Structural Activity and HAD Inhibition Efficiency of Pelargonidin and Its Glucoside—A Theoretical Approach
Abstract
:1. Introduction
2. Computation Detail
3. Discussion
3.1. Frontier Molecular Orbital (FMO) Analysis
3.2. Molecular Electrostatic Potential (MEP) Analysis
3.3. Molecular Descriptive Parameters
3.4. Insilico Analysis
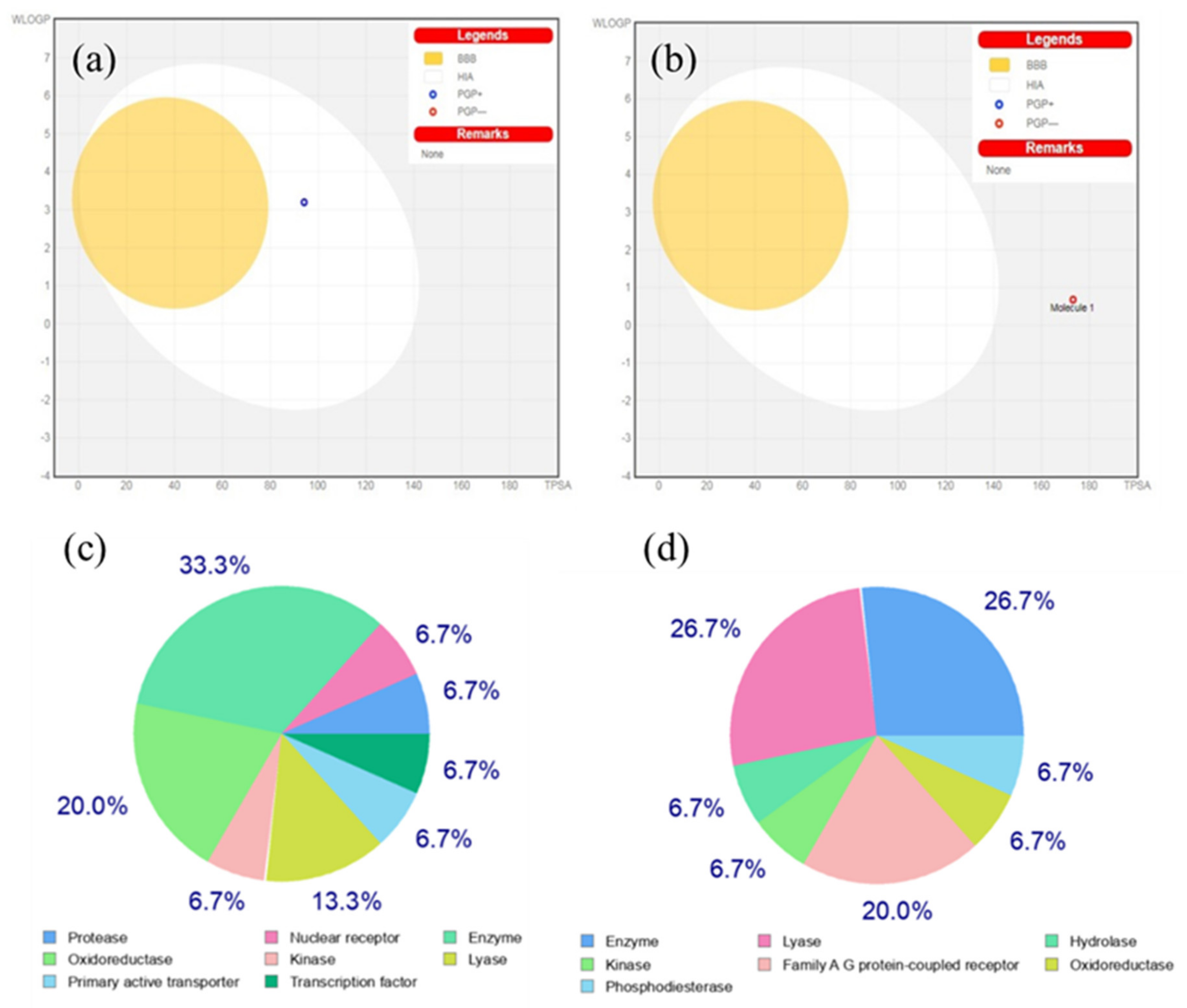
3.4.1. BOILED-Egg Analysis
3.4.2. Molecular Target Prediction and Docking Analysis
3.5. Molecular Dynamics Analysis
3.5.1. Root Mean Square Deviation
3.5.2. Radius of Gyration
3.5.3. Root Mean Square Fluctuation (RMSF)
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Tsuda, T. Dietary anthocyanin-rich plants: Biochemical basis and recent progress in health benefits studies. Mol. Nutr. Food Res. 2012, 56, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.X. Potential mechanisms of cancer chemoprevention by anthocyanins. Curr. Mol. Med. 2003, 3, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Saito, N.; Tatsuzawa, F.; Yokoi, M.; Kasahara, K.; Iida, S.; Shigihara, A.; Honda, T. Acylated Pelargonidin glycosides in red-purple flowers of Ipomoea purpurea. Phytochemistry 1996, 43, 1365–1370. [Google Scholar] [CrossRef]
- Fartzov, K.; Drenska, D. Effect of anthocyanins on phagocytic activity of mice peritoneal macrophages. J. Wine Res. 1994, 5, 237–240. [Google Scholar] [CrossRef]
- Monobe, M.; Ema, K.; Tokuda, Y.; Maeda-Yamamoto, M. Enhancement of phagocytic activity of macrophage-like cells by pyrogallol-type green tea polyphenols through caspase signaling pathways. Cytotechnology 2010, 62, 201–203. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, P.; Alvarado, C.; Puerto, M.; Schlumberger, A.; Jimenez, L.; De la Fuente, M. Improvement of leukocyte functions in prematurely aging mice after five weeks of diet supplementation with polyphenol-rich cereals. Nutrition 2006, 22, 913–921. [Google Scholar] [CrossRef]
- Denev, P.; Kratchanova, M.; Ciz, M.; Lojek, A.; Vasicek, O.; Nedelcheva, P. Biological activities of selected polyphenol-rich fruits related to immunity and gastrointestinal health. Food Chem. 2014, 157, 37–44. [Google Scholar] [CrossRef]
- Roth, S.; Spalinger, M.R.; Muller, I.; Lang, S.; Rogler, G.; Scharl, M. Bilberry-derived anthocyanins prevent IFN-γ-induced pro-inflammatory signalling and cytokine secretion in human THP-1 monocytic cells. Digestion 2014, 90, 179–189. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Li, Y.; Xie, J.; Xie, L.; Mo, J.; Chen, W. Bioavailability, Absorption, and Metabolism of Pelargonidin-Based Anthocyanins Using Sprague–Dawley Rats and Caco-2 Cell Monolayers. J. Agric. Food Chem. 2021, 69, 7841–7850. [Google Scholar] [CrossRef]
- Shishir, M.R.I.; Karim, N.; Xie, J.; Rashwan, A.K.; Chen, W. Colonic delivery of Pelargonidin-3-O-glucoside using pectin-chitosan-nanoliposome: Transport mechanism and bioactivity retention. Int. J. Biol. Macromol. 2020, 159, 341–355. [Google Scholar] [CrossRef]
- Samadder, A.; Abraham, S.K.; Khuda-Bukhsh, A.R. Nanopharmaceutical approach using Pelargonidin towards enhancement of efficacy for prevention of alloxan-induced DNA damage in L6 cells via activation of PARP and p53. Environ. Toxicol. Pharmacol. 2016, 43, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Kang, J.S.; Kang, N.J.; Je, B.I.; Lee, Y.J.; Park, Y.H.; Choi, Y.W. Pelargonidin suppresses adipogenesis in 3T3-L1 cells through inhibition of PPAR-γ signaling pathway. Arch. Biochem. Biophys. 2020, 686, 108365. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Pittman, H.E., 3rd; Prior, R.L. Pelargonidin is absorbed and metabolized differently than cyanidin after marionberry consumption in pigs. J. Nutr. 2004, 134, 2603–2610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noratto, G.D.; Angel-Morales, G.; Talcott, S.T.; Mertens-Talcott, S.U. Polyphenolics from acai (Euterpe oleracea Mart.) and red muscadine grape (Vitis rotundifolia) protect human umbilical vascular endothelial cells (HUVEC) from glucose- and lipopolysaccharide (LPS)-induced inflammation and target MicroRNA-126. J. Agric. Food Chem. 2011, 59, 7999–8012. [Google Scholar] [CrossRef]
- Esposito, D.; Chen, A.; Grace, M.H.; Komarnytsky, S.; Lila, M.A. Inhibitory effects of wild blueberry anthocyanins and other flavonoids on biomarkers of acute and chronic inflammation in vitro. J. Agric. Food Chem. 2014, 62, 7022–7028. [Google Scholar] [CrossRef]
- Wang, J.; Mazza, G. Effects of anthocyanins and other phenolic compounds on the production of tumor necrosis factor α in LPS/IFN-γ-activated RAW 264.7 macrophages. J. Agric. Food Chem. 2002, 50, 4183–4189. [Google Scholar] [CrossRef]
- Herath, H.M.T.; Takano-Ishikawa, Y.; Yamaki, K. Inhibitory effect of some flavonoids on tumor necrosis factor-α production in lipopolysaccharide-stimulated mouse macrophage cell line J774.1. J.Med. Food. 2003, 6, 365–370. [Google Scholar] [CrossRef]
- Jin, X.H.; Ohgami, K.; Shiratori, K.; Suzuki, Y.; Koyama, Y.; Yoshida, K. Effects of blue honeysuckle (Lonicera caerulea L.) extract on lipopolysaccharide-induced inflammation in vitro and in vivo. Exp. Eye Res. 2006, 82, 860–867. [Google Scholar] [CrossRef]
- Bauer, P.; Hess, B.; Lindahl, E. GROMACS 2022.3 Manual (2022.3). Zenodo 2022. [Google Scholar] [CrossRef]
- Bognar, E.; Sarszegi, Z.; Szabo, A.; Debreceni, B.; Kalman, N.; Tucsek, Z. Antioxidant and anti-inflammatory effects in RAW264.7 macrophages of malvidin, a major red wine polyphenol. PLoS ONE 2013, 8, e65355. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Lian, F.Z.; Zhu, Y.N.; Xia, M.; Wang, Q.; Ling, W.H. Cyanidin-3-O-β-glucoside inhibits LPS-induced expression of inflammatory mediators through decreasing IκBα phosphorylation in THP-1 cells. Inflamm. Res. 2010, 59, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, A.; Retterstol, L.; Laake, P.; Paur, I.; Kjolsrud-Bohn, S.; Sandvik, L. Anthocyanins inhibit nuclear factor-κB activation in monocytes and reduce plasma concentrations of pro-inflammatory mediators in healthy adults. J. Nutr. 2007, 137, 1951–1954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlsen, A.; Paur, I.; Bohn, S.K.; Sakhi, A.K.; Borge, G.I.; Serafini, M. Bilberry juice modulates plasma concentration of NF-κB related inflammatory markers in subjects at increased risk of CVD. Eur. J. Clin. Nutr. 2010, 49, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Mossalayi, M.D.; Rambert, J.; Renouf, E.; Micouleau, M.; Merillon, J.M. Grape polyphenols and propolis mixture inhibits inflammatory mediator release from human leukocytes and reduces clinical scores in experimental arthritis. Phytomedicine 2014, 21, 290–297. [Google Scholar] [CrossRef]
- Del Corno, M.; Varano, B.; Scazzocchino, B.; Filesi, C.; Masella, R.; Gessani, S. Protocatechuic acid inhibits human dentritic cell functional activation: Role of PPARy up-modulation. Immunobiology 2014, 219, 416–424. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Revision A.02; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Jeevitha, D.; Sadasivam, K.; Praveena, R.; Jayaprakasam, R. DFT study of glycosyl group reactivity in quercetin derivatives. J. Mol. Struct. 2016, 1120, 15–24. [Google Scholar] [CrossRef]
- Praveena, R.; Sadasivam, K.; Deepha, V.; Sivakumar, R. Antioxidant potential of orientin: A combined experimental and DFT approach. J. Mol. Struct. 2014, 1061, 114–123. [Google Scholar] [CrossRef]
- Sadasivam, K.; Kumaresan, R. Antioxidant behavior of mearnsetin and myricetin flavonoid compounds—A DFT study. Spectrochim. Acta Part A 2011, 79, 282–293. [Google Scholar] [CrossRef]
- Zdarilova, A.; Svobodova, A.R.; Chytilova, K.; Simanek, V.; Ulrichova, J. Polyphenolic fraction of Lonicera caerulea L. fruits reduces oxidative stress and inflammatory markers induced by lipopolysaccharide in gingival fibroblasts. Food Chem. Toxicol. 2010, 48, 1555–1561. [Google Scholar] [CrossRef]
- Galli, R.L.; Shukitt-Hale, B.; Youdim, K.A.; Joseph, J.A. Fruit polyphenolics and brain aging: Nutritional interventions targeting age-related neuronal and behavioral deficits. Ann. N. Y. Acad. Sci. 2002, 959, 128–132. [Google Scholar] [CrossRef]
- Dey, R.; Nandi, S.; Samadder, A. Pelargonidin mediated selective activation of p53 and PARP proteins in preventing food additive induced genotoxicity: An coupled molecular docking study. Eur. J. Pharm. Sci. 2020, 156, 105586. [Google Scholar] [CrossRef] [PubMed]
- Bub, A.; Watzl, B.; Blockhaus, M.; Briviba, K.; Liegibel, U.; Muller, H. Fruit juice consumption modulates antioxidative status, immune status and DNA damage. J. Nutr. Biochem. 2003, 14, 90–98. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg to Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barycki, J.J.; O’Brien, L.K.; Strauss, A.W.; Banaszak, L.J. Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase. J. Biol. Chem. 2000, 275, 27186–27196. [Google Scholar] [CrossRef]
- Gfeller, D.; Michielin, O.; Zoete, V. Shaping the interaction landscape of bioactive molecules. Bioinformatics 2013, 29, 3073–3079. [Google Scholar] [CrossRef] [Green Version]
- Kapoor, R.R.; James, C.; Flanagan, S.E.; Ellard, S.; Eaton, S.; Hussain, K. 3-Hydroxyacyl-Coenzyme a Dehydrogenase Deficiency and Hyper insulinemic Hypoglycemia: Characterization of a Novel Mutation and Severe Dietary Protein Sensitivity. J. Clin. Endocrinol. Metab. 2009, 94, 2221–2225. [Google Scholar] [CrossRef]





| Molecular Descriptors (eV) | Eo (eV) of Pelargonidin | Eo (eV) of Pelargonidin-3–O–Glucoside |
|---|---|---|
| IP (eV) | 4.07 | 4.19 |
| EA (eV) | 2.15 | 2.32 |
| ω (eV) | 0.95 | 0.93 |
| S (eV) | 0.52 | 0.53 |
| Χ (eV) | 3.11 | 3.25 |
| η (eV) | 5.07 | 5.68 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Praveena, R.; Balasankar, A.; Aruchamy, K.; Oh, T.; Polisetti, V.; Ramasundaram, S.; Anbazhakan, K. Structural Activity and HAD Inhibition Efficiency of Pelargonidin and Its Glucoside—A Theoretical Approach. Molecules 2022, 27, 8016. https://doi.org/10.3390/molecules27228016
Praveena R, Balasankar A, Aruchamy K, Oh T, Polisetti V, Ramasundaram S, Anbazhakan K. Structural Activity and HAD Inhibition Efficiency of Pelargonidin and Its Glucoside—A Theoretical Approach. Molecules. 2022; 27(22):8016. https://doi.org/10.3390/molecules27228016
Chicago/Turabian StylePraveena, Rangasamy, Athinarayanan Balasankar, Kanakaraj Aruchamy, Taehwan Oh, Veerababu Polisetti, Subramaniyan Ramasundaram, and Kandasamy Anbazhakan. 2022. "Structural Activity and HAD Inhibition Efficiency of Pelargonidin and Its Glucoside—A Theoretical Approach" Molecules 27, no. 22: 8016. https://doi.org/10.3390/molecules27228016
APA StylePraveena, R., Balasankar, A., Aruchamy, K., Oh, T., Polisetti, V., Ramasundaram, S., & Anbazhakan, K. (2022). Structural Activity and HAD Inhibition Efficiency of Pelargonidin and Its Glucoside—A Theoretical Approach. Molecules, 27(22), 8016. https://doi.org/10.3390/molecules27228016






