Krasteva, George Miloshev, Nadezhda Tabakova, Zhanina Petkova, and Snezhana M. Bakalova. 2022. "Small Heterocyclic Ligands as Anticancer Agents: QSAR with a Model G-Quadruplex" Molecules 27, no. 21: 7577. https://doi.org/10.3390/molecules27217577
APA StyleKaneti, J., Kurteva, V., Georgieva, M., Krasteva, N., Miloshev, G., Tabakova, N., Petkova, Z., & Bakalova, S. M. (2022). Small Heterocyclic Ligands as Anticancer Agents: QSAR with a Model G-Quadruplex. Molecules, 27(21), 7577. https://doi.org/10.3390/molecules27217577









