Separation and Structural Characterization of a Novel Exopolysaccharide from Rhizopus nigricans
Abstract
:1. Introduction
2. Results and Discussion
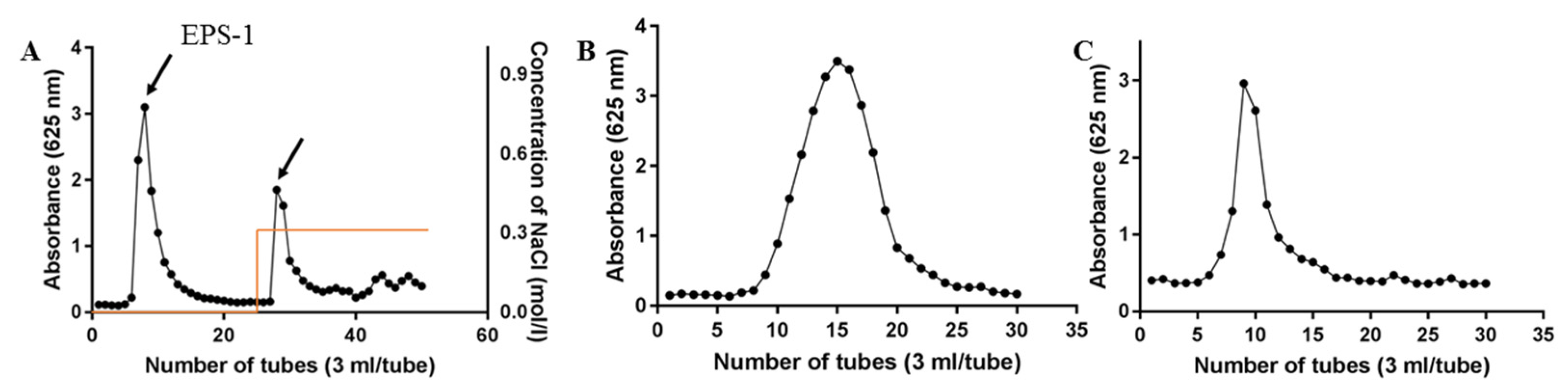
2.1. Isolation and Purification of EPS2-1
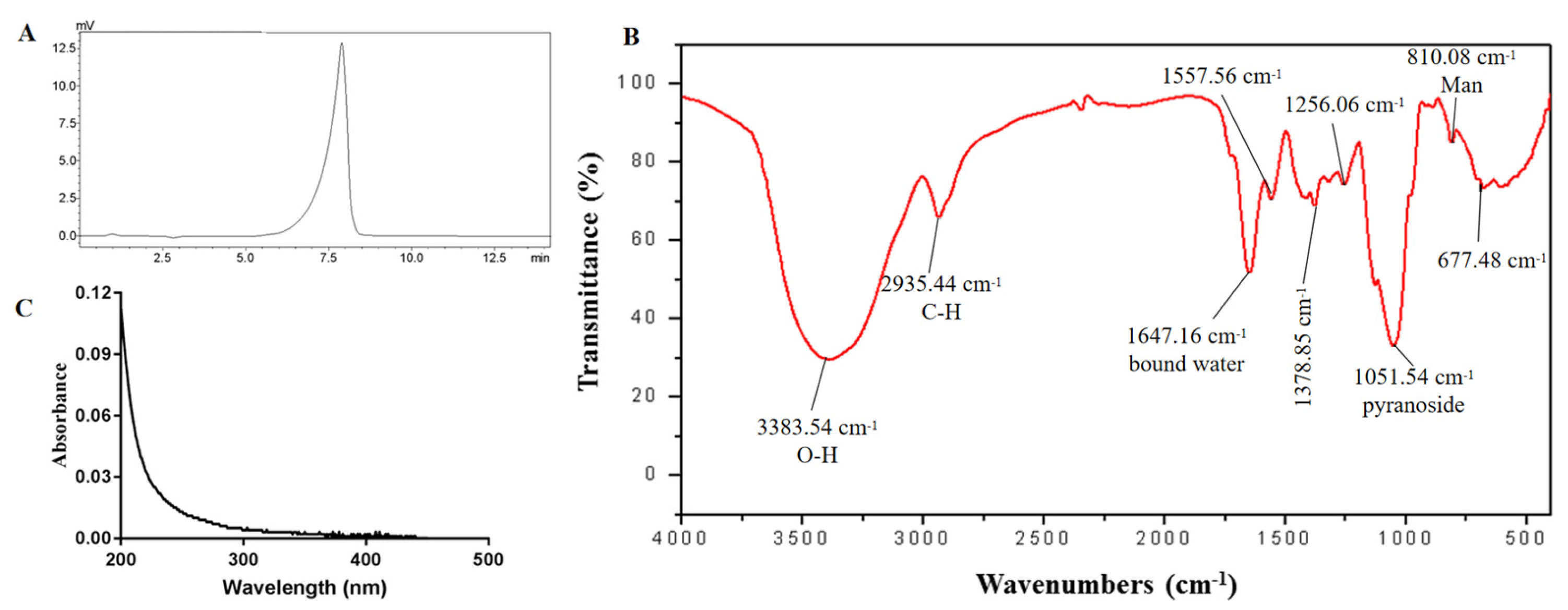
2.2. Molecular Weight and Monosaccharide Composition
2.3. FT-IR and UV Spectrum
2.4. Methylation Analysis
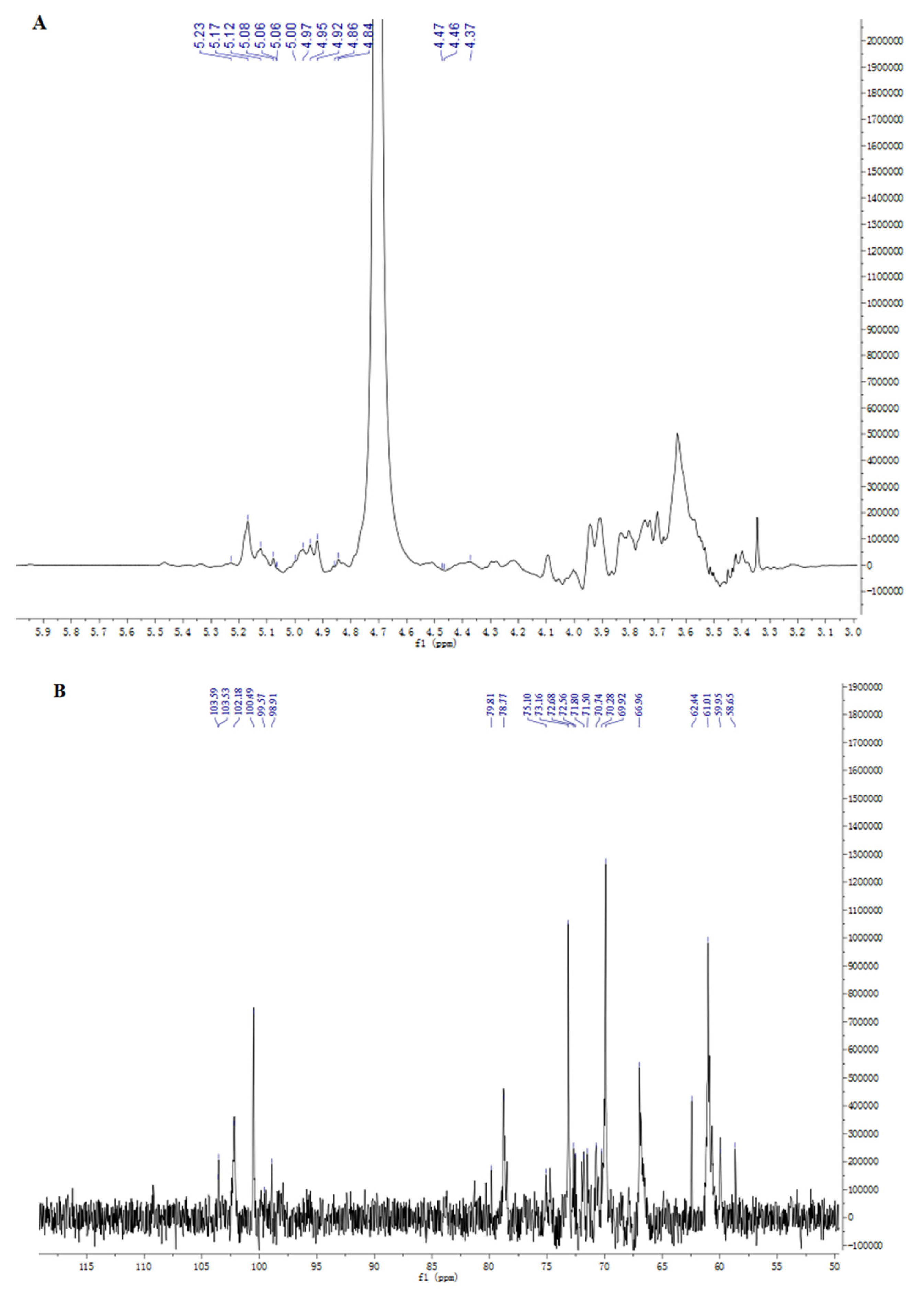
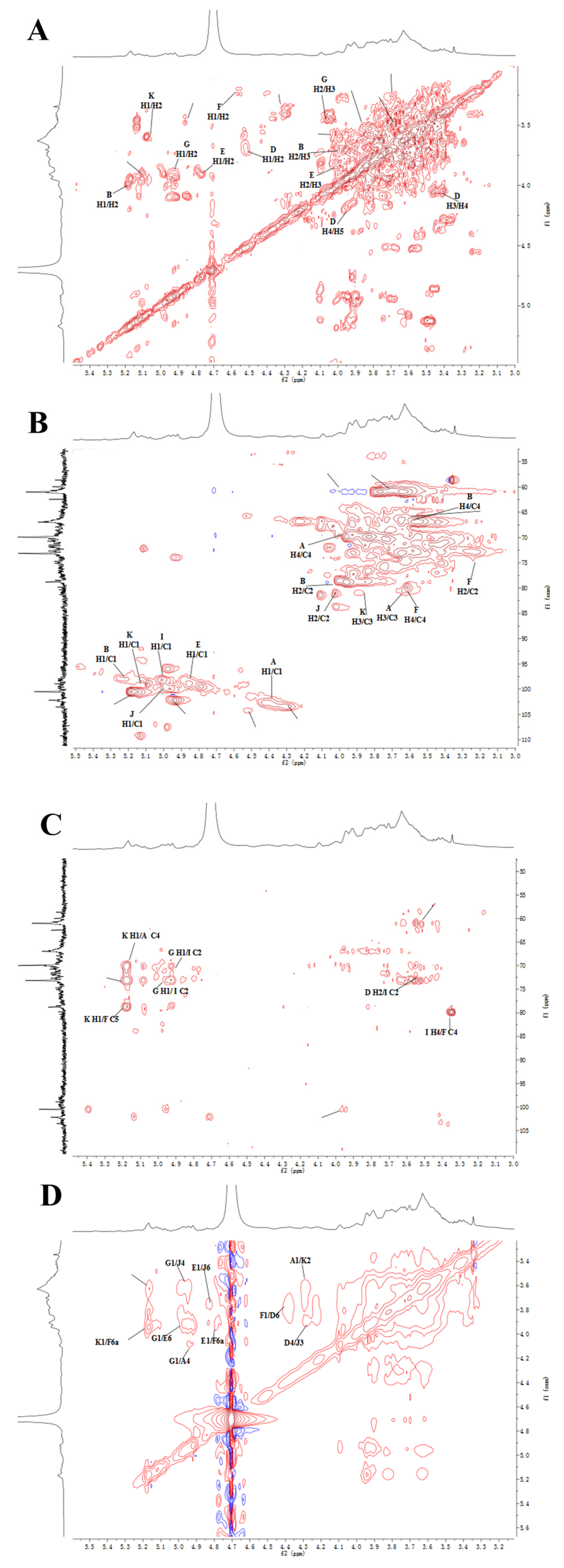
2.5. NMR Analysis
2.6. Molecular Morphology
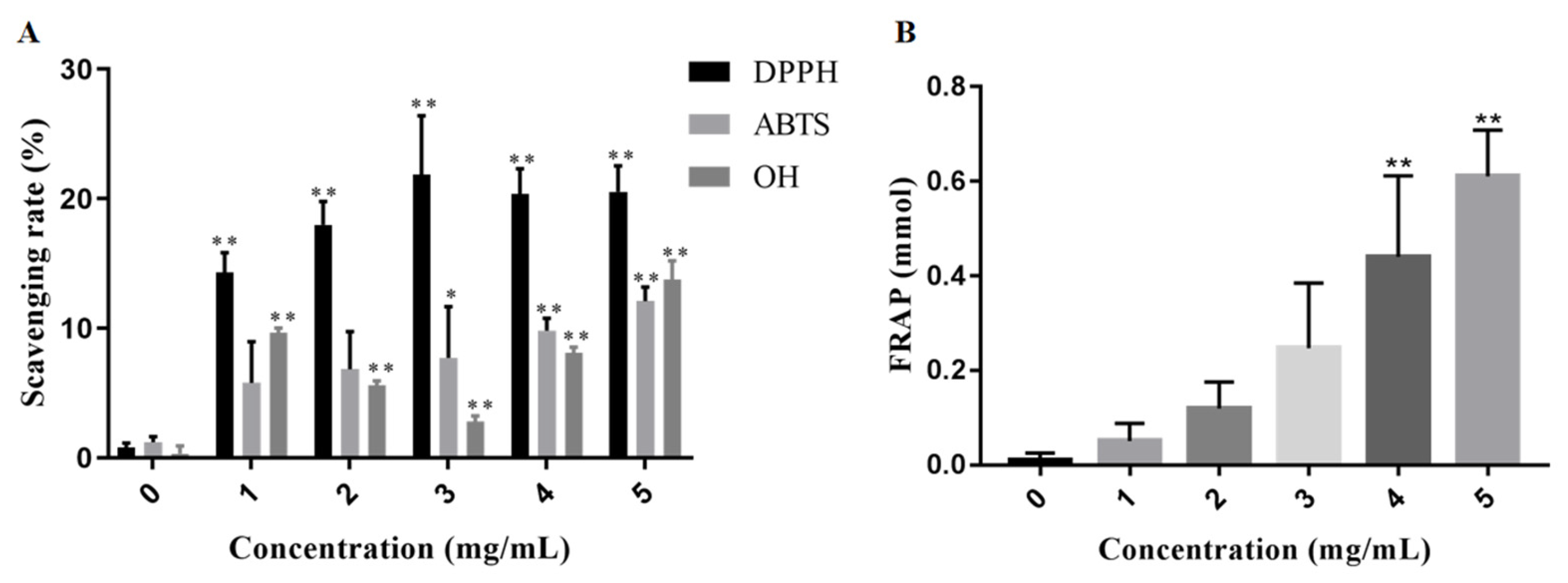
2.7. In Vitro Antioxidant Activities of EPS2-1
2.8. Effect of EPS2-1 in the Proliferation of Tumor and RAW264.7 Cells
3. Materials and Methods
3.1. Materials and Reagents
3.2. Isolation and Purification of Exopolysaccharides from Rhizopus Nigricans
3.3. Molecular Weight Determination
3.4. Monosaccharide Composition Analysis
3.5. FT-IR and UV Spectroscopy
3.6. Methylation and GC-MS Analysis
3.7. NMR Analysis
3.8. Morphological Analysis
3.9. Antioxidant Activity In Vitro
3.10. Cell Culture
3.11. Cell Viability Assay
3.12. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wei, M.; Hu, Y.; Zou, W.; Li, Y.; Cao, Y.; Li, S.; Huang, J.; Xing, L.; Huang, B.; Wang, X. Physicochemical property and antioxidant activity of polysaccharide from the seed cakes of Camellia oleifera Abel. Food. Sci. Nutr. 2022, 10, 1667–1682. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Z.; Zhong, C.; Pu, Y.; Yang, Z.; Bao, Y. Structure characteristics and immunomodulatory activities of a polysaccharide RGRP-1b from radix ginseng Rubra. Int. J. Biol. Macromol. 2021, 189, 980–992. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Shan, S.; Gao, X.; Zeng, D.; Lu, W. Structure characterization and lipid-lowering activity of a homogeneous heteropolysaccharide from sweet tea (Rubus Suavissmus S. Lee). Carbohydr. Polym. 2022, 277, 118757. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Tan, J.; Nima, L.; Sang, Y.; Cai, X.; Xue, H. Polysaccharides from fungi: A review on their extraction, purification, structural features, and biological activities. Food Chem. X 2022, 15, 100414. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.X.; Zhu, H.; Hu, Q.M.; Liu, Y.Y.; Zhao, S.M.; Peng, N.; Liang, Y.X. A novel polysaccharide from mycelia of cultured Phellinus linteus displays antitumor activity through apoptosis. Carbohydr. Polym. 2015, 124, 90–97. [Google Scholar] [CrossRef]
- Wan, X.; Jin, X.; Wu, X.; Yang, X.; Lin, D.; Li, C.; Fu, Y.; Liu, Y.; Liu, X.; Lv, J.; et al. Structural characterisation and antitumor activity against non-small cell lung cancer of polysaccharides from Sanghuangporus vaninii. Carbohydr. Polym. 2022, 276, 118798. [Google Scholar] [CrossRef]
- Ren, L.; Zhang, J.; Zhang, T.H. Immunomodulatory activities of polysaccharides from Ganoderma on immune effector cells. Food. Chem. 2021, 340, 127933. [Google Scholar] [CrossRef]
- Liu, G.K.; Yang, T.X.; Wang, J.R. Polysaccharides from Polyporus umbellatus: A review on their extraction, modification, structure, and bioactivities. Int. J. Biol. Macromol. 2021, 189, 124–134. [Google Scholar] [CrossRef]
- Wang, Y.F.; Jia, J.X.; Ren, X.J.; Li, B.H.; Zhang, Q. Extraction, preliminary characterization and in vitro antioxidant activity of polysaccharides from Oudemansiella radicata mushroom. Int. J. Biol. Macromol. 2018, 120, 1760–1769. [Google Scholar] [CrossRef]
- Zhang, G.Y.; Yin, Q.S.; Han, T.; Zhao, Y.X.; Su, J.J.; Li, M.Z.; Ling, J.Y. Purification and antioxidant effect of novel fungal polysaccharides from the stroma of Cordyceps kyushuensis. Ind. Crop. Prod. 2015, 69, 485–491. [Google Scholar] [CrossRef]
- Yan, H.X.; Ma, X.Q.; Mi, Z.; He, Z.H.; Rong, P.F. Extracellular Polysaccharide from Rhizopus nigricans Inhibits Hepatocellular Carcinoma via miR-494-3p/TRIM36 Axis and Cyclin E Ubiquitination. J. Clin. Transl. Hepatol. 2022, 10, 608–619. [Google Scholar] [CrossRef] [PubMed]
- Xiang, A.Z.; Ling, C.; Zhang, W.; Chen, H.G. Effects of Rhizopus Nigricans Exopolysaccharide on Proliferation, Apoptosis, and Migration of Breast Cancer MCF-7 Cells and Akt Signaling Pathway. Int. J. Polym. Sci. 2021, 2021, 5621984. [Google Scholar] [CrossRef]
- Yu, W.; Chen, G.; Zhang, P.; Chen, K. Purification, partial characterization and antitumor effect of an exopolysaccharide from Rhizopus nigricans. Int. J. Biol. Macromol. 2016, 82, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Lu, Y.; Yu, Z.; Xu, L.; Liu, J.; Chen, K.; Zhang, P. The inhibitory effect of polysaccharide from Rhizopus nigricans on colitis-associated colorectal cancer. Biomed. Pharmacother. 2019, 112, 108593. [Google Scholar] [CrossRef] [PubMed]
- Chu, G.; Miao, Y.; Huang, K.; Song, H.; Liu, L. Role and mechanism of Rhizopus Nigrum polysaccharide EPS1-1 as pharmaceutical for therapy of hepatocellular carcinoma. Front. Bioeng. Biotechnol. 2020, 8, 509. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, X.; Wang, J.; Chen, K. Exopolysaccharides isolated from Rhizopus nigricans induced colon cancer cell apoptosis in vitro and in vivo via activating the AMPK pathway. Biosci. Rep. 2020, 40, 1. [Google Scholar] [CrossRef] [Green Version]
- Masuko, T.; Minami, A.; Iwasaki, N.; Majima, T.; Nishimura, S.I.; Lee, Y.C. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal. Biochem. 2005, 339, 69–72. [Google Scholar] [CrossRef]
- Liu, S.; Yang, Y.; Qu, Y.; Guo, X.; Yang, X.; Cui, X.; Wang, C. Structural characterization of a novel polysaccharide from Panax notoginseng residue and its immunomodulatory activity on bone marrow dendritic cells. Int. J. Biol. Macromol. 2020, 161, 797–809. [Google Scholar] [CrossRef]
- Liu, J.; Zeng, Y.; Bi, S.; Zhou, J.; Cheng, R.; Li, J.; Jia, A. Characterization and chemical modification of PLN-1, an exopolysaccharide from Phomopsis liquidambari NJUSTb1. Carbohydr. Polym. 2021, 253, 117197. [Google Scholar] [CrossRef]
- Bac, V.H.; Paulsen, B.S.; Truong, L.V.; Koschella, A.; Trinh, T.C.; Wold, C.W.; Yogarajah, S.; Heinze, T. Neutral polysaccharide from the leaves of Pseuderanthemum carruthersii: Presence of 3-O-Methyl galactose and anti-inflammatory activity in LPS-stimulated RAW 264.7 cells. Polymers 2019, 11, 1219. [Google Scholar] [CrossRef]
- Tian, J.; Wang, X.; Zhang, X.; Zhang, C.; Chen, X.; Dong, M.; Rui, X.; Zhang, Q.; Fang, Y.; Li, W. Isolation, structural characterization and neuroprotective activity of exopolysaccharide from Paecilomyces cicada TJJ1213. Int. J. Biol. Macromol. 2021, 183, 1034–1046. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, S.; Park, J.; Mun, S.; Kwak, M.; Ko, H.; Baek, S. Structure and antiviral activity of a pectic polysaccharide from the root of Sanguisorba officinalis against enterovirus 71 in vitro/vivo. Carbohydr. Polym. 2022, 281, 119057. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Song, H.; Wang, L.; Peng, H.; Sun, Y.; Ai, C.; Wen, C.; Zhu, B.; Song, S. Structural characterization and SARS-CoV-2 inhibitory activity of a sulfated polysaccharide from Caulerpa lentillifera. Carbohydr. Polym. 2022, 280, 119006. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.W.; Luo, G.F.; Zheng, F.; Wu, K.; Yang, H.N.; Chen, L.; Tian, W. Structural characterization and evaluation the elicitors activity of polysaccharides from Chrysanthemum indicum. Carbohydr. Polym. 2021, 263, 117994. [Google Scholar] [CrossRef]
- Sun, H.; Yu, X.; Li, T.; Zhu, Z. Structure and hypoglycemic activity of a novel exopolysaccharide of Cordyceps militaris. Int. J. Biol. Macromol. 2021, 166, 496–508. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, X.; Zhao, P.; Ren, L.; Li, X.; Gao, W. Physicochemical characteristics and immunoregulatory activities of polysaccharides from five cultivars of Chrysanthemi Flos. Food. Sci. Nutr. 2022, 10, 1391–1400. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Z.; Zhang, B.; Ge, M.; Ng, H.; Niu, Y.; Liu, L. Production, structure and morphology of exopolysaccharides yielded by submerged fermentation of Antrodia cinnamomea. Carbohydr. Polym. 2019, 205, 271–278. [Google Scholar] [CrossRef]
- He, Y.; Peng, H.; Zhang, H.; Liu, Y.; Sun, H. Structural characteristics and immunopotentiation activity of two polysaccharides from the petal of Crocus sativus. Int. J. Biol. Macromol. 2021, 180, 129–142. [Google Scholar] [CrossRef]
- Zhang, X.; Li, J.; Li, M.; Chen, Y.; He, H.; Liu, S.; Cheng, F.; Liu, C.; Zou, K. Isolation, structure identification and hepatoprotective activity of a polysaccharide from Sabia parviflora. Bioorg. Med. Chem. Lett. 2021, 32, 127719. [Google Scholar] [CrossRef]
- Song, J.; Geng, X.; Su, Y.; Zhang, X.; Tu, L.; Zheng, Y.; Wang, M. Structure feature and antidepressant-like activity of a novel exopolysaccharide isolated from Marasmius androsaceus fermentation broth. Int. J. Biol. Macromol. 2020, 165, 1646–1655. [Google Scholar] [CrossRef]
- Islam, M.S.; Wang, H.; Mahdi, A.A.; Ahamed, M.I.; Lou, Z.; Wei, F.A. Effect of heat-treatment duration on antioxidant activities of muscle, liver and other parts of grass turtle (Chinemys reevesii). Pak. J. Zool. 2022, 54, 2003–2009. [Google Scholar] [CrossRef]
- Kim, J.S.; Cuong, D.M.; Bae, Y.B.; Cho, S.K. Antioxidant and antiproliferative activities of solvent fractions of broccoli (Brassica oleracea L.) sprout. Appl. Biol. Chem. 2022, 65, 34. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, T.; Zhang, X.; Zhang, F.; Linhardt, R.J. Structural and immunological studies on the polysaccharide from spores of a medicinal entomogenous fungus Paecilomyces cicadae. Carbohydr. Polym. 2021, 254, 117462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Qin, H.; An, R.; Zhang, W.; Liu, J.; Yu, Q.; Liu, W.; Huang, X. Isolation, purification, structural characterization and antitumor activities of a polysaccharide from Lilium davidii var. unicolor Cotton. J. Mol. Struct. 2022, 1261, 132941. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, L.; Qi, X.; Zhou, T.; Li, Y.; Cai, M.; Yan, Y.; Qian, J.Y.; Peng, D. Immunostimulatory and antioxidant activities of the selenized polysaccharide from edible Grifola frondosa. Food. Sci. Nutr. 2022, 10, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Huang, G. Extraction, characterization and antioxidant activities of pumpkin polysaccharide. Int. J. Biol. Macromol. 2018, 118, 770–774. [Google Scholar] [CrossRef]
- Tao, A.; Feng, X.; Sheng, Y.; Song, Z. Optimization of the Artemisia polysaccharide fermentation process by Aspergillus niger. Front. Nutr. 2022, 9, 842766. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Z.; Yang, L.; Yang, X.; Wang, X.; Zhang, Z. In vitro antioxidant activities of sulfated derivatives of polysaccharides extracted from Auricularia auricular. Int. J. Mol. Sci. 2011, 12, 3288–3302. [Google Scholar] [CrossRef]
- Shi, S.Y.; Chang, M.L.; Liu, H.P.; Ding, S.Y.; Yan, Z.Q.; Si, K.; Gong, T.T. The Structural Characteristics of an Acidic Water-Soluble Polysaccharide from Bupleurum chinense DC and Its In Vivo Anti-Tumor Activity on H22 Tumor-Bearing Mice. Polymers 2022, 14, 1119. [Google Scholar] [CrossRef]
- Chen, F.; Huang, G. Antioxidant activity of polysaccharides from different sources of ginseng. Int. J. Biol. Macromol. 2019, 125, 906–908. [Google Scholar] [CrossRef]
- Ji, X.; Hou, C.; Yan, Y.; Shi, M.; Liu, Y. Comparison of structural characterization and antioxidant activity of polysaccharides from jujube (Ziziphus jujuba Mill.) fruit. Int. J. Biol. Macromol. 2020, 149, 1008–1018. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Chen, X.; Qi, H. Characterization and bioactivity of phlorotannin loaded protein-polysaccharide nanocomplexes. LWT-Food. Sci. Technol. 2022, 155, 112998. [Google Scholar] [CrossRef]
- Li, W.; Wu, D.; Li, F.; Gan, R.; Hu, Y.; Zou, L. Structural and biological properties of water soluble polysaccharides from lotus leaves: Effects of Drying Techniques. Molecules 2021, 26, 4395. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zeng, Y.; Luo, D. Structure elucidation of a non-branched and entangled heteropolysaccharide from Tremella sanguinea Peng and its antioxidant activity. Carbohydr. Polym. 2016, 152, 33–40. [Google Scholar] [CrossRef]
- Wang, S.; Li, G.; Zhang, X.; Wang, Y.; Qiang, Y.; Wang, B.; Zou, J.; Niu, J.; Wang, Z. Structural characterization and antioxidant activity of Polygonatum sibiricum polysaccharides. Carbohydr. Polym. 2022, 291, 119524. [Google Scholar] [CrossRef]
- Lian, K.; Zhu, X.; Chen, J.; Liu, G.; Gu, X. Selenylation modification: Enhancement of the antioxidant activity of a Glycyrrhiza uralensis polysaccharide. Glycoconj. J. 2018, 35, 243–253. [Google Scholar] [CrossRef]
- Liu, C.; Ji, H.; Wu, P.; Yu, J.; Liu, A. The preparation of a cold-water soluble polysaccharide from Grifola frondosa and its inhibitory effects on MKN-45 cells. Glycoconj. J. 2020, 37, 413–422. [Google Scholar] [CrossRef]
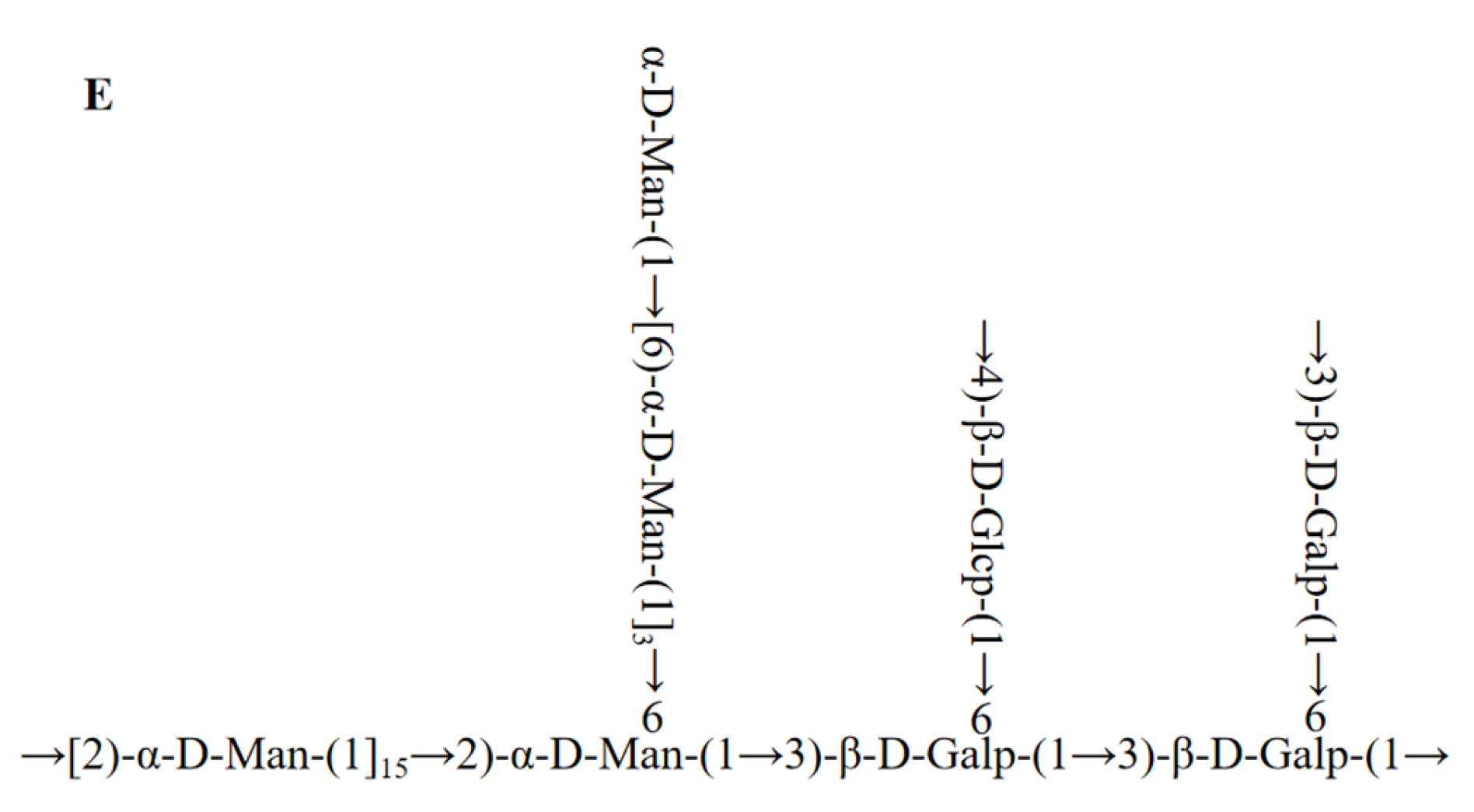


| Retention Time (min) | Monosaccharide Composition | Relative Molar Ratio |
|---|---|---|
| 5.844 | fucose | 0.029 |
| 12.592 | arabinose | 0.031 |
| 15.775 | galactose | 0.204 |
| 17.875 | glucose | 0.065 |
| 22.084 | mannose | 0.519 |
| 11.567 | rhamnose | 0.000 |
| 20.867 | xylose | 0.000 |
| 25.159 | fructose | 0.000 |
| 27.95 | ribose | 0.000 |
| 44.25 | galacturonic acid | 0.000 |
| 44.942 | guluronic acid | 0.000 |
| 47.017 | glucuronic acid | 0.000 |
| 49.742 | mannuronic acid | 0.000 |
| RT | Methylated Sugar | Mass Fragments (m/z) | Molar Ratios | Type of Linkage |
|---|---|---|---|---|
| 16.253 | 2,3,4,6-Me4-Manp | 43,71,87,101,117,129,145,161,205 | 3.42 | Manp-(1→ |
| 20.580 | 3,4,6-Me3-Manp | 43,71,87,99,101,129,145,161,189 | 64.37 | →2)-Manp-(1→ |
| 20.843 | 2,3,6-Me3-Glcp | 43,71,87,99,101,113,117,129,131,161,173,233 | 4.35 | →4)-Glcp-(1→ |
| 21.471 | 2,4,6-Me3-Manp | 43,71,87,99,101,117,129,161,201,233 | 2.02 | →3)-Manp-(1→ |
| 21.983 | 2,4,6-Me3-Galp | 43,71,87,101,117,129,143,161,173,233 | 1.68 | →3)-Galp-(1→ |
| 22.181 | 2,3,4-Me3-Glcp | 43,71,87,99,101,117,129,161,173,189,233 | 1.69 | →6)-Glcp-(1→ |
| 22.372 | 2,3,4-Me3-Manp | 43,71,87,99,101,117,129,161,173,189 | 12.09 | →6)-Manp-(1→ |
| 24.228 | 3,4–Me2-Manp | 43,71,87,99,129,159,173,189,233 | 1.41 | →2,6)-Manp-(1→ |
| 29.312 | 2,4-Me2-Galp | 43,87,101,117,129,139,159,173,189,233 | 8.97 | →3,6)-Galp-(1→ |
| Glycosyl Residues | C1/H1 | C2/H2 | C3/H3 | C4/H4 | C5/H5 | C6/H6a | H6b |
|---|---|---|---|---|---|---|---|
| (A):→3)-β-D-Galp-(1→ | 104.48/4.37 | 71.97/3.47 | 81.58/3.60 | 70.25/3.97 | 76.40/3.64 | 62.26/3.69 | |
| (B):→2)-α-D-Manp-(1→ | 101.95/5.23 | 80.21/4.01 | 71.32/3.88 | 68.19/3.6 | 74.5/3.7 | 62.3/3.69 | 3.81 |
| (D):→3,6)-β-D-Galp-(1→ | 104.49/4.47 | 71.31/3.57 | 81.5/3.68 | 69.82/4.05 | 74.81/3.87 | 70.76/3.96 | 3.86 |
| (E):→6)-α-D-Manp-(1→ | 100.83/4.85 | 71.42/3.94 | 71.95/3.8 | 68.03/3.65 | 74.53/3.78 | 67.2/3.78 | 3.71 |
| (F):→4)-β-D-Glcp-(1→ | 103.84/4.46 | 74.17/3.3 | 71.58/3.63 | 80.16/3.64 | 76.69/3.47 | 61.67/3.95 | 3.79 |
| (G):α-D-Manp-(1→ | 103.6/4.99 | 71.44/4.01 | 71.95/3.78 | 68.66/3.58 | 74.67/3.71 | 62.4/3.68 | 3.82 |
| (I):→6)-α-D-Glcp-(1→ | 99.1/4.91 | 72.73/3.53 | 74.63/3.67 | 70.87/3.47 | 71.47/3.86 | 66.8/3.7 | 3.93 |
| (J):→2,6)-α-D-Manp-(1→ | 99.62/5.05 | 80.12/3.98 | 71.58/3.86 | 68.06/3.6 | 74.52/3.7 | 67.2/3.77 | 3.72 |
| (K):→3)-α-D-Manp-(1→ | 101.1/5.15 | 71.55/3.65 | 79.7/3.88 | 68.4/3.6 | 74.75/3.78 | 62.16/3.69 | 3.82 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Li, J.; Xu, X.; Luo, Z.; Sun, J.; Wang, H.; Liu, C.; Ni, X.; Sun, J.; Xu, J.; et al. Separation and Structural Characterization of a Novel Exopolysaccharide from Rhizopus nigricans. Molecules 2022, 27, 7756. https://doi.org/10.3390/molecules27227756
Li Z, Li J, Xu X, Luo Z, Sun J, Wang H, Liu C, Ni X, Sun J, Xu J, et al. Separation and Structural Characterization of a Novel Exopolysaccharide from Rhizopus nigricans. Molecules. 2022; 27(22):7756. https://doi.org/10.3390/molecules27227756
Chicago/Turabian StyleLi, Zhang, Jianhua Li, Xuan Xu, Zhen Luo, Jiayi Sun, Hongyun Wang, Chunyan Liu, Xiuwen Ni, Jianqi Sun, Jun Xu, and et al. 2022. "Separation and Structural Characterization of a Novel Exopolysaccharide from Rhizopus nigricans" Molecules 27, no. 22: 7756. https://doi.org/10.3390/molecules27227756
APA StyleLi, Z., Li, J., Xu, X., Luo, Z., Sun, J., Wang, H., Liu, C., Ni, X., Sun, J., Xu, J., & Chen, K. (2022). Separation and Structural Characterization of a Novel Exopolysaccharide from Rhizopus nigricans. Molecules, 27(22), 7756. https://doi.org/10.3390/molecules27227756





