A Background-Free SERS Strategy for Sensitive Detection of Hydrogen Peroxide
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Characterization Conditions
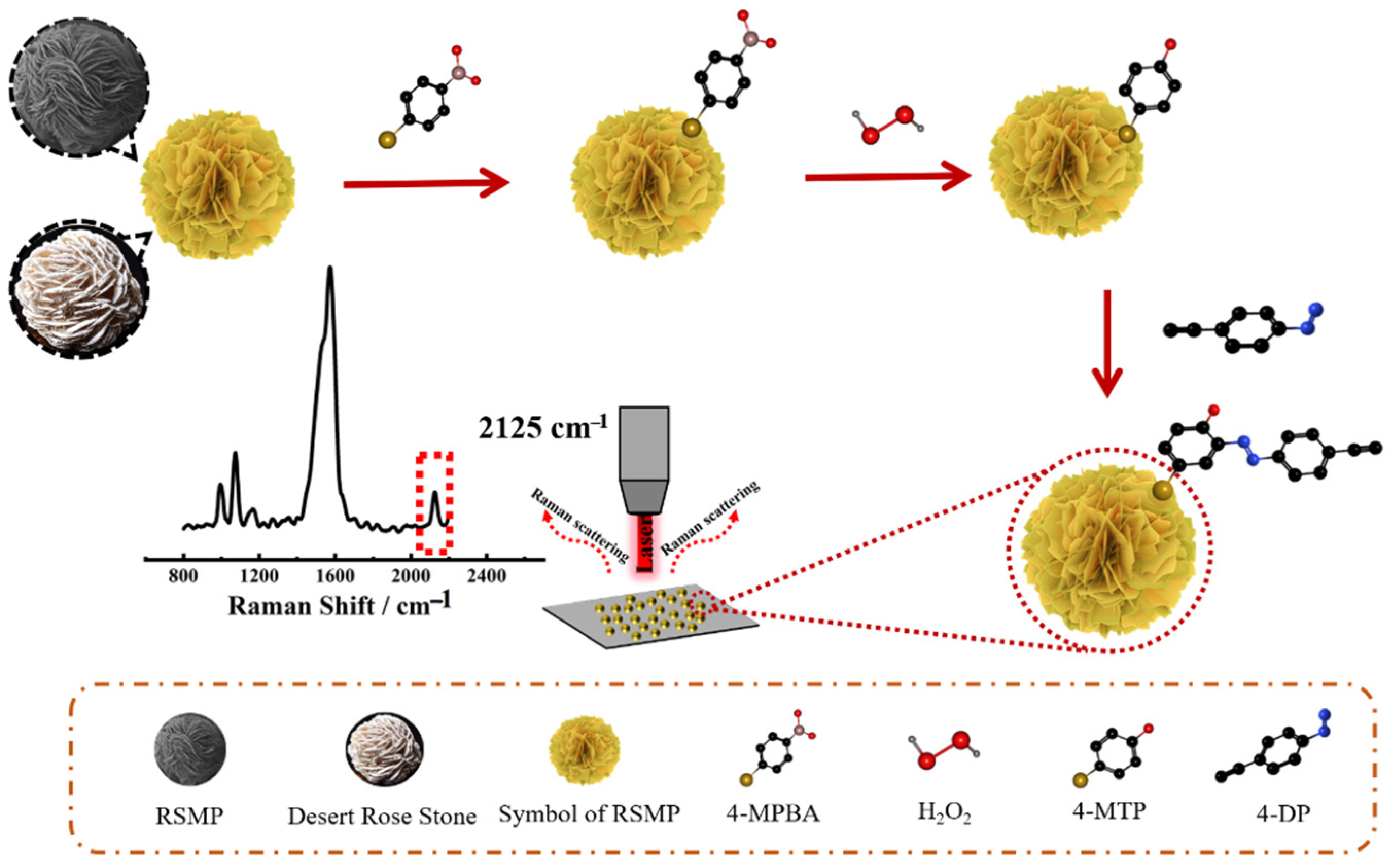
2.3. Synthesis of RSMPs
2.4. Synthesis of 4-Diazonium-Phenylaklyne (4-DP)
2.5. Preparation of SERS Probe
2.6. H2O2 Detection
2.7. Interference Experiment
2.8. Real Sample Test
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Langer, J.; Jimenez de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguié, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.J.; Boisen, A.; Brolo, A.G.; et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2020, 14, 28–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, B.; Frontiera, R.R.; Henry, A.-I.; Ringe, E.; Van Duyne, R.P. SERS: Materials, applications, and the future. Mater. Today 2012, 15, 16–25. [Google Scholar] [CrossRef]
- Yin, Y.; Li, Q.; Ma, S.; Liu, H.; Dong, B.; Yang, J.; Liu, D. Prussian Blue as a Highly Sensitive and Background-Free Resonant Raman Reporter. Anal. Chem. 2017, 89, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gao, X.; Xu, C.; Liu, D. SERS Tags for Biomedical Detection and Bioimaging. Theranostics 2022, 12, 1870–1903. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.A.; Dina, N.E.; Cheng, H.; Valev, V.K.; Zhang, L. Surface-enhanced Raman spectroscopy for bioanalysis and diagnosis. Nanoscale 2021, 13, 11593–11634. [Google Scholar] [CrossRef]
- Jain, M.; Nilsson, R.; Sharma, S.; Madhusudhan, N.; Kitami, T.; Souza Amanda, L.; Kafri, R.; Kirschner Marc, W.; Clish Clary, B.; Mootha Vamsi, K. Metabolite Profiling Identifies a Key Role for Glycine in Rapid Cancer Cell Proliferation. Science 2012, 336, 1040–1044. [Google Scholar] [CrossRef] [Green Version]
- Fernie, A.R.; Trethewey, R.N.; Krotzky, A.J.; Willmitzer, L. Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004, 5, 763–769. [Google Scholar] [CrossRef]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448–453. [Google Scholar] [CrossRef] [Green Version]
- Asiago, V.M.; Alvarado, L.Z.; Shanaiah, N.; Gowda, G.A.N.; Owusu-Sarfo, K.; Ballas, R.A.; Raftery, D. Early Detection of Recurrent Breast Cancer Using Metabolite Profiling. Cancer Res. 2010, 70, 8309–8318. [Google Scholar] [CrossRef] [Green Version]
- Gong, J.; Tang, H.; Wang, M.; Lin, X.; Wang, K.; Liu, J. Novel three-dimensional graphene nanomesh prepared by facile electro-etching for improved electroanalytical performance for small biomolecules. Mater. Des. 2022, 215, 110506. [Google Scholar] [CrossRef]
- Gong, J.; Zhang, T.; Luo, T.; Luo, X.; Yan, F.; Tang, W.; Liu, J. Bipolar silica nanochannel array confined electrochemiluminescence for ultrasensitive detection of SARS-CoV-2 antibody. Biosens. Bioelectron. 2022, 215, 114563. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ma, X.; Sailjoi, A.; Zou, Y.; Lin, X.; Yan, F.; Su, B.; Liu, J. Vertical silica nanochannels supported by nanocarbon composite for simultaneous detection of serotonin and melatonin in biological fluids. Sens. Actuators B Chem. 2022, 353, 131101. [Google Scholar] [CrossRef]
- Gong, J.; Zhang, T.; Chen, P.; Yan, F.; Liu, J. Bipolar silica nanochannel array for dual-mode electrochemiluminescence and electrochemical immunosensing platform. Sens. Actuators B Chem. 2022, 368, 132086. [Google Scholar] [CrossRef]
- Hong, S.; Chen, T.; Zhu, Y.; Li, A.; Huang, Y.; Chen, X. Live-Cell Stimulated Raman Scattering Imaging of Alkyne-Tagged Biomolecules. Angew. Chem. Int. Ed. 2014, 53, 5827–5831. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Fan, X.; Chen, C.; Huang, L.; Wang, J.; Tang, X. Multicolor Raman Beads for Multiplexed Tumor Cell and Tissue Imaging and in Vivo Tumor Spectral Detection. Anal. Chem. 2019, 91, 3784–3789. [Google Scholar] [CrossRef] [Green Version]
- Tian, S.; Li, H.; Li, Z.; Tang, H.; Yin, M.; Chen, Y.; Wang, S.; Gao, Y.; Yang, X.; Meng, F.; et al. Polydiacetylene-based ultrastrong bioorthogonal Raman probes for targeted live-cell Raman imaging. Nat. Commun. 2020, 11, 81. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, D.C.; Hoop, K.A.; Tay, L.-L.; Pezacki, J.P. Development of nanoparticle probes for multiplex SERS imaging of cell surface proteins. Nanoscale 2010, 2, 1413–1416. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Gao, M.-Y.; Zhu, Q.; Chi, B.; Zeng, L.-W.; Hu, J.-M.; Shen, A.-G. Monodispersed plasmonic Prussian blue nanoparticles for zero-background SERS/MRI-guided phototherapy. Nanoscale 2020, 12, 3292–3301. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, R.; Gao, M.; Zhang, X. Facile synthesis of thiol and alkynyl contained SERS reporter molecular and its usage in assembly of polydopamine protected bioorthogonal SERS tag for live cell imaging. Talanta 2016, 158, 315–321. [Google Scholar] [CrossRef]
- Li, N.; Chen, H.L.; Zhang, M.Y.; Zha, Y.C.; Mu, Z.X.; Ma, Y.; Chen, P. A universal ultrasensitive platform for enzyme-linked immunoassay based on responsive surface-enhanced Raman scattering. Sens. Actuators B Chem. 2020, 315, 128135. [Google Scholar] [CrossRef]
- Lin, L.; Tian, X.; Hong, S.; Dai, P.; You, Q.; Wang, R.; Feng, L.; Xie, C.; Tian, Z.-Q.; Chen, X. A Bioorthogonal Raman Reporter Strategy for SERS Detection of Glycans on Live Cells. Angew. Chem. Int. Ed. 2013, 52, 7266–7271. [Google Scholar] [CrossRef] [PubMed]
- Hanif, S.; Liu, H.-L.; Ahmed, S.A.; Yang, J.-M.; Zhou, Y.; Pang, J.; Ji, L.-N.; Xia, X.-H.; Wang, K. Nanopipette-Based SERS Aptasensor for Subcellular Localization of Cancer Biomarker in Single Cells. Anal. Chem. 2017, 89, 9911–9917. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Wang, Y.; Wang, L.; Lu, Y.; Li, J.; Lu, D.; Zhou, T.; Huang, Z.; Huang, J.; Huang, H.; et al. Interference-free and high precision biosensor based on surface enhanced Raman spectroscopy integrated with surface molecularly imprinted polymer technology for tumor biomarker detection in human blood. Biosens. Bioelectron. 2019, 143, 111599. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yung, L.Y.L. Gold Nanoplate-Based 3D Hierarchical Microparticles: A Single Particle with High Surface-Enhanced Raman Scattering Enhancement. Langmuir 2016, 32, 7854–7859. [Google Scholar] [CrossRef]
- Alvarez-Puebla, R.A.; Liz-Marzán, L.M. SERS-Based Diagnosis and Biodetection. Small 2010, 6, 604–610. [Google Scholar] [CrossRef]
- Xu, L.; Yan, W.; Ma, W.; Kuang, H.; Wu, X.; Liu, L.; Zhao, Y.; Wang, L.; Xu, C. SERS Encoded Silver Pyramids for Attomolar Detection of Multiplexed Disease Biomarkers. Adv. Mater. 2015, 27, 1706–1711. [Google Scholar] [CrossRef]
- Li, N.; Zhang, M.; Zha, Y.; Cao, Y.; Ma, Y. π-π stacking-directed self-assembly of nanoplatelets into diversified three-dimensional superparticles for high surface-enhanced Raman scattering. J. Colloid Interface Sci. 2020, 575, 54–60. [Google Scholar] [CrossRef]
- Yamakoshi, H.; Dodo, K.; Palonpon, A.; Ando, J.; Fujita, K.; Kawata, S.; Sodeoka, M. Alkyne-Tag Raman Imaging for Visualization of Mobile Small Molecules in Live Cells. J. Am. Chem. Soc. 2012, 134, 20681–20689. [Google Scholar] [CrossRef]
- Ma, Y.; Yung, L.Y.L. Synthesis of Self-Stabilized Poly(N-(3-Amidino)-Aniline) Particles and their CO2-Responsive Properties. Part Part Syst. Char. 2015, 32, 743–748. [Google Scholar] [CrossRef]
- Cheng, W.; Tang, P.; He, X.; Xing, X.; Xing, M.; Lu, L.; Ju, P.; Wang, Y.; Liu, S.; Lu, X.; et al. Dynamic monitoring and quantitative characterization of intracellular H2O2 content by using SERS based boric acid nanoprobe. Talanta 2020, 214, 120863. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Yan, B.; Chen, L.X. SERS Tags: Novel Optical Nanoprobes for Bioanalysis. Chem Rev 2013, 113, 1391–1428. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef] [PubMed]
- Nordberg, J.; Arnér, E.S.J. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic. Biol. Med. 2001, 31, 1287–1312. [Google Scholar] [CrossRef]
- Belousov, V.V.; Fradkov, A.F.; Lukyanov, K.A.; Staroverov, D.B.; Shakhbazov, K.S.; Terskikh, A.V.; Lukyanov, S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods 2006, 3, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Bienert, G.P.; Møller, A.L.B.; Kristiansen, K.A.; Schulz, A.; Møller, I.M.; Schjoerring, J.K.; Jahn, T.P. Specific Aquaporins Facilitate the Diffusion of Hydrogen Peroxide across Membranes*. J. Biol. Chem. 2007, 282, 1183–1192. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Ren, J.; Bao, X.; Gao, W.; Wu, C.; Zhao, Y. pH-Switchable Fluorescent Probe for Spatially-Confined Visualization of Intracellular Hydrogen Peroxide. Anal. Chem. 2016, 88, 5865–5870. [Google Scholar] [CrossRef]
- Huang, X.; Zhou, H.; Huang, Y.; Jiang, H.; Yang, N.; Shahzad, S.A.; Meng, L.; Yu, C. Silver nanoparticles decorated and tetraphenylethene probe doped silica nanoparticles: A colorimetric and fluorometric sensor for sensitive and selective detection and intracellular imaging of hydrogen peroxide. Biosens. Bioelectron. 2018, 121, 236–242. [Google Scholar] [CrossRef]
- Wu, W.-S. The signaling mechanism of ROS in tumor progression. Cancer Metastasis Rev. 2006, 25, 695–705. [Google Scholar] [CrossRef]
- Cai, H. Hydrogen peroxide regulation of endothelial function: Origins, mechanisms, and consequences. Cardiovasc. Res. 2005, 68, 26–36. [Google Scholar] [CrossRef] [Green Version]
- Tabner, B.J.; El-Agnaf, O.M.A.; Turnbull, S.; German, M.J.; Paleologou, K.E.; Hayashi, Y.; Cooper, L.J.; Fullwood, N.J.; Allsop, D. Hydrogen Peroxide Is Generated during the Very Early Stages of Aggregation of the Amyloid Peptides Implicated in Alzheimer Disease and Familial British Dementia *. J. Biol. Chem. 2005, 280, 35789–35792. [Google Scholar] [CrossRef]
- Zhao, S.; Zang, G.; Zhang, Y.; Liu, H.; Wang, N.; Cai, S.; Durkan, C.; Xie, G.; Wang, G. Recent advances of electrochemical sensors for detecting and monitoring ROS/RNS. Biosens. Bioelectron. 2021, 179, 113052. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Li, Y.; Li, Y.; Ye, Z.; Zhang, J.; Zhu, T.; Li, F. An L012@PAni-PAAm hydrogel composite based-electrochemiluminescence biosensor for in situ detection of H2O2 released from cardiomyocytes. Electrochim. Acta 2020, 354, 136763. [Google Scholar] [CrossRef]
- Ma, J.; Wang, X.; Li, N.; Cheng, Y. A bifunctional probe that allows dual-channel fluorescence turn-on detection of protein aggregates and hydrogen peroxide in neurodegenerative diseases. Sens. Actuators B Chem. 2021, 346, 130536. [Google Scholar] [CrossRef]
- Ren, M.; Dong, D.; Xu, Q.; Yin, J.; Wang, S.; Kong, F. A biotin-guided two-photon fluorescent probe for detection of hydrogen peroxide in cancer cells ferroptosis process. Talanta 2021, 234, 122684. [Google Scholar] [CrossRef]
- Xu, M.; Han, J.M.; Zhang, Y.; Yang, X.; Zang, L. A selective fluorescence turn-on sensor for trace vapor detection of hydrogen peroxide. Chem. Commun. 2013, 49, 11779–11781. [Google Scholar] [CrossRef]
- Tsaplev, Y.B. Chemiluminescence determination of hydrogen peroxide. J. Anal. Chem. 2012, 67, 506–514. [Google Scholar] [CrossRef]
- He, S.; Shi, W.; Zhang, X.; Li, J.; Huang, Y. β-Cyclodextrins-based inclusion complexes of CoFe2O4 magnetic nanoparticles as catalyst for the luminol chemiluminescence system and their applications in hydrogen peroxide detection. Talanta 2010, 82, 377–383. [Google Scholar] [CrossRef]
- Ramos, M.C.; Torijas, M.C.; Navas Díaz, A. Enhanced chemiluminescence biosensor for the determination of phenolic compounds and hydrogen peroxide. Sens. Actuators B Chem. 2001, 73, 71–75. [Google Scholar] [CrossRef]
- Hu, X.; Han, H.; Hua, L.; Sheng, Z. Electrogenerated chemiluminescence of blue emitting ZnSe quantum dots and its biosensing for hydrogen peroxide. Biosens. Bioelectron. 2010, 25, 1843–1846. [Google Scholar] [CrossRef]
- Li, Q.; Ge, X.; Ye, J.; Li, Z.; Su, L.; Wu, Y.; Yang, H.; Song, J. Dual Ratiometric SERS and Photoacoustic Core–Satellite Nanoprobe for Quantitatively Visualizing Hydrogen Peroxide in Inflammation and Cancer. Angew. Chem. Int. Ed. 2021, 60, 7323–7332. [Google Scholar] [CrossRef]
- Bai, L.; Wang, X.; Zhang, K.; Tan, X.; Zhang, Y.; Xie, W. Etchable SERS nanosensor for accurate pH and hydrogen peroxide sensing in living cells. Chem. Commun. 2019, 55, 12996–12999. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Si, Y.; Deng, T.; Zheng, J.; Li, J.; Yang, R.; Tan, W. A novel SERS nanoprobe for the ratiometric imaging of hydrogen peroxide in living cells. Chem. Commun. 2016, 52, 8553–8556. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.-L.; Liu, Y.-Y.; He, S.-H.; Chen, J.-Q.; Liang, Y.; Li, H.-T. Highly selective and sensitive surface enhanced Raman scattering nanosensors for detection of hydrogen peroxide in living cells. Biosens. Bioelectron. 2016, 77, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, X.; Rong, P.; Liu, D. Recent advances in background-free Raman scattering for bioanalysis. TrAC Trends Anal. Chem. 2020, 123, 115765. [Google Scholar] [CrossRef]
- Plakas, K.; Rosch, L.E.; Clark, M.D.; Adbul-Rashed, S.; Shaffer, T.M.; Harmsen, S.; Gambhir, S.S.; Detty, M.R. Design and evaluation of Raman reporters for the Raman-silent region. Nanotheranostics 2022, 6, 1–9. [Google Scholar] [CrossRef]
- Qiu, C.; Cheng, Z.; Lv, C.; Wang, R.; Yu, F. Development of bioorthogonal SERS imaging probe in biological and biomedical applications. Chin. Chem. Lett. 2021, 32, 2369–2379. [Google Scholar] [CrossRef]
- Zimmermann, F.L.T.; Beyer, C.; Stebani, J.; Nuyken, O.; Wokaun, A. N=N Vibrational Frequencies and Fragmentation Patterns of Substituted 1-Aryl-3,3-Dialkyl-Triazenes: Comparison with Other High-Nitrogen Compounds. Appl. Spectrosc. 1993, 47, 986–993. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, D.; Chen, H.-Y. Electrochemiluminescence Analysis of Hydrogen Peroxide Using L012 Modified Electrodes. J. Anal. Test. 2020, 4, 122–127. [Google Scholar] [CrossRef]
- Gao, J.; Liu, H.; Tong, C.; Pang, L.; Feng, Y.; Zuo, M.; Wei, Z.; Li, J. Hemoglobin-Mn3(PO4)2 hybrid nanoflower with opulent electroactive centers for high-performance hydrogen peroxide electrochemical biosensor. Sens. Actuators B Chem. 2020, 307, 127628. [Google Scholar] [CrossRef]
- Manavalan, S.; Ganesamurthi, J.; Chen, S.-M.; Veerakumar, P.; Murugan, K. A robust Mn@FeNi-S/graphene oxide nanocomposite as a high-efficiency catalyst for the non-enzymatic electrochemical detection of hydrogen peroxide. Nanoscale 2020, 12, 5961–5972. [Google Scholar] [CrossRef]
- Ye, S.; Hu, J.J.; Zhao, Q.A.; Yang, D. Fluorescent probes for in vitro and in vivo quantification of hydrogen peroxide. Chem. Sci. 2020, 11, 11989–11997. [Google Scholar] [CrossRef]
- Dervisevic, E.; Voelcker, N.H.; Risbridger, G.; Tuck, K.L.; Cadarso, V.J. Colorimetric Detection of Extracellular Hydrogen Peroxide Using an Integrated Microfluidic Device. Anal. Chem. 2022, 94, 1726–1732. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhong, Q.; Wang, Y.; Yuan, C.; Qin, X.; Xu, Y. Colorimetric detection of hydrogen peroxide and glucose by exploiting the peroxidase-like activity of papain. RSC Adv. 2019, 9, 16566–16570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, X.; Wang, H.; Schultz, Z.D.; Camden, J.P. Sensing Glucose in Urine and Serum and Hydrogen Peroxide in Living Cells by Use of a Novel Boronate Nanoprobe Based on Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2016, 88, 7191–7197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, Y.; Fu, C.; Wu, Y.; Cao, H.; Shi, W.; Jung, Y.M. A simple enzyme-free SERS sensor for the rapid and sensitive detection of hydrogen peroxide in food. Analyst 2020, 145, 607–612. [Google Scholar] [CrossRef]
- Li, X.; Duan, X.; Yang, P.; Li, L.; Tang, B. Accurate In Situ Monitoring of Mitochondrial H2O2 by Robust SERS Nanoprobes with a Au–Se Interface. Anal. Chem. 2021, 93, 4059–4065. [Google Scholar] [CrossRef]
- Lyu, Q.; Zhai, Q.; Dyson, J.; Gong, S.; Zhao, Y.; Ling, Y.; Chandrasekaran, R.; Dong, D.; Cheng, W. Real-Time and In-Situ Monitoring of H2O2 Release from Living Cells by a Stretchable Electrochemical Biosensor Based on Vertically Aligned Gold Nanowires. Anal. Chem. 2019, 91, 13521–13527. [Google Scholar] [CrossRef]








| Method | Material | Linear Range (M) | Limit of Detection (M) | Reference |
|---|---|---|---|---|
| Electrochemiluminescence | Hydrogel composite | 1.0 × 10−8–5.0 × 10−5 | 2.9 × 10−9 | [42] |
| Electrochemiluminescence | Modified Electrodes | 1.0 × 10−5–5.0 × 10−3 | 4.3 × 10−6 | [58] |
| Electrochemistry | Hybrid nanoflower | 2.0 × 10−8–3.6 × 10−6 | 7.0 × 10−9 | [59] |
| Electrochemistry | Graphene oxide nanocomposite | 5.5 × 10−7–5.2 × 10−4 | 8.8 × 10−9 | [60] |
| Fluorescence | Arylboronate-pyridinium | 0–1.5 × 10−5 | 5.6 × 10−6 | [43] |
| Fluorescence | HKPerox-Red | 0–1.0 × 10−4 | 4.8 × 10−9 | [61] |
| Colorimetry | Oxidized HRP and ABTS | 5.0 × 10−7–6.5 × 10−5 | 1.7 × 10−9 | [62] |
| Colorimetry | Papain and TMB | 55.0 × 10−6–9.0 × 10−5 | 2.1 × 10−6 | [63] |
| SERS | Gold Nanorod | 1.0 × 10−6–10−4 | 3.0 × 10−7 | [52] |
| SERS | AuNPs | 1.0 × 10−7–2.5 × 10−6 | 7.0 × 10−8 | [64] |
| SERS | AuNPs | 1.0 × 10−7–2.0 × 10−5 | 8.0 × 10−8 | [53] |
| SERS | AgNPs | 1.0 × 10−6–10−2 | 1.0 × 10−6 | [65] |
| SERS | ZIF-8 | 1.0 × 10−9–10−3 | 3.6 × 10−10 | [66,67] |
| SERS | AuNPs | 1.0 × 10−6–10−4 | 2.0 × 10−7 | [66] |
| SERS | RSMPs | 1.0 × 10−10–1.5 × 10−3 | 1.0 × 10−10 | this work |
| Sample | Spiking Conc. (mM) | Detected Conc. (mM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| 1 | 1.200 | 1.180 ± 0.240 | 98.640 | 6.630 |
| 2 | 0.500 | 0.580 ± 0.120 | 116.620 | 9.220 |
| 3 | 0.010 | 0.009 ± 0.003 | 96.520 | 1.760 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, K.; Chen, H.; Liang, S.; Wu, J.; Zhou, P.; Li, N. A Background-Free SERS Strategy for Sensitive Detection of Hydrogen Peroxide. Molecules 2022, 27, 7918. https://doi.org/10.3390/molecules27227918
Chen K, Chen H, Liang S, Wu J, Zhou P, Li N. A Background-Free SERS Strategy for Sensitive Detection of Hydrogen Peroxide. Molecules. 2022; 27(22):7918. https://doi.org/10.3390/molecules27227918
Chicago/Turabian StyleChen, Kaixin, Haoling Chen, Songxian Liang, Jindan Wu, Ping Zhou, and Nan Li. 2022. "A Background-Free SERS Strategy for Sensitive Detection of Hydrogen Peroxide" Molecules 27, no. 22: 7918. https://doi.org/10.3390/molecules27227918
APA StyleChen, K., Chen, H., Liang, S., Wu, J., Zhou, P., & Li, N. (2022). A Background-Free SERS Strategy for Sensitive Detection of Hydrogen Peroxide. Molecules, 27(22), 7918. https://doi.org/10.3390/molecules27227918





