Molecular Characterization of an Isoflavone 2′-Hydroxylase Gene Revealed Positive Insights into Flavonoid Accumulation and Abiotic Stress Tolerance in Safflower
Abstract
:1. Introduction
2. Results
2.1. Identification and Physicochemical Properties CtCYP81E Subfamily in Safflower
2.2. Phylogenetic Analysis of CtCYP81E-Encoding Genes
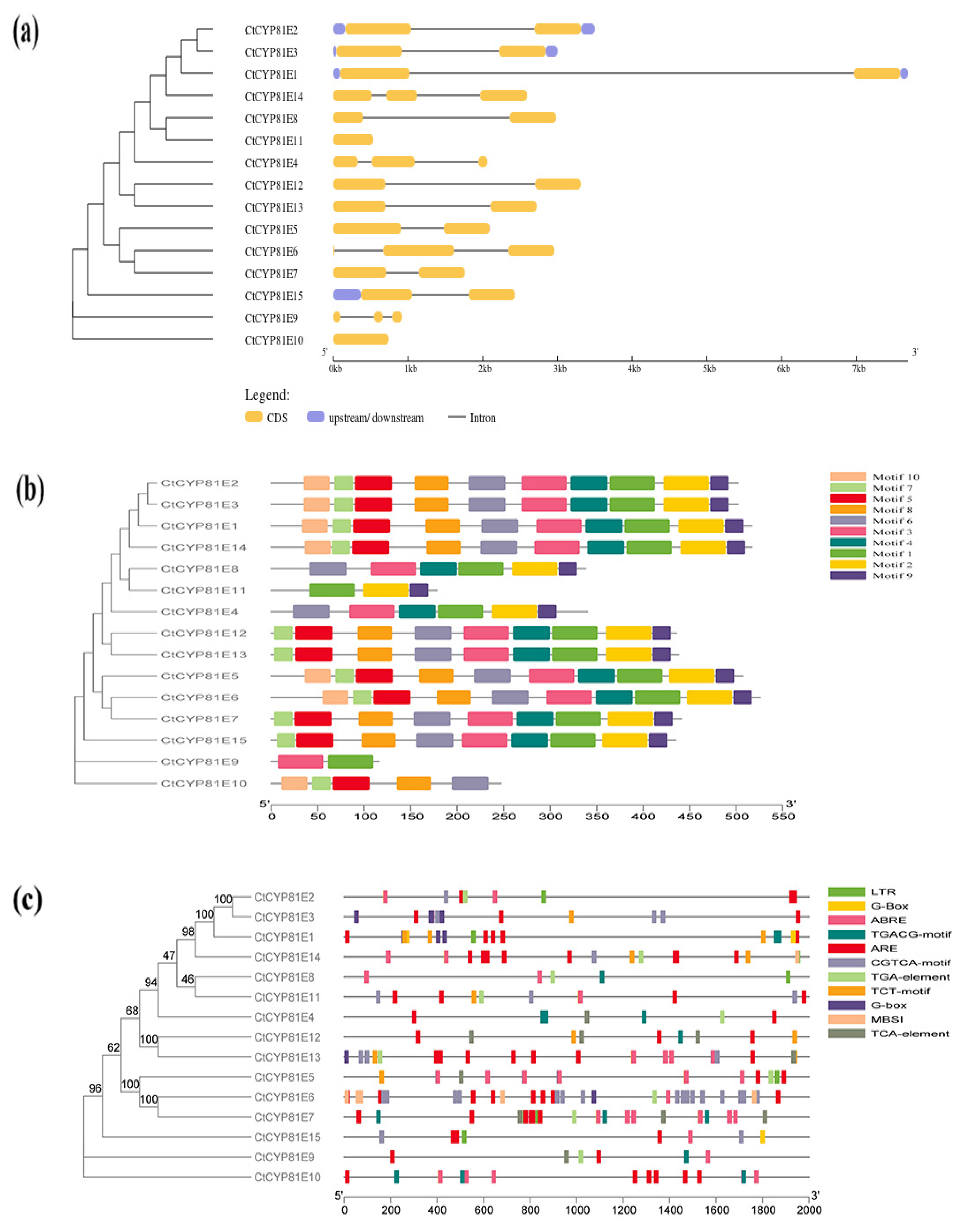
2.3. Analysis of CtCYP81E Gene Structure, Protein Motifs, and Cis-Elements
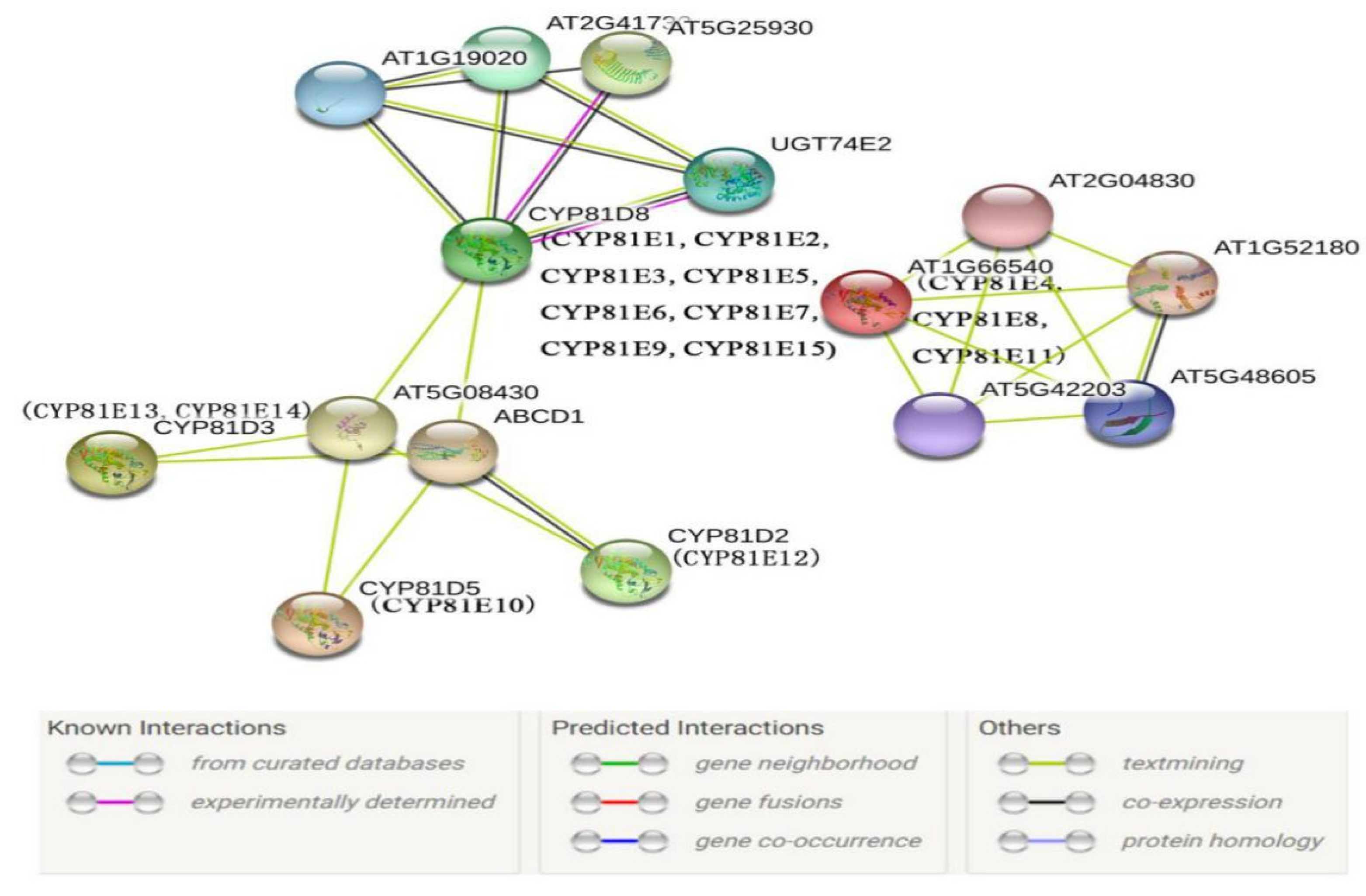
2.4. Protein Interaction Network Prediction
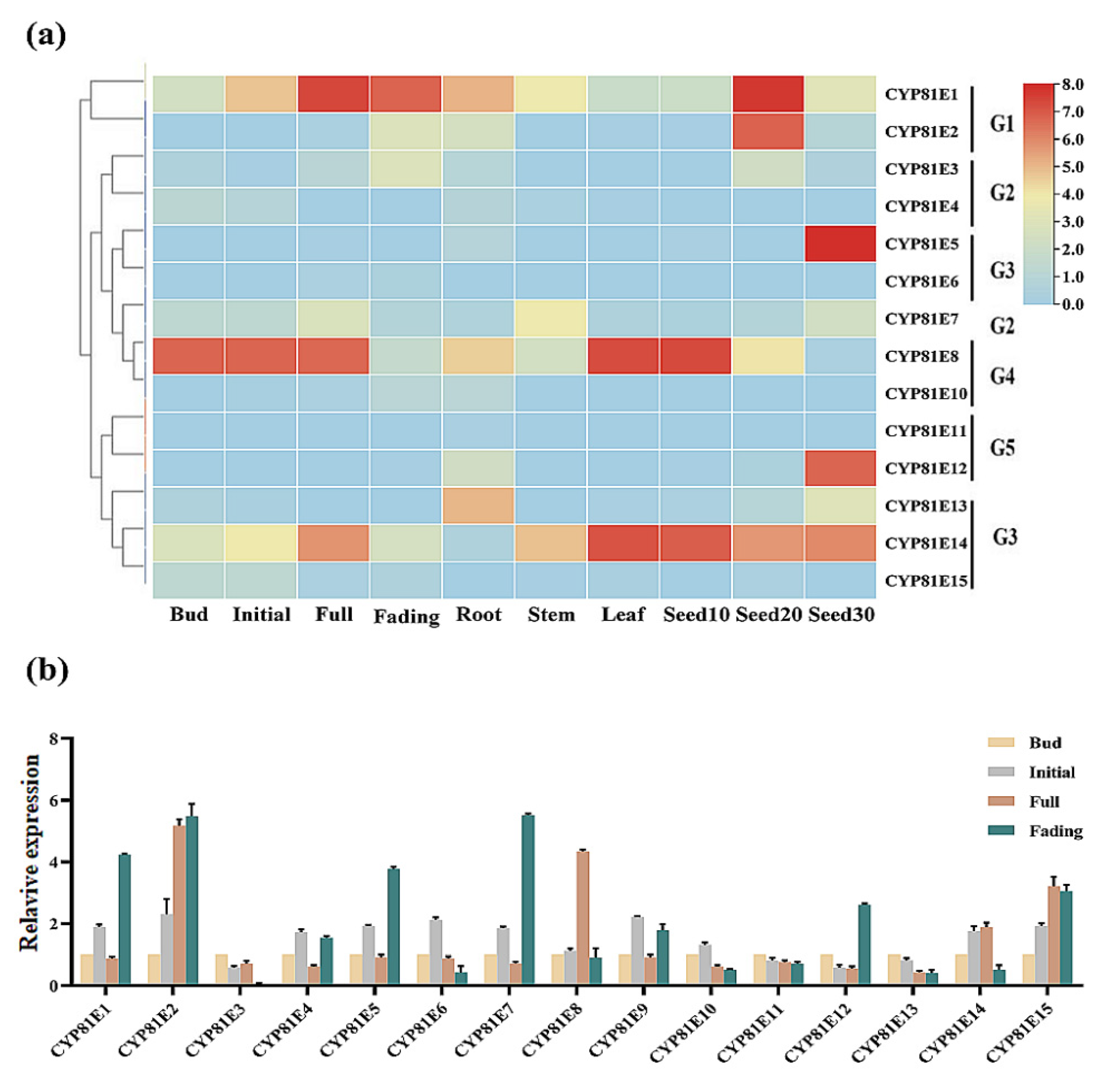
2.5. Tissue-Specific Expression of CtCYP81E-Encoding Genes in Safflower
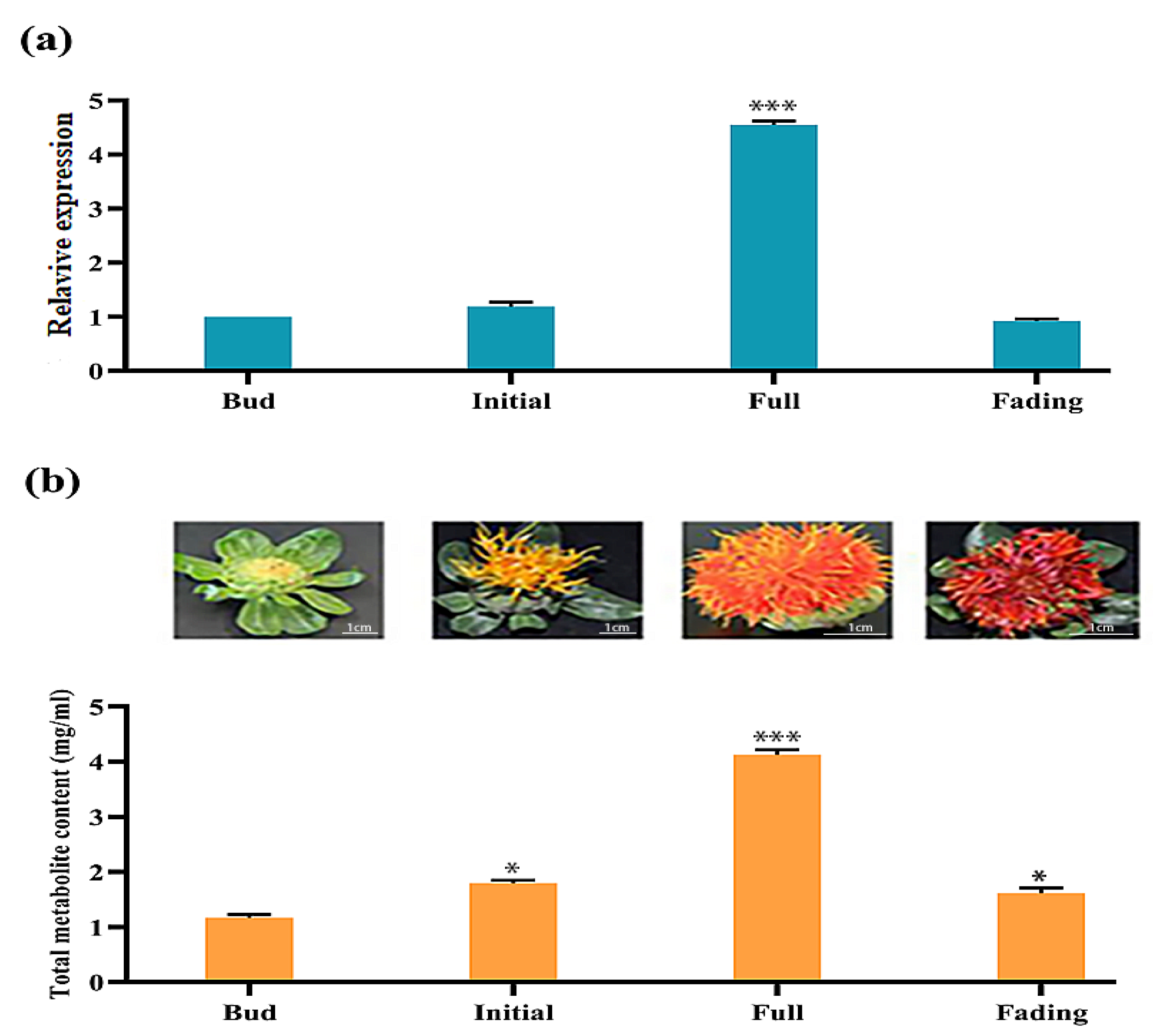
2.6. Differential Expression of CtCYP81E8 Encoding a Putative Isoflavone 2′-Hydroxylase Correlates with Metabolite Accumulation in Different Flowering Stages
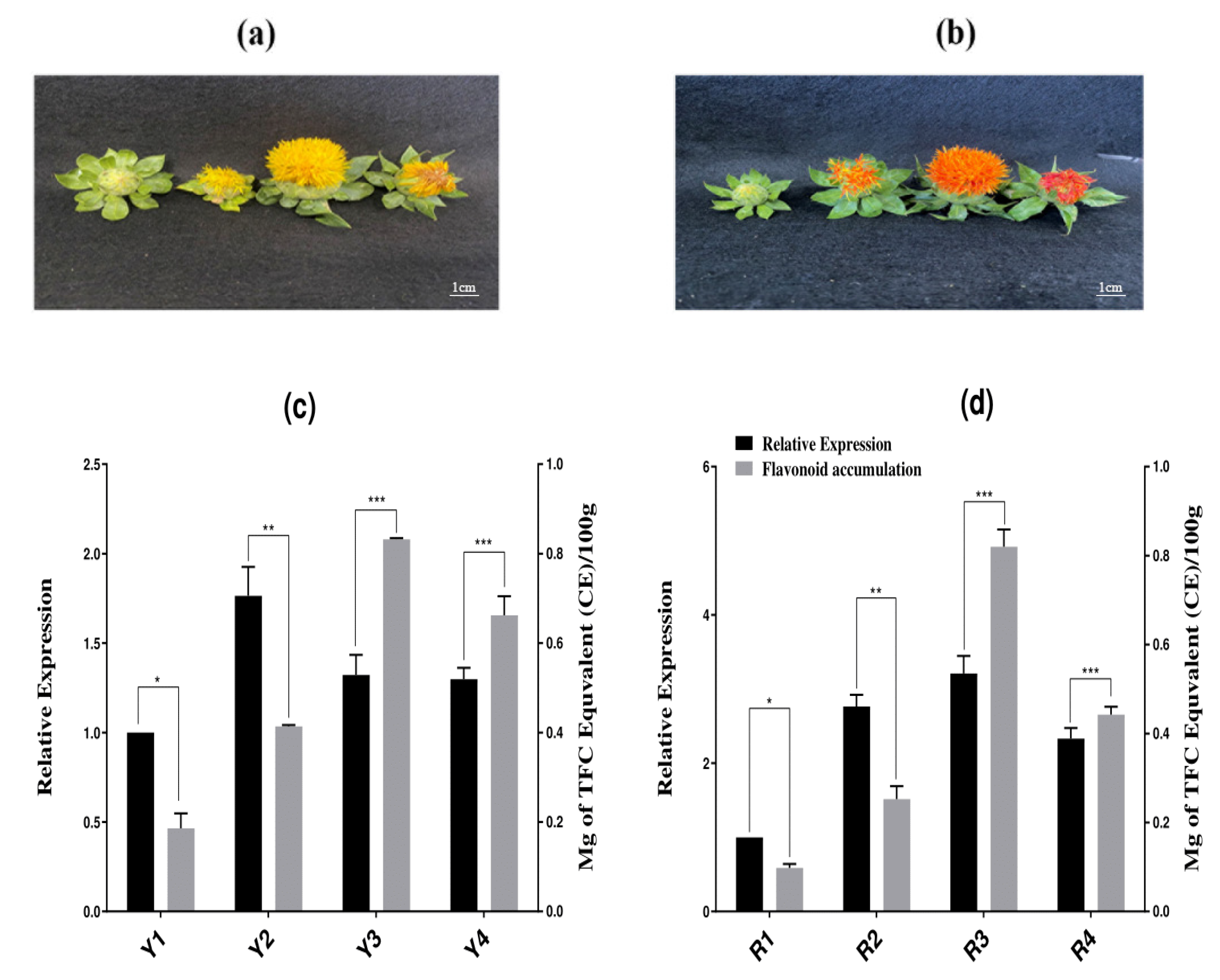
2.7. The Orchestrated Link between Transcriptional Regulation of CtCYP81E8 Encoding a Putative Isoflavone 2′-Hydroxylase and Flavonoid Accumulation in the Yellow and Red Flowering of Safflower
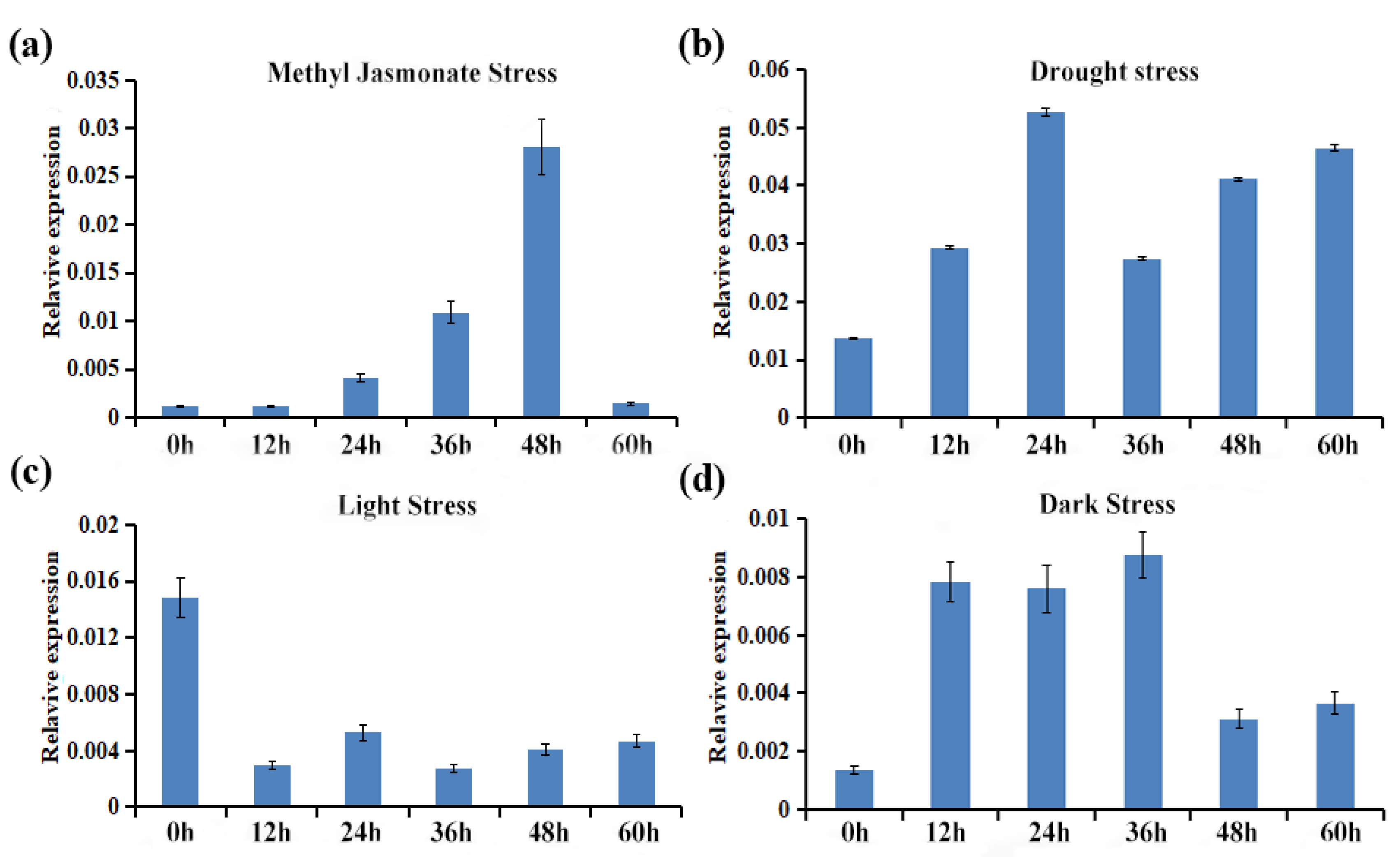
2.8. Expression Profiling of CtCYP81E8 Encoding a Putative Isoflavone 2′-Hydroxylase under Different Abiotic Stress Conditions
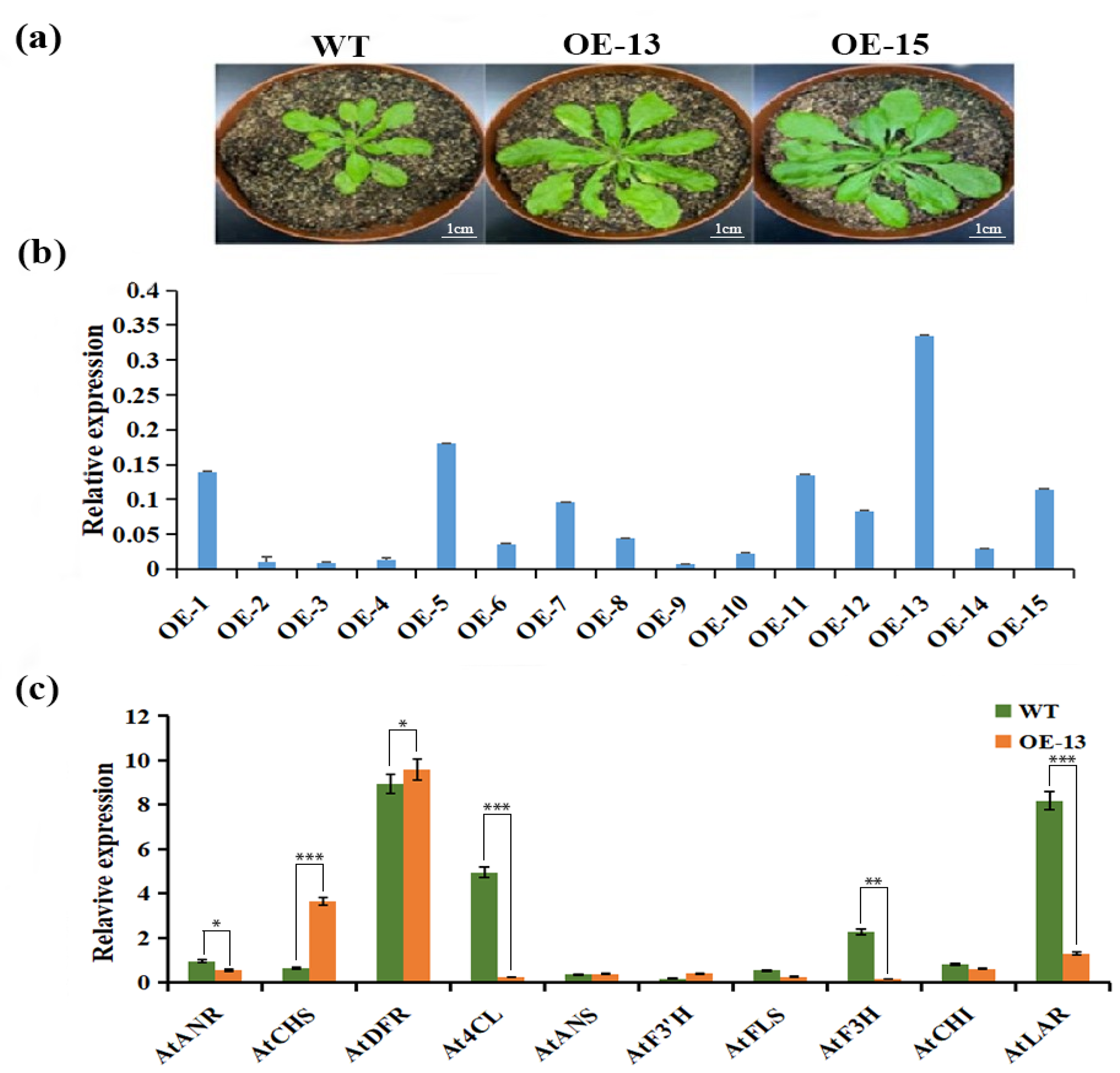
2.9. Expression Analysis of CtCYP81E8 Encoding a Putative Isoflavone 2′-Hydroxylase and Other Key Flavonoid Biosynthetic Genes in Transgenic Arabidopsis
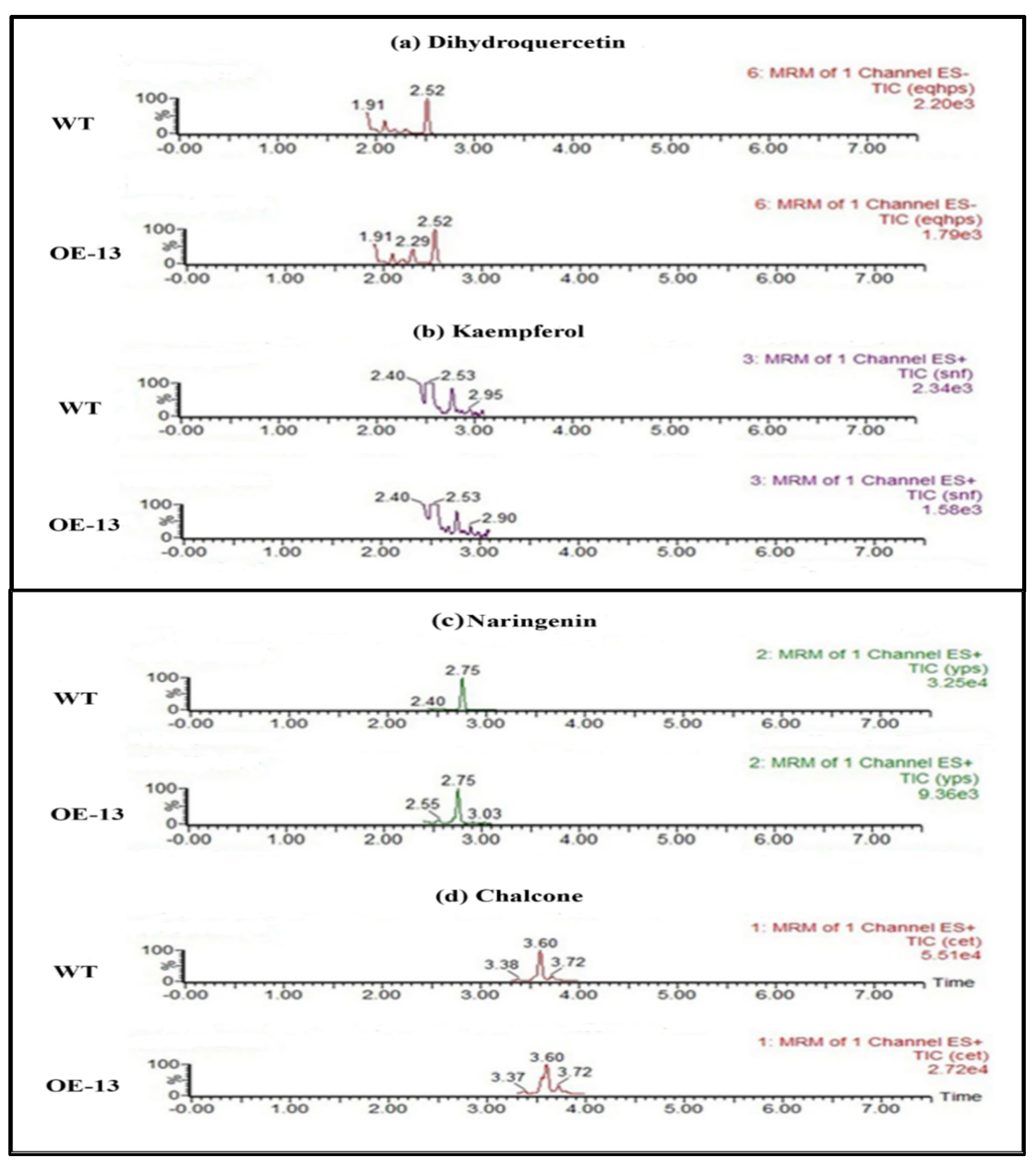
2.10. Determination of Flavonoid Contents in CtCYP81E8 Overexpressed Transgenic Arabidopsis Using HPLC-MS/MS
3. Discussion
4. Materials and Methods
4.1. Identification and Phylogenetic Analysis of CtCYP81E Subfamily in Safflower
4.2. Gene Structure, Protein Motifs, Promoter Analysis and Protein–Protein Interaction
4.3. Experimental Materials, Vectors, and Strains
4.4. Differential Expression of CtCYP81E-Encoding Genes in Safflower
4.5. CtCYP81E8 Expression and Metabolite Accumulation in Different Flowering Stages
4.6. Plant Material and Stress Conditions
4.7. Generation of CtCYP81E8 Overexpressed Transgenic Arabidopsis Lines
4.8. Expression Analysis of CtCYP81E8 and Other Key Flavonoid Biosynthetic Genes in Transgenic Lines
4.9. Determination of Flavonoids via HPLC-MS/MS Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China; Part 1; Beijing Chemical Industry Press: Beijing, China, 2015; pp. 188–189.
- Dai, Y.; Witkamp, G.-J.; Verpoorte, R.; Choi, Y.H. Natural deep eutectic solvents as a new extraction media for phenolic metabolites in Carthamus tinctorius L. Anal. Chem. 2013, 85, 6272–6278. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.; Wang, L.-Y.; Jin, W.-T.; Tang, Y.-P.; Jin, Y.; Shi, X.-Q.; Shang, L.-L.; Shang, E.-X.; Duan, J.-A. Comparative analysis of the effects of Hydroxysafflor yellow a and Anhydrosafflor yellow B in safflower series of herb pairs using prep-HPLC and a selective knock-out approach. Molecules 2016, 21, 1480. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Liu, H.; Sun, X.; Fu, F.; Zhang, X.; Wang, J.; An, J.; Ding, H. Hydroxysafflor yellow A protects rat brains against ischemia-reperfusion injury by antioxidant action. Neurosci. Lett. 2005, 386, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Tang, Y.; Li, S.; Duan, J.-A. Chemical and biological properties of quinochalcone C-glycosides from the florets of Carthamus tinctorius. Molecules 2013, 18, 15220–15254. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wang, Z.; Ma, C.; Tian, J.; Fu, F.; Li, C.; Guo, D.; Roeder, E.; Liu, K. Neuroprotective effects of hydroxysafflor yellow A: In Vivo and In Vitro studies. Planta Med. 2003, 69, 429–433. [Google Scholar] [PubMed]
- Zhang, Y.; Guo, J.; Dong, H.; Zhao, X.; Zhou, L.; Li, X.; Liu, J.; Niu, Y. Hydroxysafflor yellow A protects against chronic carbon tetrachloride-induced liver fibrosis. Eur. J. Pharmacol. 2011, 660, 438–444. [Google Scholar] [CrossRef]
- ul Hassan, S.S.; Jin, H.-Z.; Abu-Izneid, T.; Rauf, A.; Ishaq, M.; Suleria, H.A.R. Stress-driven discovery in the natural products: A gateway towards new drugs. Biomed. Pharmacother. 2019, 109, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Dong, Y.; Yao, N.; Zhang, Y.; Wang, N.; Cui, X.; Li, X.; Wang, Y.; Wang, F.; Yang, J. De Novo Sequencing and Analysis of the Safflower Transcriptome to Discover Putative Genes Associated with Safflor Yellow in Carthamus tinctorius L. Int. J. Mol. Sci. 2015, 16, 25657–25677. [Google Scholar] [CrossRef] [Green Version]
- Golkar, P.; Arzani, A.; Rezaei, A.M. Genetic variation in safflower (Carthamus tinctorious L.) for seed quality-related traits and inter-simple sequence repeat (ISSR) markers. Int. J. Mol. Sci. 2011, 12, 2664–2677. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, Z.; Chang, H.; Wang, Y.; Guo, M. Expression of CT-wpr, screened by cDNA-AFLP approach, associated with hydroxysafflor yellow A in Carthamus tinctorius L. Biochem. Syst. Ecol. 2010, 38, 1148–1155. [Google Scholar] [CrossRef]
- Chen, J.; Wang, J.; Wang, R.; Xian, B.; Ren, C.; Liu, Q.; Wu, Q.; Pei, J. Integrated metabolomics and transcriptome analysis on flavonoid biosynthesis in safflower (Carthamus tinctorius L.) under MeJA treatment. BMC Plant Biol. 2020, 20, 1–12. [Google Scholar] [CrossRef]
- De Montellano, P.R.O. Cytochrome P450: Structure, Mechanism, and Biochemistry; Springer Science & Business Media: Berlin, Germany, 2005. [Google Scholar]
- Yan, Q.; Cui, X.; Lin, S.; Gan, S.; Xing, H.; Dou, D. GmCYP82A3, a soybean cytochrome P450 family gene involved in the jasmonic acid and ethylene signaling pathway, enhances plant resistance to biotic and abiotic stresses. PLoS ONE 2016, 11, e0162253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, M.J.; Xu, Y.; Tang, X.; Huang, Y.; Liu, J.; Deng, X.; Xu, Q. CsCYT75B1, a citrus cytochrome P450 gene, is involved in accumulation of antioxidant flavonoids and induces drought tolerance in transgenic Arabidopsis. Antioxidants 2020, 9, 161. [Google Scholar] [CrossRef] [Green Version]
- Fischer, M.; Knoll, M.; Sirim, D.; Wagner, F.; Funke, S.; Pleiss, J. The Cytochrome P450 Engineering Database: A navigation and prediction tool for the cytochrome P450 protein family. Bioinformatics 2007, 23, 2015–2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andre, C.M.; Legay, S.; Deleruelle, A.; Nieuwenhuizen, N.; Punter, M.; Brendolise, C.; Cooney, J.M.; Lateur, M.; Hausman, J.F.; Larondelle, Y. Multifunctional oxidosqualene cyclases and cytochrome P450 involved in the biosynthesis of apple fruit triterpenic acids. New Phytol. 2016, 211, 1279–1294. [Google Scholar] [CrossRef] [Green Version]
- Hassan, S.S.U.; Muhammad, I.; Abbas, S.Q.; Hassan, M.; Majid, M.; Jin, H.-Z.; Bungau, S. Stress driven discovery of natural products from actinobacteria with anti-oxidant and cytotoxic activities including docking and admet properties. Int. J. Mol. Sci. 2021, 22, 11432. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. It takes a garden: How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol. 2001, 127, 1399–1404. [Google Scholar] [CrossRef]
- Akashi, T.; Aoki, T.; Ayabe, S.-I. Cloning and functional expression of a cytochrome P450 cDNA encoding 2-hydroxyisoflavanone synthase involved in biosynthesis of the isoflavonoid skeleton in licorice. Plant Physiol. 1999, 121, 821–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akashi, T.; Aoki, T.; Ayabe, S.-I. Identification of a cytochrome P450 cDNA encoding (2S)-flavanone 2-hydroxylase of licorice (Glycyrrhiza echinata L.; Fabaceae) which represents licodione synthase and flavone synthase II. FEBS Lett. 1998, 431, 287–290. [Google Scholar] [CrossRef] [Green Version]
- Akashi, T.; Aoki, T.; Ayabe, S.-I. CYP81E1, a cytochrome P450 cDNA of licorice (Glycyrrhiza echinata L.), encodes Isoflavone 2′-hydroxylase. Biochem. Biophys. Res. Commun. 1998, 251, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Kochs, G.; Grisebach, H. Enzymic synthesis of isoflavones. Eur. J. Biochem. 1986, 155, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Kessmann, H.; Choudhary, A.D.; Dixon, R.A. Stress responses in alfalfa (Medicago sativa L.) III. Induction of medicarpin and cytochrome P450 enzyme activities in elicitor-treated cell suspension cultures and protoplasts. Plant Cell Rep. 1990, 9, 38–41. [Google Scholar] [CrossRef] [PubMed]
- Clemens, S.; Hinderer, W.; Wittkampf, U.; Barz, W. Characterization of cytochrome P450-dependent isoflavone hydroxylases from chickpea. Phytochemistry 1993, 32, 653–657. [Google Scholar] [CrossRef]
- Chen, J.; Tang, X.; Ren, C.; Wei, B.; Wu, Y.; Wu, Q.; Pei, J. Full-length transcriptome sequences and the identification of putative genes for flavonoid biosynthesis in safflower. BMC Genom. 2018, 19, 548. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Dong, Y.; Yang, J.; Liu, X.; Wang, Y.; Yao, N.; Guan, L.; Wang, N.; Wu, J.; Li, X. De novo transcriptome of safflower and the identification of putative genes for oleosin and the biosynthesis of flavonoids. PLoS ONE 2012, 7, e30987. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chi, M.; Li, L.; Li, H.; Noman, M.; Yang, Y.; Ji, K.; Lan, X.; Qiang, W.; Du, L. Genome-Wide Identification, Expression Profiling, and Functional Validation of Oleosin Gene Family in Carthamus tinctorius L. Front. Plant Sci. 2018, 9, 1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Otani, K.; Takahashi, T.; Furuya, T.; Ayabe, S. Licodione synthase, a cytochrome P450 monooxygenase catalyzing 2-hydroxylation of 5-deoxyflavanone, in cultured Glycyrrhiza echinata L. cells. Plant Physiol. 1994, 105, 1427–1432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paquette, S.M.; Jensen, K.; Bak, S. A web-based resource for the Arabidopsis P450, cytochromes b5, NADPH-cytochrome P450 reductases, and family 1 glycosyltransferases (http://www.P450.kvl.dk). Phytochemistry 2009, 70, 1940–1947. [Google Scholar] [CrossRef]
- Gotoh, O. Divergent structures of Caenorhabditis elegans cytochrome P450 genes suggest the frequent loss and gain of introns during the evolution of nematodes. Mol. Biol. Evol. 1998, 15, 1447–1459. [Google Scholar] [CrossRef] [PubMed]
- Mani, V.; Lee, S.-K.; Yeo, Y.; Hahn, B.-S. A metabolic perspective and opportunities in pharmacologically important safflower. Metabolites 2020, 10, 253. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.R. Plant cytochrome P450s from moss to poplar. Phytochem. Rev. 2006, 5, 193–204. [Google Scholar] [CrossRef] [Green Version]
- Cramer, C.L.; Edwards, K.; Dron, M.; Liang, X.; Dildine, S.L.; Bolwell, G.P.; Dixon, R.A.; Lamb, C.J.; Schuch, W. Phenylalanine ammonia-lyase gene organization and structure. Plant Mol. Biol. 1989, 12, 367–383. [Google Scholar] [CrossRef]
- Roy, S.W.; Gilbert, W. Rates of intron loss and gain: Implications for early eukaryotic evolution. Proc. Natl. Acad. Sci. USA 2005, 102, 5773–5778. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Luo, Y.; Jia, L.; Qi, X.; Zeng, Q.; Xiang, Z.; He, N. Genome-wide identification and expression analyses of cytochrome P450 genes in mulberry (Morus notabilis). J. Integr. Plant Biol. 2014, 56, 887–901. [Google Scholar] [CrossRef] [PubMed]
- Barvkar, V.T.; Pardeshi, V.C.; Kale, S.M.; Kadoo, N.Y.; Gupta, V.S. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genom. 2012, 13, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paquette, S.M.; Bak, S.; Feyereisen, R. Intron–exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol. 2000, 19, 307–317. [Google Scholar] [CrossRef]
- Hassan, S.S.; Shah, S.A.A.; Pan, C.; Fu, L.; Cao, X.; Shi, Y.; Wu, X.; Wang, K.; Wu, B. Production of an antibiotic enterocin from a marine actinobacteria strain H1003 by metal-stress technique with enhanced enrichment using response surface methodology. Pak. J. Pharm. Sci. 2017, 30, 313–324. [Google Scholar]
- Bak, S.; Beisson, F.; Bishop, G.; Hamberger, B.; Höfer, R.; Paquette, S.; Werck-Reichhart, D. Cytochromes P450. Arab. Book Am. Soc. Plant Biol. 2011, 9, e0144. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Chu, H.; Chu, I.K.; Lo, C. CYP93G2 is a flavanone 2-hydroxylase required for C-glycosylflavone biosynthesis in rice. Plant Physiol. 2010, 154, 324–333. [Google Scholar] [CrossRef] [Green Version]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, X.; Zhu, L.; Liu, W.; Jiang, L. Programming an Orthogonal Self-Assembling Protein Cascade Based on Reactive Peptide–Protein Pairs for In Vitro Enzymatic Trehalose Production. J. Agric. Food Chem. 2022, 70, 4690–4700. [Google Scholar] [CrossRef]
- Guttikonda, S.K.; Trupti, J.; Bisht, N.C.; Chen, H.; An, Y.-Q.C.; Pandey, S.; Xu, D.; Yu, O. Whole genome co-expression analysis of soybean cytochrome P450 genes identifies nodulation-specific P450 monooxygenases. BMC Plant Biol. 2010, 10, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, L.; Wang, K.; Tan, J.; Li, W.; Li, S. Putative cytochrome P450 genes in rice genome (Oryza sativa L. ssp. indica) and their EST evidence. Sci. China Ser. C Life Sci. 2002, 45, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Jianyu, L.; Xu, T.; Noman, M.; Jameel, A.; Na, Y.; Yuanyuan, D.; Nan, W.; Xiaowei, L.; Fawei, W. Overexpression of a novel cytochrome P450 promotes flavonoid biosynthesis and osmotic stress tolerance in transgenic Arabidopsis. Genes 2019, 10, 756. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, N.; Li, T.; Liu, Y.; Hoang, N.Q.V.; Ma, X.; Zhang, X.; Liu, J.; Yao, N.; Liu, X.; Li, H. Molecular and biochemical rhythms in dihydroflavonol 4-reductase-mediated regulation of leucoanthocyanidin biosynthesis in Carthamus tinctorius L. Ind. Crops Prod. 2020, 156, 112838. [Google Scholar] [CrossRef]
- Hamberger, B.; Bak, S. Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120426. [Google Scholar] [CrossRef] [Green Version]
- Katsumata, T.; Fukazawa, J.; Magome, H.; Jikumaru, Y.; Kamiya, Y.; Natsume, M.; Kawaide, H.; Yamaguchi, S. Involvement of the CYP78A subfamily of cytochrome P450 monooxygenases in protonema growth and gametophore formation in the moss Physcomitrella patens. Biosci. Biotechnol. Biochem. 2011, 75, 331–336. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Liu, Y.; Cai, Y.; Zhang, F.; Xia, G.; Xiang, F. Cloning and functional analysis of geraniol 10-hydroxylase, a cytochrome P450 from Swertia mussotii Franch. Biosci. Biotechnol. Biochem. 2010, 74, 1583–1590. [Google Scholar] [CrossRef]
- Pang, C.H.; Li, K.; Wang, B. Overexpression of SsCHLAPXs confers protection against oxidative stress induced by high light in transgenic Arabidopsis thaliana. Physiol. Plant. 2011, 143, 355–366. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, H.; Liu, Y.; Tang, C.; Xu, X.; Liu, J. Rh (iii)-catalyzed synthesis of dibenzo [b, d] pyran-6-ones from aryl ketone O-acetyl oximes and quinones via C–H activation and C–C bond cleavage. RSC Adv. 2022, 12, 14435–14438. [Google Scholar] [CrossRef]
- Hong, Y.; Ahmad, N.; Zhang, J.; Lv, Y.; Zhang, X.; Ma, X.; Xiuming, L.; Na, Y. Genome-wide analysis and transcriptional reprogrammings of MYB superfamily revealed positive insights into abiotic stress responses and anthocyanin accumulation in Carthamus tinctorius L. Mol. Genet. Genom. 2022, 297, 125–145. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liu, X.; Ahmad, N.; Yang, L.; Fu, T.; Kong, J.; Yao, N.; Dong, Y.; Wang, N.; Li, X.; Wang, F. Molecular cloning and functional characterization of chalcone isomerase from Carthamus tinctorius. AMB Express 2019, 9, 132. [Google Scholar] [CrossRef] [PubMed]
- Mimmi, S.F.H.; Islam, A.; Islam, A. A comparative study of environment risk assessment guidelines for genetically engineered plants of developing and developed countries including Bangladesh. Sci. Herit. J. 2021, 5, 21–28. [Google Scholar] [CrossRef]

| Gene Name | Gene ID | Protein Length | PI | MW | Subcellular Localization | IS Index | Arabidopsis Homology | GRAVY |
|---|---|---|---|---|---|---|---|---|
| CYP81E1 | CCG011365.1 | 516 | 8.38 | 58.2 | Plasma membrane | 49.02 | AT4G37370.1 | −0.111 |
| CYP81E2 | CCG011366.1 | 501 | 8.96 | 57.29 | Plasma membrane | 46.06 | AT4G37370.1 | −0.125 |
| CYP81E3 | CCG011367.1 | 501 | 8.73 | 57.29 | Plasma membrane | 43.78 | AT4G37370.1 | −0.162 |
| CYP81E4 | CCG011368.1 | 339 | 5.61 | 38.28 | Plasma membrane | 40.66 | AT1G66540.1 | −0.332 |
| CYP81E5 | CCG007304.1 | 506 | 6.78 | 57.57 | Plasma membrane | 53.61 | AT4G37370.1 | −0.173 |
| CYP81E6 | CCG007305.1 | 506 | 6.78 | 57.57 | Plasma membrane | 53.61 | AT4G37370.1 | −0.173 |
| CYP81E7 | CCG007306.1 | 440 | 6.45 | 50 | Plasma membrane | 54.2 | AT4G37370.1 | −0.255 |
| CYP81E8 | CCG007309.1 | 337 | 6.49 | 38.48 | Plasma membrane | 44.14 | AT1G66540.1 | −0.32 |
| CYP81E9 | CCG007984.1 | 115 | 4.74 | 12.97 | Plasma membrane | 70.48 | AT4G37370.1 | −0.64 |
| CYP81E10 | CCG008862.1 | 246 | 9.35 | 27.85 | Plasma membrane | 47.65 | AT4G37320.1 | −0.147 |
| CYP81E11 | CCG008863.1 | 177 | 6.83 | 19.94 | Endoplasmic reticulum | 47.57 | AT1G66540.1 | −0.194 |
| CYP81E12 | CCG013393.1 | 435 | 8.49 | 49.49 | Plasma membrane | 44.98 | AT4G37360.1 | −0.143 |
| CYP81E13 | CCG013395.1 | 437 | 8.84 | 49.74 | Plasma membrane | 42.55 | AT4G37340.1 | −0.127 |
| CYP81E14 | CCG021962.1 | 516 | 7.68 | 59 | Plasma membrane | 45.29 | AT4G37340.1 | −0.156 |
| CYP81E15 | CCG004067.1 | 434 | 8.41 | 50.12 | Plasma membrane | 46.92 | AT4G37370.1 | −0.286 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Ahmad, N.; Hong, Y.; Zhu, M.; Zaman, S.; Wang, N.; Yao, N.; Liu, X. Molecular Characterization of an Isoflavone 2′-Hydroxylase Gene Revealed Positive Insights into Flavonoid Accumulation and Abiotic Stress Tolerance in Safflower. Molecules 2022, 27, 8001. https://doi.org/10.3390/molecules27228001
Liu J, Ahmad N, Hong Y, Zhu M, Zaman S, Wang N, Yao N, Liu X. Molecular Characterization of an Isoflavone 2′-Hydroxylase Gene Revealed Positive Insights into Flavonoid Accumulation and Abiotic Stress Tolerance in Safflower. Molecules. 2022; 27(22):8001. https://doi.org/10.3390/molecules27228001
Chicago/Turabian StyleLiu, Jianyu, Naveed Ahmad, Yingqi Hong, Meihua Zhu, Shah Zaman, Nan Wang, Na Yao, and Xiuming Liu. 2022. "Molecular Characterization of an Isoflavone 2′-Hydroxylase Gene Revealed Positive Insights into Flavonoid Accumulation and Abiotic Stress Tolerance in Safflower" Molecules 27, no. 22: 8001. https://doi.org/10.3390/molecules27228001
APA StyleLiu, J., Ahmad, N., Hong, Y., Zhu, M., Zaman, S., Wang, N., Yao, N., & Liu, X. (2022). Molecular Characterization of an Isoflavone 2′-Hydroxylase Gene Revealed Positive Insights into Flavonoid Accumulation and Abiotic Stress Tolerance in Safflower. Molecules, 27(22), 8001. https://doi.org/10.3390/molecules27228001








