Investigating the Function of Human Jumping Translocation Breakpoint Protein (hJTB) and Its Interacting Partners through In-Solution Proteomics of MCF7 Cells
Abstract
1. Introduction
2. Results
2.1. JTB Dysregulation Is Associated with the EMT Process
2.2. JTB Dysregulation Is Associated with Mitochondrial Organization and Function
2.3. JTB-Related Proteins Are Involved in Oxidative Stress (OS)
2.4. JTB-Related Proteins Are Involved in Apoptotic Pathway
2.5. JTB-Related Proteins Are Involved in Interferon Alpha and Gamma Signaling Pathways
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Plasmids for Upregulation
4.3. Plasmids for Downregulation
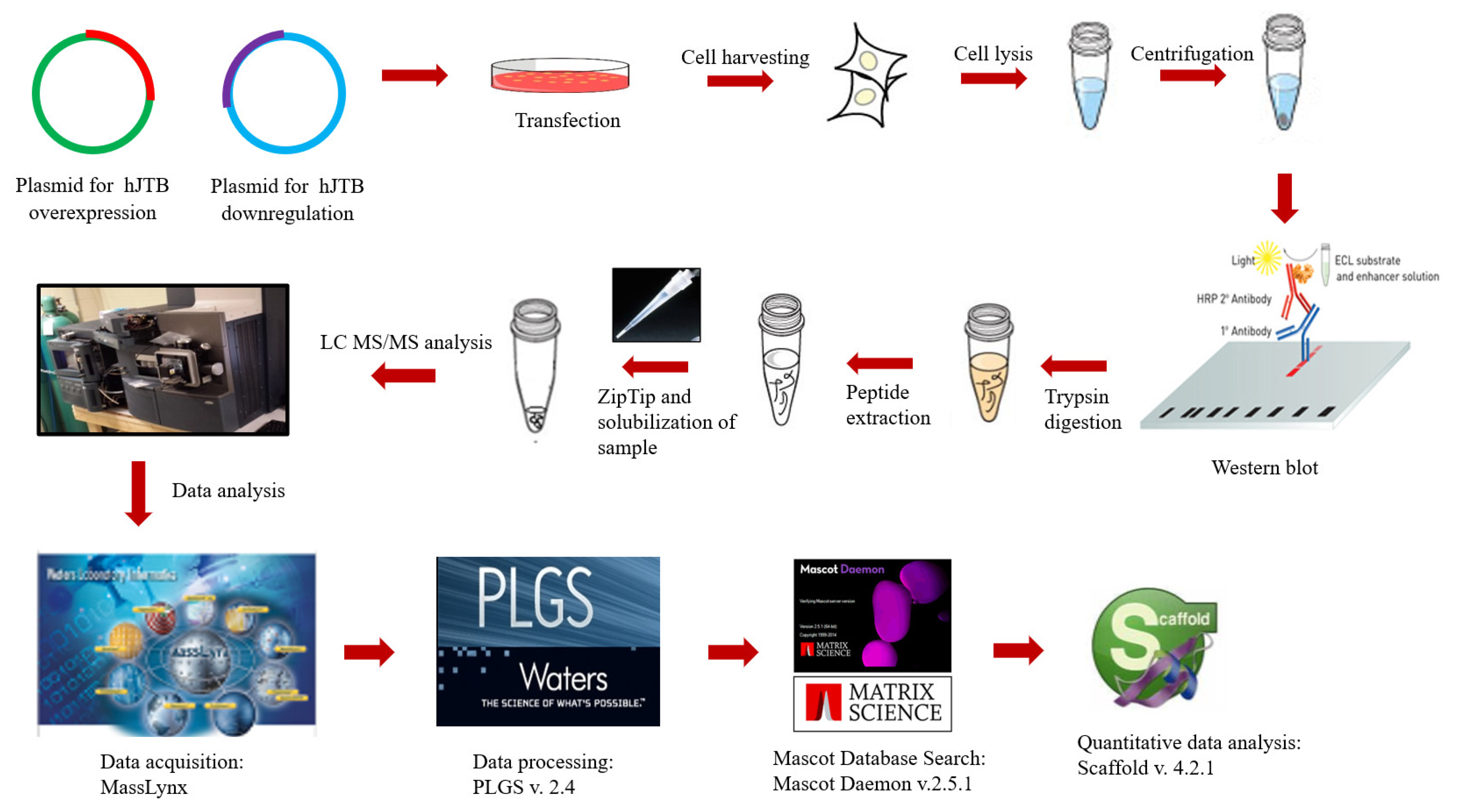
4.4. Transfection into MCF7 Cells
4.5. Western Blot Analysis
4.6. In-Solution Digestion
4.7. MS Analysis
4.8. Data Processing and Protein Identification
4.9. Data Sharing
4.10. Statistical Analysis
4.11. Gene Set Enrichment Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Qu, X. Cancer biomarker detection: Recent achievements and challenges. Chem. Soc. Rev. 2015, 44, 2963–2997. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.H.; Ladame, S. Chapter 1.1—Diagnostic, prognostic, and predictive biomarkers for cancer. In Bioengineering Innovative Solutions for Cancer; Ladame, S., Chang, J.Y.H., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 3–21. [Google Scholar] [CrossRef]
- Goossens, N.; Nakagawa, S.; Sun, X.; Hoshida, Y. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015, 4, 256–269. [Google Scholar] [PubMed]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Spellman, D.S.; Deinhardt, K.; Darie, C.C.; Chao, M.V.; Neubert, T.A. Stable isotopic labeling by amino acids in cultured primary neurons: Application to brain-derived neurotrophic factor-dependent phosphotyrosine-associated signaling. Mol. Cell. Proteom. 2008, 7, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Kanome, T.; Itoh, N.; Ishikawa, F.; Mori, K.; Kim-Kaneyama, J.; Nose, K.; Shibanuma, M. Characterization of Jumping translocation breakpoint (JTB) gene product isolated as a TGF-β1-inducible clone involved in regulation of mitochondrial function, cell growth and cell death. Oncogene 2007, 26, 5991–6001. [Google Scholar] [CrossRef]
- Jayathirtha, M.; Channaveerappa, D.; Darie, C. Investigation and Characterization of the Jumping Translocation Breakpoint (JTB) Protein using Mass Spectrometry based Proteomics. FASEB J. 2021, 35. [Google Scholar] [CrossRef]
- Xu, X.F.; Zhou, S.W.; Zhang, X.; Ye, Z.Q.; Zhang, J.H.; Ma, X.; Zheng, T.; Li, H.Z. Prostate androgen-regulated gene: A novel potential target for androgen-independent prostate cancer therapy. Asian J. Androl. 2006, 8, 455–462. [Google Scholar] [CrossRef]
- Hatakeyama, S.; Osawa, M.; Omine, M.; Ishikawa, F. JTB: A novel membrane protein gene at 1q21 rearranged in a jumping translocation. Oncogene 1999, 18, 2085–2090. [Google Scholar] [CrossRef]
- Rousseau, F.; Pan, B.; Fairbrother, W.J.; Bazan, J.F.; Lingel, A. The Structure of the Extracellular Domain of the Jumping Translocation Breakpoint Protein Reveals a Variation of the Midkine Fold. J. Mol. Biol. 2012, 415, 22–28. [Google Scholar] [CrossRef]
- Platica, O.; Chen, S.; Ivan, E.; Lopingco, M.; Holland, J.; Platica, M. PAR, a novel androgen regulated gene, ubiquitously expressed in normal and malignant cells. Int. J. Oncol. 2000, 16, 1055–1116. [Google Scholar] [CrossRef] [PubMed]
- Jayathirtha, M.; Neagu, A.-N.; Whitham, D.; Alwine, S.; Darie, C. Investigation of the effects of overexpression of jumping translocation breakpoint (JTB) protein in MCF7 cells for potential use as a biomarker in breast cancer. Am. J. Cancer Res. 2022, 12, 1784–1823. [Google Scholar] [PubMed]
- Jayathirtha, M.; Neagu, A.-N.; Whitham, D.; Alwine, S.; Darie, C.C. Investigation of the effects of downregulation of jumping translocation breakpoint (JTB) protein expression in MCF7 cells for potential use as a biomarker in breast cancer. Am. J. Cancer Res. 2022, 12, 4373–4398. [Google Scholar] [PubMed]
- Aslebagh, R.; Channaveerappa, D.; Pentecost, B.T.; Arcaro, K.F.; Darie, C.C. Combinatorial Electrophoresis and Mass Spectrometry-Based Proteomics in Breast Milk for Breast Cancer Biomarker Discovery. Adv. Exp. Med. Biol. 2019, 1140, 451–467. [Google Scholar] [CrossRef] [PubMed]
- Cox, H.D.; Chao, C.-K.; Patel, S.A.; Thompson, C.M. Efficient digestion and mass spectral analysis of vesicular glutamate transporter 1: A recombinant membrane protein expressed in yeast. J. Proteome Res. 2008, 7, 570–578. [Google Scholar] [CrossRef][Green Version]
- Coumans, J.V.F.; Gau, D.; Poljak, A.; Wasinger, V.; Roy, P.; Moens, P.D.J. Profilin-1 overexpression in MDA-MB-231 breast cancer cells is associated with alterations in proteomics biomarkers of cell proliferation, survival, and motility as revealed by global proteomics analyses. Omics 2014, 18, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.-H.; Shu, C.-W.; Chao, J.-K.; Lee, C.-H.; Fu, T.-Y.; Liou, H.-H.; Ger, L.-P.; Liu, P.-F. HSPD1 repressed E-cadherin expression to promote cell invasion and migration for poor prognosis in oral squamous cell carcinoma. Sci. Rep. 2019, 9, 8932. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Z.; Huang, Y.; Wei, W.; Ning, S.; Li, J.; Liang, X.; Liu, K.; Zhang, L. Plasma HSP90AA1 Predicts the Risk of Breast Cancer Onset and Distant Metastasis. Front. Cell Dev. Biol. 2021, 9, 639596. [Google Scholar] [CrossRef]
- Condelli, V.; Crispo, F.; Pietrafesa, M.; Lettini, G.; Matassa, D.S.; Esposito, F.; Landriscina, M.; Maddalena, F. HSP90 Molecular Chaperones, Metabolic Rewiring, and Epigenetics: Impact on Tumor Progression and Perspective for Anticancer Therapy. Cells 2019, 8, 532. [Google Scholar] [CrossRef]
- Liu, K.; Chen, J.; Yang, F.; Zhou, Z.; Liu, Y.; Guo, Y.; Hu, H.; Gao, H.; Li, H.; Zhou, W.; et al. BJ-B11, an Hsp90 Inhibitor, Constrains the Proliferation and Invasion of Breast Cancer Cells. Front. Oncol. 2019, 9, 1447. [Google Scholar] [CrossRef]
- de Freitas, G.B.; Penteado, L.; Miranda, M.M.; Filassi, J.R.; Baracat, E.C.; Linhares, I.M. The circulating 70kDa heat shock protein (HSPA1A) level is a potential biomarker for breast carcinoma and its progression. Sci. Rep. 2022, 12, 13012. [Google Scholar] [CrossRef] [PubMed]
- Nikotina, A.D.; Vladimirova, S.A.; Komarova, E.Y.; Alexeev, D.; Efremov, S.; Leonova, E.; Pavlov, R.; Kartsev, V.G.; Polonik, S.G.; Margulis, B.A.; et al. Prevention of High Glucose-Mediated EMT by Inhibition of Hsp70 Chaperone. Int. J. Mol. Sci. 2021, 22, 6902. [Google Scholar] [CrossRef] [PubMed]
- Sheng, C.; Qiu, J.; Wang, Y.; He, Z.; Wang, H.; Wang, Q.; Huang, Y.; Zhu, L.; Shi, F.; Chen, Y.; et al. Knockdown of Ran GTPase expression inhibits the proliferation and migration of breast cancer cells. Mol. Med. Rep. 2018, 18, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yao, S.; Luo, G.; Zhou, Y.; Fang, Q. Downregulation of RPS14 inhibits the proliferation and metastasis of estrogen receptor-positive breast cancer cells. Anti-Cancer Drugs 2021, 32, 1019–1028. [Google Scholar] [CrossRef]
- Wu, Q.; Gou, Y.; Wang, Q.; Jin, H.; Cui, L.; Zhang, Y.; He, L.; Wang, J.; Nie, Y.; Shi, Y.; et al. Downregulation of RPL6 by siRNA inhibits proliferation and cell cycle progression of human gastric cancer cell lines. PLoS ONE 2011, 6, e26401. [Google Scholar] [CrossRef]
- Yang, N.; Liu, Z.; Pang, S.; Wu, J.; Liang, J.; Sun, L. Predicative value of IFITM2 in renal clear cell carcinoma: IFITM2 is associated with lymphatic metastasis and poor clinical outcome. Biochem. Biophys. Res. Commun. 2021, 534, 157–164. [Google Scholar] [CrossRef]
- Yao, B.; Qu, S.; Hu, R.; Gao, W.; Jin, S.; Ju, J.; Zhao, Q. Delivery of platelet TPM3 mRNA into breast cancer cells via microvesicles enhances metastasis. FEBS Open Bio 2019, 9, 2159–2169. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Shen, Z.; Gao, L.; Yu, S.; Zhang, P.; Han, Z.; Kang, M. TPM3 mediates epithelial-mesenchymal transition in esophageal cancer via MMP2/MMP9. Ann. Transl. Med. 2021, 9, 1338. [Google Scholar] [CrossRef]
- Choi, H.-S.; Yim, S.-H.; Xu, H.-D.; Jung, S.-H.; Shin, S.-H.; Hu, H.-J.; Jung, C.-K.; Choi, J.Y.; Chung, Y.-J. Tropomyosin3 overexpression and a potential link to epithelial-mesenchymal transition in human hepatocellular carcinoma. BMC Cancer 2010, 10, 122. [Google Scholar] [CrossRef]
- Gao, S.; Wang, S.; Zhao, Z.; Zhang, C.; Liu, Z.; Ye, P.; Xu, Z.; Yi, B.; Jiao, K.; Naik, G.A.; et al. TUBB4A interacts with MYH9 to protect the nucleus during cell migration and promotes prostate cancer via GSK3β/β-catenin signalling. Nat. Commun. 2022, 13, 2792. [Google Scholar] [CrossRef]
- Patsialou, A.; Wang, Y.; Lin, J.; Whitney, K.; Goswami, S.; Kenny, P.; Condeelis, J. Selective gene-expression profiling of migratory tumor cells in vivo predicts clinical outcome in breast cancer patients. Breast Cancer Res. BCR 2012, 14, R139. [Google Scholar] [CrossRef] [PubMed]
- Kallergi, G.; Aggouraki, D.; Zacharopoulou, N.; Stournaras, C.; Georgoulias, V.; Martin, S.S. Evaluation of α-tubulin, detyrosinated α-tubulin, and vimentin in CTCs: Identification of the interaction between CTCs and blood cells through cytoskeletal elements. Breast Cancer Res. 2018, 20, 67. [Google Scholar] [CrossRef]
- Shin, D.; Park, J.; Han, D.; Moon, J.H.; Ryu, H.S.; Kim, Y. Identification of TUBB2A by quantitative proteomic analysis as a novel biomarker for the prediction of distant metastatic breast cancer. Clin. Proteom. 2020, 17, 16. [Google Scholar] [CrossRef] [PubMed]
- Saha, T. LAMP2A overexpression in breast tumors promotes cancer cell survival via chaperone-mediated autophagy. Autophagy 2012, 8, 1643–1656. [Google Scholar] [CrossRef]
- Hao, Y.; Kacal, M.; Ouchida, A.T.; Zhang, B.; Norberg, E.; Vakifahmetoglu-Norberg, H. Targetome analysis of chaperone-mediated autophagy in cancer cells. Autophagy 2019, 15, 1558–1571. [Google Scholar] [CrossRef] [PubMed]
- Shin, G.-C.; Moon, S.U.; Kang, H.S.; Choi, H.-S.; Han, H.D.; Kim, K.-H. PRKCSH contributes to tumorigenesis by selective boosting of IRE1 signaling pathway. Nat. Commun. 2019, 10, 3185. [Google Scholar] [CrossRef] [PubMed]
- Nigro, P.; Pompilio, G.; Capogrossi, M.C. Cyclophilin A: A key player for human disease. Cell Death Dis. 2013, 4, e888. [Google Scholar] [CrossRef]
- Cheng, S.; Luo, M.; Ding, C.; Peng, C.; Lv, Z.; Tong, R.; Xiao, H.; Xie, H.; Zhou, L.; Wu, J.; et al. Downregulation of Peptidylprolyl isomerase A promotes cell death and enhances doxorubicin-induced apoptosis in hepatocellular carcinoma. Gene 2016, 591, 236–244. [Google Scholar] [CrossRef]
- Guo, Y.; Jiang, M.; Zhao, X.; Gu, M.; Wang, Z.; Xu, S.; Yue, W. Cyclophilin A promotes non-small cell lung cancer metastasis via p38 MAPK. Thorac. Cancer 2018, 9, 120–128. [Google Scholar] [CrossRef]
- Liu, C.-c.; Dsaa, A.; Wang, W.; Wang, L.; Liu, W.-j.; Wang, J.-h.; Geng, Q.-r.; Lu, Y. ENO2 Promotes Cell Proliferation, Glycolysis, and Glucocorticoid-Resistance in Acute Lymphoblastic Leukemia. Cell. Physiol. Biochem. 2018, 46, 1525–1535. [Google Scholar] [CrossRef]
- Liu, D.; Mao, Y.; Chen, C.; Zhu, F.; Lu, W.; Ma, H. Expression patterns and clinical significances of ENO2 in lung cancer: An analysis based on Oncomine database. Ann. Transl. Med. 2020, 8, 639. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Cebrián, N.; Domingo-Ortí, I.; Poveda, J.L.; Vicent, M.J.; Puchades-Carrasco, L.; Pineda-Lucena, A. Multi-Omic Approaches to Breast Cancer Metabolic Phenotyping: Applications in Diagnosis, Prognosis, and the Development of Novel Treatments. Cancers 2021, 13, 4544. [Google Scholar] [CrossRef] [PubMed]
- Lytovchenko, O.; Kunji, E.R.S. Expression and putative role of mitochondrial transport proteins in cancer. Biochim. Biophys. Acta (BBA)-Bioenerg. 2017, 1858, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.-Y.; Choi, Y.; Jeon, Y.-K.; Kim, C.-W. Suppression of adenine nucleotide translocase-2 by vector-based siRNA in human breast cancer cells induces apoptosis and inhibits tumor growth in vitro and in vivo. Breast Cancer Res. 2008, 10, R11. [Google Scholar] [CrossRef] [PubMed]
- Abbas, W.; Kumar, A.; Herbein, G. The eEF1A Proteins: At the Crossroads of Oncogenesis, Apoptosis, and Viral Infections. Front. Oncol. 2015, 5, 75. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Beattie, A.; Baradaran, B.; Dray, E.; Duijf, P.H.G. Contradictory mRNA and protein misexpression of EEF1A1 in ductal breast carcinoma due to cell cycle regulation and cellular stress. Sci. Rep. 2018, 8, 13904. [Google Scholar] [CrossRef]
- Eigentler, A.; Tymoszuk, P.; Zwick, J.; Schmitz, A.A.; Pircher, A.; Kocher, F.; Schlicker, A.; Lesche, R.; Schäfer, G.; Theurl, I.; et al. The Impact of Cand1 in Prostate Cancer. Cancers 2020, 12, 428. [Google Scholar] [CrossRef]
- Alhammad, R. Bioinformatics Identification of TUBB as Potential Prognostic Biomarker for Worse Prognosis in ERα-Positive and Better Prognosis in ERα-Negative Breast Cancer. Diagnostics 2022, 12, 2067. [Google Scholar] [CrossRef]
- López-Mateo, I.; Villaronga, M.Á.; Llanos, S.; Belandia, B. The transcription factor CREBZF is a novel positive regulator of p53. Cell Cycle 2012, 11, 3887–3895. [Google Scholar] [CrossRef]
- Fang, J.; Jiang, G.; Mao, W.; Huang, L.; Huang, C.; Wang, S.; Xue, H.; Ke, J.; Ni, Q. Up-regulation of long noncoding RNA MBNL1-AS1 suppresses breast cancer progression by modulating miR-423-5p/CREBZF axis. Bioengineered 2022, 13, 3707–3723. [Google Scholar] [CrossRef]
- Luo, X.; Cheng, C.; Tan, Z.; Li, N.; Tang, M.; Yang, L.; Cao, Y. Emerging roles of lipid metabolism in cancer metastasis. Mol. Cancer 2017, 16, 76. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Chen, T.; Dong, L.; Li, T.; Xue, H.; Gao, B.; Ding, X.; Wang, H.; Li, H. Fatty acid synthase promotes breast cancer metastasis by mediating changes in fatty acid metabolism. Oncol. Lett. 2020, 21, 27. [Google Scholar] [CrossRef]
- Zhang, W.; Huang, J.; Tang, Y.; Yang, Y.; Huaidong, H. Inhibition of Fatty Acid Synthase (FASN) Affects the Proliferation and Apoptosis of HepG2 Hepatoma Carcinoma Cells via the β-catenin/C-myc Signaling Pathway. Ann. Hepatol. 2020, 19, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Wang, D.; Lei, M.; Guo, Y.; Cui, Y.; Chen, F.; Sun, W.; Chen, X. TPI1 activates the PI3K/AKT/mTOR signaling pathway to induce breast cancer progression by stabilizing CDCA5. J. Transl. Med. 2022, 20, 191. [Google Scholar] [CrossRef]
- Ding, C.; Fan, X.; Wu, G. Peroxiredoxin 1—An antioxidant enzyme in cancer. J. Cell. Mol. Med. 2017, 21, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Bajor, M.; Zych, A.O.; Graczyk-Jarzynka, A.; Muchowicz, A.; Firczuk, M.; Trzeciak, L.; Gaj, P.; Domagala, A.; Siernicka, M.; Zagozdzon, A.; et al. Targeting peroxiredoxin 1 impairs growth of breast cancer cells and potently sensitises these cells to prooxidant agents. Br. J. Cancer 2018, 119, 873–884. [Google Scholar] [CrossRef]
- Guo, Q.J.; Mills, J.N.; Bandurraga, S.G.; Nogueira, L.M.; Mason, N.J.; Camp, E.R.; Larue, A.C.; Turner, D.P.; Findlay, V.J. MicroRNA-510 promotes cell and tumor growth by targeting peroxiredoxin1 in breast cancer. Breast Cancer Res. BCR 2013, 15, R70. [Google Scholar] [CrossRef] [PubMed]
- Jezierska-Drutel, A.; Attaran, S.; Hopkins, B.L.; Skoko, J.J.; Rosenzweig, S.A.; Neumann, C.A. The peroxidase PRDX1 inhibits the activated phenotype in mammary fibroblasts through regulating c-Jun N-terminal kinases. BMC Cancer 2019, 19, 812. [Google Scholar] [CrossRef]
- Powell, L.E.; Foster, P.A. Protein disulphide isomerase inhibition as a potential cancer therapeutic strategy. Cancer Med. 2021, 10, 2812–2825. [Google Scholar] [CrossRef]
- Gomez, M.L.; Shah, N.; Kenny, T.C.; Jenkins, E.C.; Germain, D. SOD1 is essential for oncogene-driven mammary tumor formation but dispensable for normal development and proliferation. Oncogene 2019, 38, 5751–5765. [Google Scholar] [CrossRef]
- Cancemi, P.; Buttacavoli, M.; Roz, E.; Feo, S. Expression of Alpha-Enolase (ENO1), Myc Promoter-Binding Protein-1 (MBP-1) and Matrix Metalloproteinases (MMP-2 and MMP-9) Reflect the Nature and Aggressiveness of Breast Tumors. Int. J. Mol. Sci. 2019, 20, 3952. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, D.; Wei, Z.; Zhang, X.; Hu, Z.; Fu, H.; Xu, J.; Wang, W. The Antitumor Effect of TPD52L2 Silencing on Oxaliplatin-Resistant Gastric Carcinoma Is Related to Endoplasmic Reticulum Stress In Vitro. Evid.-Based Complement. Altern. Med. Ecam 2022, 2022, 4451178. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.-S.; Kao, H.-Y. Alpha-actinin 4 and tumorigenesis of breast cancer. Vitam. Horm. 2013, 93, 323–351. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Patel, S.A.; Hassan, M.K.; Mohapatra, N.; Pattanaik, N.; Dixit, M. Reduced IQGAP2 expression promotes EMT and inhibits apoptosis by modulating the MEK-ERK and p38 signaling in breast cancer irrespective of ER status. Cell Death Dis. 2021, 12, 389. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Han, X.; Zheng, S.; Liu, Q.; Tuerxun, A.; Zhang, Q.; Yang, L.; Lu, X. CALM1 promotes progression and dampens chemosensitivity to EGFR inhibitor in esophageal squamous cell carcinoma. Cancer Cell Int. 2021, 21, 121. [Google Scholar] [CrossRef]
- Ochieng, J.; Nangami, G.; Sakwe, A.; Moye, C.; Alvarez, J.; Whalen, D.; Thomas, P.; Lammers, P. Impact of Fetuin-A (AHSG) on Tumor Progression and Type 2 Diabetes. Int. J. Mol. Sci. 2018, 19, 2211. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhou, Z.; Wei, R.; Xiao, C.; Zhang, H.; Fan, T.; Zheng, B.; Li, C.; He, J. The RNA-binding protein PCBP1 represses lung adenocarcinoma progression by stabilizing DKK1 mRNA and subsequently downregulating β-catenin. J. Transl. Med. 2022, 20, 343. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, K.; Zhang, J.; Liu, S.S.; Dai, L.; Zhang, J.-Y. Using proteomic approach to identify tumor-associated proteins as biomarkers in human esophageal squamous cell carcinoma. J. Proteome Res. 2011, 10, 2863–2872. [Google Scholar] [CrossRef]
- Sanchez-Martin, D.; Martinez-Torrecuadrada, J.; Teesalu, T.; Sugahara, K.; Alvarez de Cienfuegos, A.; Ximénez-Embún, P.; Fernández-Periáñez, R.; Martín, M.; Molina-Privado, I.; Ruppen, I.; et al. Proteasome activator complex PA28 identified as an accessible target in prostate cancer by in vivo selection of human antibodies. Proc. Natl. Acad. Sci. USA 2013, 110, 13791–13796. [Google Scholar] [CrossRef]
- Wang, Q.; Pan, F.; Li, S.; Huang, R.; Wang, X.; Wang, S.; Liao, X.; Li, D.; Zhang, L. The prognostic value of the proteasome activator subunit gene family in skin cutaneous melanoma. J. Cancer 2019, 10, 2205–2219. [Google Scholar] [CrossRef]
- Gu, Y.; Barwick, B.G.; Shanmugam, M.; Hofmeister, C.C.; Kaufman, J.; Nooka, A.; Gupta, V.; Dhodapkar, M.; Boise, L.H.; Lonial, S. Downregulation of PA28α induces proteasome remodeling and results in resistance to proteasome inhibitors in multiple myeloma. Blood Cancer J. 2020, 10, 125. [Google Scholar] [CrossRef] [PubMed]
- Ananthi, S.; Lakshmi, N.; Paul, A.; Kumaraswamy, A.; Mahalingam, S. Global Quantitative Proteomics reveal Deregulation of Cytoskeletal and Apoptotic Signalling Proteins in Oral Tongue Squamous Cell Carcinoma. Sci. Rep. 2018, 8, 1567. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Lim, S.G.; Koay, E.S.-C. Proteomic identification of down-regulation of oncoprotein DJ-1 and proteasome activator subunit 1 in hepatitis B virus-infected well-differentiated hepatocellular carcinoma. Int. J. Oncol. 2007, 31, 577–584. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fu, D.; He, C.; Wei, J.; Zhang, Z.; Luo, Y.; Tan, H.; Ren, C. PGK1 is a Potential Survival Biomarker and Invasion Promoter by Regulating the HIF-1α–Mediated Epithelial-Mesenchymal Transition Process in Breast Cancer. Cell. Physiol. Biochem. 2018, 51, 2434–2444. [Google Scholar] [CrossRef] [PubMed]
- Husi, H.; Skipworth, R.J.E.; Cronshaw, A.; Fearon, K.C.H.; Ross, J.A. Proteomic identification of potential cancer markers in human urine using subtractive analysis. Int. J. Oncol. 2016, 48, 1921–1932. [Google Scholar] [CrossRef]
- Barger, C.J.; Zhang, W.; Sharma, A.; Chee, L.; James, S.R.; Kufel, C.N.; Miller, A.; Meza, J.; Drapkin, R.; Odunsi, K.; et al. Expression of the POTE gene family in human ovarian cancer. Sci. Rep. 2018, 8, 17136. [Google Scholar] [CrossRef]
- Hsu, M.-T.; Wang, Y.-K.; Tseng, Y.J. Exosomal Proteins and Lipids as Potential Biomarkers for Lung Cancer Diagnosis, Prognosis, and Treatment. Cancers 2022, 14, 732. [Google Scholar] [CrossRef]
- Nami, B.; Wang, Z. Genetics and Expression Profile of the Tubulin Gene Superfamily in Breast Cancer Subtypes and Its Relation to Taxane Resistance. Cancers 2018, 10, 274. [Google Scholar] [CrossRef]
- Wang, D.; Jiao, Z.; Ji, Y.; Zhang, S. Elevated TUBA1A Might Indicate the Clinical Outcomes of Patients with Gastric Cancer, Being Associated with the Infiltration of Macrophages in the Tumor Immune Microenvironment. J. Gastrointest. Liver Dis. 2020, 29, 509–522. [Google Scholar] [CrossRef]
- Zoppino, F.C.M.; Guerrero-Gimenez, M.E.; Castro, G.N.; Ciocca, D.R. Comprehensive transcriptomic analysis of heat shock proteins in the molecular subtypes of human breast cancer. BMC Cancer 2018, 18, 700. [Google Scholar] [CrossRef]
- Xiao, R.; Shen, S.; Yu, Y.; Pan, Q.; Kuang, R.; Huang, H. TMSB10 promotes migration and invasion of cancer cells and is a novel prognostic marker for renal cell carcinoma. Int. J. Clin. Exp. Pathol. 2019, 12, 305–312. [Google Scholar] [PubMed]
- Zhang, X.; Ren, D.; Guo, L.; Wang, L.; Wu, S.; Lin, C.; Ye, L.; Zhu, J.; Li, J.; Song, L.; et al. Thymosin beta 10 is a key regulator of tumorigenesis and metastasis and a novel serum marker in breast cancer. Breast Cancer Res. BCR 2017, 19, 15. [Google Scholar] [CrossRef] [PubMed]
- Jin, W. Novel Insights into PARK7 (DJ-1), a Potential Anti-Cancer Therapeutic Target, and Implications for Cancer Progression. J.Clin. Med. 2020, 9, 1256. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.H.; Peters, M.; Jang, Y.; Shi, W.; Pintilie, M.; Fletcher, G.C.; DeLuca, C.; Liepa, J.; Zhou, L.; Snow, B.; et al. DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell 2005, 7, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wei, J.; Zhang, H.; Zheng, X.; Zhou, H.; Luo, Y.; Yang, J.; Deng, Q.; Huang, S.; Fu, Z. PRDX2 promotes the proliferation of colorectal cancer cells by increasing the ubiquitinated degradation of p53. Cell Death Dis. 2021, 12, 605. [Google Scholar] [CrossRef] [PubMed]
- Nicolussi, A.; D’Inzeo, S.; Capalbo, C.; Giannini, G.; Coppa, A. The role of peroxiredoxins in cancer. Mol. Clin. Oncol. 2017, 6, 139–153. [Google Scholar] [CrossRef]
- Li, H.; Yang, H.; Wang, D.; Zhang, L.; Ma, T. Peroxiredoxin2 (Prdx2) Reduces Oxidative Stress and Apoptosis of Myocardial Cells Induced by Acute Myocardial Infarction by Inhibiting the TLR4/Nuclear Factor kappa B (NF-κB) Signaling Pathway. Med. Sci. Monit. 2020, 26, e926281. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Zhang, F.; Hong, C.-Q.; Giuliano, A.E.; Cui, X.-J.; Zhou, G.-J.; Zhang, G.-J.; Cui, Y.-K. Critical protein GAPDH and its regulatory mechanisms in cancer cells. Cancer Biol. Med. 2015, 12, 10–22. [Google Scholar] [CrossRef]
- Révillion, F.; Pawlowski, V.; Hornez, L.; Peyrat, J. Glyceraldhyde-3-phosphate dehydrogenase gene expression in human breast cancer. Eur. J. Cancer 2000, 36, 1038–1042. [Google Scholar] [CrossRef]
- Liu, K.; Tang, Z.; Huang, A.; Chen, P.; Liu, P.; Yang, J.; Lu, W.; Liao, J.; Sun, Y.; Wen, S.; et al. Glyceraldehyde-3-phosphate dehydrogenase promotes cancer growth and metastasis through upregulation of SNAIL expression. Int. J. Oncol. 2016, 50, 252–262. [Google Scholar] [CrossRef]
- Tarrado-Castellarnau, M.; Diaz-Moralli, S.; Polat, I.H.; Sanz-Pamplona, R.; Alenda, C.; Moreno, V.; Castells, A.; Cascante, M. Glyceraldehyde-3-phosphate dehydrogenase is overexpressed in colorectal cancer onset. Transl. Med. Commun. 2017, 2, 6. [Google Scholar] [CrossRef]
- Suresh, R.; Diaz, R.J. The remodelling of actin composition as a hallmark of cancer. Transl. Oncol. 2021, 14, 101051. [Google Scholar] [CrossRef]
- Suresh, R.; Picard, D.; Lo, R.; Beaulieu, J.; Remke, M.; Diaz, R.J. Expression of cell type incongruent alpha-cardiac actin 1 subunit in medulloblastoma reveals a novel mechanism for cancer cell survival and control of migration. Neuro-Oncol. Adv. 2021, 3, vdab064. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.-W.; Ambe, C.M.; Miller, T.C.; Chen, J.-Q.; Wiegand, G.W.; Anderson, A.J.; Ray, S.; Mullinax, J.E.; Hari, D.M.; Koizumi, T.; et al. Liver Label Retaining Cancer Cells Are Relatively Resistant to the Reported Anti-Cancer Stem Cell Drug Metformin. J. Cancer 2016, 7, 1142–1151. [Google Scholar] [CrossRef]
- Li, J.; Ge, Z. High HSPA8 expression predicts adverse outcomes of acute myeloid leukemia. BMC Cancer 2021, 21, 475. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-Y.; Liao, P.; Yan, L.-Y.; Zhao, Q.-Y.; Xie, Z.-Y.; Dong, J.; Sun, H.-T. Correlation of MKI67 with prognosis, immune infiltration, and T cell exhaustion in hepatocellular carcinoma. BMC Gastroenterol. 2021, 21, 416. [Google Scholar] [CrossRef] [PubMed]
- Mrouj, K.; Andrés-Sánchez, N.; Dubra, G.; Singh, P.; Sobecki, M.; Chahar, D.; Al Ghoul, E.; Aznar, A.B.; Prieto, S.; Pirot, N.; et al. Ki-67 regulates global gene expression and promotes sequential stages of carcinogenesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2026507118. [Google Scholar] [CrossRef]
- Shi, Y.; Du, L.; Lv, D.; Li, H.; Shang, J.; Lu, J.; Zhou, L.; Bai, L.; Tang, H. Exosomal Interferon-Induced Transmembrane Protein 2 Transmitted to Dendritic Cells Inhibits Interferon Alpha Pathway Activation and Blocks Anti-Hepatitis B Virus Efficacy of Exogenous Interferon Alpha. Hepatology 2019, 69, 2396–2413. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Boku, S.; Kobayashi, K.; Kurumida, Y.; Sukeno, M.; Masuda, M.; Mizushima, K.; Kato, C.; Iizumi, Y.; Hirota, K.; et al. A chemoproteoinformatics approach demonstrates that aspirin increases sensitivity to MEK inhibition by directly binding to RPS5. PNAS Nexus 2022, 1, pgac059. [Google Scholar] [CrossRef]
- Yang, C.-M.; Ji, S.; Li, Y.; Fu, L.-Y.; Jiang, T.; Meng, F.-D. β-Catenin promotes cell proliferation, migration, and invasion but induces apoptosis in renal cell carcinoma. Onco Targets Ther. 2017, 10, 711–724. [Google Scholar] [CrossRef]
- Kim, W.K.; Kwon, Y.; Jang, M.; Park, M.; Kim, J.; Cho, S.; Jang, D.G.; Lee, W.-B.; Jung, S.H.; Choi, H.J.; et al. β-catenin activation down-regulates cell-cell junction-related genes and induces epithelial-to-mesenchymal transition in colorectal cancers. Sci. Rep. 2019, 9, 18440. [Google Scholar] [CrossRef] [PubMed]
- Desai, S.D. ISG15: A double edged sword in cancer. Oncoimmunology 2015, 4, e1052935. [Google Scholar] [CrossRef] [PubMed]
- Bolado-Carrancio, A.; Lee, M.; Ewing, A.; Muir, M.; Macleod, K.G.; Gallagher, W.M.; Nguyen, L.K.; Carragher, N.O.; Semple, C.A.; Brunton, V.G.; et al. ISGylation drives basal breast tumour progression by promoting EGFR recycling and Akt signalling. Oncogene 2021, 40, 6235–6247. [Google Scholar] [CrossRef] [PubMed]
- Bektas, N.; Noetzel, E.; Veeck, J.; Press, M.F.; Kristiansen, G.; Naami, A.; Hartmann, A.; Dimmler, A.; Beckmann, M.W.; Knüchel, R.; et al. The ubiquitin-like molecule interferon-stimulated gene 15 (ISG15) is a potential prognostic marker in human breast cancer. Breast Cancer Res. 2008, 10, R58. [Google Scholar] [CrossRef]
- Madureira, P.A.; Hill, R.; Miller, V.A.; Giacomantonio, C.A.; Lee, P.W.K.; Waisman, D.M. Annexin A2 is a novel Cellular Redox Regulatory Protein involved in Tumorigenesis. Oncotarget 2011, 2, 1075–1093. [Google Scholar] [CrossRef]
- Yan, L.-R.; Shen, S.-X.; Wang, A.; Xi, D.; Liu, Y.-N.; Yuan, Y.; Xu, Q. Comprehensive Pan-Cancer Analysis of Heat Shock Protein 110, 90, 70, and 60 Families. Front. Mol. Biosci. 2021, 8, 726244. [Google Scholar] [CrossRef]
- Zhang, Y.; Cai, H.; Liao, Y.; Zhu, Y.; Wang, F.; Hou, J. Activation of PGK1 under hypoxic conditions promotes glycolysis and increases stem cell-like properties and the epithelial-mesenchymal transition in oral squamous cell carcinoma cells via the AKT signalling pathway. Int. J. Oncol. 2020, 57, 743–755. [Google Scholar] [CrossRef]
- Conacci-Sorrell, M.; Zhurinsky, J.; Ben-Ze’ev, A. The cadherin-catenin adhesion system in signaling and cancer. J. Clin. Investig. 2002, 109, 987–991. [Google Scholar] [CrossRef]
- Leggett, S.E.; Hruska, A.M.; Guo, M.; Wong, I.Y. The epithelial-mesenchymal transition and the cytoskeleton in bioengineered systems. Cell Commun. Signal. 2021, 19, 32. [Google Scholar] [CrossRef]
- González-Mariscal, L.; Miranda, J.; Gallego-Gutiérrez, H.; Cano-Cortina, M.; Amaya, E. Relationship between apical junction proteins, gene expression and cancer. Biochim. Biophys. Acta (BBA)-Biomembr. 2020, 1862, 183278. [Google Scholar] [CrossRef]
- Gehren, A.S.; Rocha, M.R.; de Souza, W.F.; Morgado-Díaz, J.A. Alterations of the apical junctional complex and actin cytoskeleton and their role in colorectal cancer progression. Tissue Barriers 2015, 3, e1017688. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Peng, H.; Yan, M.; Chen, S. Silencing ACTG1 Expression Induces Prostate Cancer Epithelial Mesenchymal Transition Through MAPK/ERK Signaling Pathway. DNA Cell Biol. 2021, 40, 1445–1455. [Google Scholar] [CrossRef] [PubMed]
- Tentler, D.; Lomert, E.; Novitskaya, K.; Barlev, N.A. Role of ACTN4 in Tumorigenesis, Metastasis, and EMT. Cells 2019, 8, 1427. [Google Scholar] [CrossRef]
- Honda, K. The biological role of actinin-4 (ACTN4) in malignant phenotypes of cancer. Cell Biosci. 2015, 5, 41. [Google Scholar] [CrossRef]
- Tanabe, S.; Kawabata, T.; Aoyagi, K.; Yokozaki, H.; Sasaki, H. Gene expression and pathway analysis of CTNNB1 in cancer and stem cells. World J. Stem Cells 2016, 8, 384–395. [Google Scholar] [CrossRef]
- Ferreira, L.T.; Figueiredo, A.C.; Orr, B.; Lopes, D.; Maiato, H. Dissecting the role of the tubulin code in mitosis. Methods Cell Biol. 2018, 144, 33–74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Fan, M.; Napolitano, F.; Gao, X.; Xu, Y.; Li, L. Transcriptomic analysis identifies organ-specific metastasis genes and pathways across different primary sites. J. Transl. Med. 2021, 19, 31. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S. Heat shock proteins and cancer: Intracellular chaperones or extracellular signalling ligands? Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20160524. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, C.; Zhou, B.P. Metabolic reprogram associated with epithelial-mesenchymal transition in tumor progression and metastasis. Genes Dis. 2020, 7, 172–184. [Google Scholar] [CrossRef]
- Kondaveeti, Y.; Guttilla Reed, I.K.; White, B.A. Epithelial–mesenchymal transition induces similar metabolic alterations in two independent breast cancer cell lines. Cancer Lett. 2015, 364, 44–58. [Google Scholar] [CrossRef]
- Luu, T. Epithelial-Mesenchymal Transition and Its Regulation Mechanisms in Pancreatic Cancer. Front. Oncol. 2021, 11, 646399. [Google Scholar] [CrossRef] [PubMed]
- Rebane-Klemm, E.; Truu, L.; Reinsalu, L.; Puurand, M.; Shevchuk, I.; Chekulayev, V.; Timohhina, N.; Tepp, K.; Bogovskaja, J.; Afanasjev, V.; et al. Mitochondrial Respiration in KRAS and BRAF Mutated Colorectal Tumors and Polyps. Cancers 2020, 12, 815. [Google Scholar] [CrossRef]
- Gill, K.S.; Fernandes, P.; O’Donovan, T.R.; McKenna, S.L.; Doddakula, K.K.; Power, D.G.; Soden, D.M.; Forde, P.F. Glycolysis inhibition as a cancer treatment and its role in an anti-tumour immune response. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2016, 1866, 87–105. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, D.; Hu, X. A Potential Fatty Acid Metabolism-Related Gene Signature for Prognosis in Clear Cell Renal Cell Carcinoma. Cancers 2022, 14, 4943. [Google Scholar] [CrossRef] [PubMed]
- Hare, S.H.; Harvey, A.J. mTOR function and therapeutic targeting in breast cancer. Am. J. Cancer Res. 2017, 7, 383–404. [Google Scholar] [PubMed]
- Paplomata, E.; O’Regan, R. The PI3K/AKT/mTOR pathway in breast cancer: Targets, trials and biomarkers. Ther. Adv. Med. Oncol. 2014, 6, 154–166. [Google Scholar] [CrossRef]
- Koundouros, N.; Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef]
- Cheng, C.-S.; Wang, Z.; Chen, J. Targeting FASN in Breast Cancer and the Discovery of Promising Inhibitors from Natural Products Derived from Traditional Chinese Medicine. Evid.-Based Complement. Altern. Med. 2014, 2014, 232946. [Google Scholar] [CrossRef]
- Rossato, F.A.; Zecchin, K.G.; La Guardia, P.G.; Ortega, R.M.; Alberici, L.C.; Costa, R.A.P.; Catharino, R.R.; Graner, E.; Castilho, R.F.; Vercesi, A.E. Fatty acid synthase inhibitors induce apoptosis in non-tumorigenic melan-a cells associated with inhibition of mitochondrial respiration. PLoS ONE 2014, 9, e101060. [Google Scholar] [CrossRef]
- Röhrig, F.; Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 2016, 16, 732–749. [Google Scholar] [CrossRef]
- Zhu, S.; Wang, W.; Zhang, J.; Ji, S.; Jing, Z.; Chen, Y.Q. Slc25a5 regulates adipogenesis by modulating ERK signaling in OP9 cells. Cell. Mol. Biol. Lett. 2022, 27, 11. [Google Scholar] [CrossRef] [PubMed]
- Bartholomeusz, C.; Gonzalez-Angulo, A.M.; Liu, P.; Hayashi, N.; Lluch, A.; Ferrer-Lozano, J.; Hortobágyi, G.N. High ERK protein expression levels correlate with shorter survival in triple-negative breast cancer patients. Oncologist 2012, 17, 766–774. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Cui, H. The Autophagy-Lysosomal Pathways and Their Emerging Roles in Modulating Proteostasis in Tumors. Cells 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Rodvold, J.J.; Chiu, K.T.; Hiramatsu, N.; Nussbacher, J.K.; Galimberti, V.; Mahadevan, N.R.; Willert, K.; Lin, J.H.; Zanetti, M. Intercellular transmission of the unfolded protein response promotes survival and drug resistance in cancer cells. Sci. Signal. 2017, 10, eaah7177. [Google Scholar] [CrossRef]
- Sannino, S.; Brodsky, J.L. Targeting protein quality control pathways in breast cancer. BMC Biol. 2017, 15, 109. [Google Scholar] [CrossRef] [PubMed]
- Santamaría, P.G.; Mazón, M.J.; Eraso, P.; Portillo, F. UPR: An Upstream Signal to EMT Induction in Cancer. J. Clin. Med. 2019, 8, 624. [Google Scholar] [CrossRef]
- Arias, E.; Cuervo, A.M. Pros and Cons of Chaperone-Mediated Autophagy in Cancer Biology. Trends Endocrinol. Metab. 2020, 31, 53–66. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. Chaperone-mediated autophagy: A unique way to enter the lysosome world. Trends Cell Biol. 2012, 22, 407–417. [Google Scholar] [CrossRef]
- Joung, E.K.; Kim, J.; Yoon, N.; Maeng, L.-S.; Kim, J.H.; Park, S.; Kang, K.; Kim, J.S.; Ahn, Y.-H.; Ko, Y.H.; et al. Expression of EEF1A1 Is Associated with Prognosis of Patients with Colon Adenocarcinoma. J. Clin. Med. 2019, 8, 1903. [Google Scholar] [CrossRef]
- Falvey, C.M.; O’Donovan, T.R.; El-Mashed, S.; Nyhan, M.J.; O’Reilly, S.; McKenna, S.L. UBE2L6/UBCH8 and ISG15 attenuate autophagy in esophageal cancer cells. Oncotarget 2017, 8, 23479–23491. [Google Scholar] [CrossRef]
- Nagaraj, N.S.; Singh, O.V.; Merchant, N.B. Proteomics: A strategy to understand the novel targets in protein misfolding and cancer therapy. Expert Rev. Proteom. 2010, 7, 613–623. [Google Scholar] [CrossRef]
- Kim, S.-K.; Kim, K.; Ryu, J.-W.; Ryu, T.-Y.; Lim, J.H.; Oh, J.-H.; Min, J.-K.; Jung, C.-R.; Hamamoto, R.; Son, M.-Y.; et al. The novel prognostic marker, EHMT2, is involved in cell proliferation via HSPD1 regulation in breast cancer. Int. J. Oncol. 2019, 54, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Elhamamsy, A.R.; Metge, B.J.; Alsheikh, H.A.; Shevde, L.A.; Samant, R.S. Ribosome Biogenesis: A Central Player in Cancer Metastasis and Therapeutic Resistance. Cancer Res. 2022, 82, 2344–2353. [Google Scholar] [CrossRef] [PubMed]
- Pecoraro, A.; Pagano, M.; Russo, G.; Russo, A. Ribosome Biogenesis and Cancer: Overview on Ribosomal Proteins. Int. J. Mol. Sci. 2021, 22, 5496. [Google Scholar] [CrossRef] [PubMed]
- Vizirianakis, I.S.; Papachristou, E.T.; Andreadis, P.; Zopounidou, E.; Matragkou, C.N.; Tsiftsoglou, A.S. Genetic manipulation of RPS5 gene expression modulates the initiation of commitment of MEL cells to erythroid maturation: Implications in understanding ribosomopathies. Int. J. Oncol. 2015, 47, 303–314. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sánchez-Aragó, M.; Formentini, L.; Cuezva, J.M. Mitochondria-mediated energy adaption in cancer: The H(+)-ATP synthase-geared switch of metabolism in human tumors. Antioxid. Redox Signal. 2013, 19, 285–298. [Google Scholar] [CrossRef]
- Polson, E.S.; Kuchler, V.B.; Abbosh, C.; Ross, E.M.; Mathew, R.K.; Beard, H.A.; da Silva, B.; Holding, A.N.; Ballereau, S.; Chuntharpursat-Bon, E.; et al. KHS101 disrupts energy metabolism in human glioblastoma cells and reduces tumor growth in mice. Sci. Transl. Med. 2018, 10, eaar2718. [Google Scholar] [CrossRef] [PubMed]
- Galai, G.; Ben-David, H.; Levin, L.; Orth, M.F.; Grünewald, T.G.P.; Pilosof, S.; Berstein, S.; Rotblat, B. Pan-Cancer Analysis of Mitochondria Chaperone-Client Co-Expression Reveals Chaperone Functional Partitioning. Cancers 2020, 12, 825. [Google Scholar] [CrossRef] [PubMed]
- Leu, J.I.J.; Barnoud, T.; Zhang, G.; Tian, T.; Wei, Z.; Herlyn, M.; Murphy, M.E.; George, D.L. Inhibition of stress-inducible HSP70 impairs mitochondrial proteostasis and function. Oncotarget 2017, 8, 45656–45669. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, S.; Zhou, H.; Su, D. PRDX2 Promotes the Proliferation and Metastasis of Non-Small Cell Lung Cancer In Vitro and In Vivo. BioMed Res. Int. 2020, 2020, 8359860. [Google Scholar] [CrossRef]
- Yuan, L.; Cai, Y.; Zhang, L.; Liu, S.; Li, P.; Li, X. Promoting Apoptosis, a Promising Way to Treat Breast Cancer With Natural Products: A Comprehensive Review. Front. Pharmacol. 2022, 12, 801662. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Zhang, Y.-J. Interference of Apoptosis by Hepatitis B Virus. Viruses 2017, 9, 230. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-P.; Yang, X.-N.; Jazag, A.; Pan, J.-S.; Hu, T.-H.; Liu, J.-J.; Guleng, B.; Ren, J.-L. HBsAg inhibits the translocation of JTB into mitochondria in HepG2 cells and potentially plays a role in HCC progression. PLoS ONE 2012, 7, e36914. [Google Scholar] [CrossRef]
- Kennedy, D.; Jäger, R.; Mosser, D.D.; Samali, A. Regulation of apoptosis by heat shock proteins. IUBMB Life 2014, 66, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Heimes, A.-S.; Härtner, F.; Almstedt, K.; Krajnak, S.; Lebrecht, A.; Battista, M.J.; Edlund, K.; Brenner, W.; Hasenburg, A.; Sahin, U.; et al. Prognostic Significance of Interferon-γ and Its Signaling Pathway in Early Breast Cancer Depends on the Molecular Subtypes. Int. J. Mol. Sci. 2020, 21, 7178. [Google Scholar] [CrossRef]
- Provance, O.K.; Lewis-Wambi, J. Deciphering the role of interferon alpha signaling and microenvironment crosstalk in inflammatory breast cancer. Breast Cancer Res. 2019, 21, 59. [Google Scholar] [CrossRef]
- Tecalco-Cruz, A.C.; Macías-Silva, M.; Ramírez-Jarquín, J.O.; Méndez-Ambrosio, B. Identification of genes modulated by interferon gamma in breast cancer cells. Biochem. Biophys. Rep. 2021, 27, 101053. [Google Scholar] [CrossRef]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-γ in tumor progression and regression: A review. Biomark. Res. 2020, 8, 49. [Google Scholar] [CrossRef]
- Karnoub, A.E.; Dash, A.B.; Vo, A.P.; Sullivan, A.; Brooks, M.W.; Bell, G.W.; Richardson, A.L.; Polyak, K.; Tubo, R.; Weinberg, R.A. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 2007, 449, 557–563. [Google Scholar] [CrossRef]
- Desai, S.; Reed, R.; Burks, J.; Wood, L.; Pullikuth, A.; Haas, A.; Liu, L.; Breslin, J.; Meiners, S.; Sankar, S. ISG15 disrupts cytoskeletal architecture and promotes motility in human breast cancer cells. Exp. Biol. Med. 2011, 237, 38–49. [Google Scholar] [CrossRef]
- Andersen, J.B.; Hassel, B.A. The interferon regulated ubiquitin-like protein, ISG15, in tumorigenesis: Friend or foe? Cytokine Growth Factor Rev. 2006, 17, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Dupree, E.J.; Manzoor, Z.; Alwine, S.; Crimmins, B.S.; Holsen, T.M.; Darie, C.C. Proteomic analysis of the lake trout (Salvelinus namaycush) heart and blood: The beginning of a comprehensive lake trout protein database. Proteomics 2022, 22, e2100146. [Google Scholar] [CrossRef] [PubMed]
- Mihăşan, M.; Babii, C.; Aslebagh, R.; Channaveerappa, D.; Dupree, E.; Darie, C.C. Proteomics based analysis of the nicotine catabolism in Paenarthrobacter nicotinovorans pAO1. Sci. Rep. 2018, 8, 16239. [Google Scholar] [CrossRef] [PubMed]
- Sokolowska, I.; Dorobantu, C.; Woods, A.G.; Macovei, A.; Branza-Nichita, N.; Darie, C.C. Proteomic analysis of plasma membranes isolated from undifferentiated and differentiated HepaRG cells. Proteome Sci. 2012, 10, 47. [Google Scholar] [CrossRef] [PubMed]
- Channaveerappa, D.; Lux, J.C.; Wormwood, K.L.; Heintz, T.A.; McLerie, M.; Treat, J.A.; King, H.; Alnasser, D.; Goodrow, R.J.; Ballard, G.; et al. Atrial electrophysiological and molecular remodelling induced by obstructive sleep apnoea. J. Cell. Mol. Med. 2017, 21, 2223–2235. [Google Scholar] [CrossRef] [PubMed]

| Pathways | NES | FDR q-Val | |
|---|---|---|---|
| Upregulated | MITOTIC SPINDLE | 1.23 | 1 |
| EPITHELIAL MESENCHYMAL TRANSITION | 1.21 | 1 | |
| FATTY ACID METABOLISM | 1.21 | 0.979 | |
| UV RESPONSE UP | 1.16 | 0.964 | |
| IL2 STAT5 SIGNALING | 1.15 | 0.82 | |
| APOPTOSIS | 1.04 | 1 | |
| Downregulated | OXIDATIVE PHOSPHORYLATION | −1.25 | 0.138 |
| APICAL JUNCTION | −1.25 | 0.127 |
| Pathways | NES | FDR q-Val | |
|---|---|---|---|
| Upregulated | COMPLEMENT | 1.23 | 1 |
| INTERFERON GAMMA RESPONSE | 1.07 | 1 | |
| UNFOLDED PROTEIN RESPONSE (UPR) | 1 | 1 | |
| Downregulated | CHOLESTEROL HOMEOSTASIS | −1.56 | 0.167 |
| GLYCOLYSIS | −1.45 | 0.239 | |
| E2F TARGETS | −1.44 | 0.187 | |
| APICAL JUNCTION | −1.32 | 0.297 | |
| HYPOXIA | −1.18 | 0.465 | |
| MYC TARGETS V2 | −1.09 | 0.534 |
| Gene Symbol | Gene Description | Expression in Malignancies and Putative Neoplastic Effects | HALLMARK_PATHWAYS | GOBP | |
|---|---|---|---|---|---|
| Overexpressed JTB Condition | |||||
| Upregulated proteins and pathways | |||||
| HSPD1 | Heat shock protein family D (HSP60) member 1 | cancer cell survival, regulation of cell death, proliferation [17], represses E-cadherin, promotes cell invasion, migration, poor prognosis [18] | PT | MYC_TARGETS_V1 | BIOLOGICAL_ADHESION; PROTEIN_FOLDING; PROTEIN_REFOLDING; PROTEIN_MATURATION; PROTEIN_CONTAINING_COMPLEX_ORGANIZATION; PROTEIN_STABILIZATION; PROTEIN_INTRACELLULAR_TRANSPORT; TRANSMEMBRANE_TRANSPORT; PROTEIN_TRANSMEMBRANE_TRANSPORT_INTO_INTRACELLULAR_ORGANELLE; MITOCHONDRION_ORGANIZATION; DNA_RECOMBINATION; IMMUNE_RESPONSE_TO_TUMOR_CELL; PROGRAMMED_CELL_DEATH |
| MTORC1_SIGNALING | |||||
| EMT [18] | |||||
| HSP90AA1 | Heat shock protein 90 alpha family class A member 1 | overexpressed in tumors [19], carcinogenesis, activation of oncogenic proteins involved in cancer cell survival, adaptation to stress, growth, proliferation, angiogenesis, signal transduction, metabolic rewiring, motility and invasiveness [20] | PT | FAM | CELL_MORPHOGENESIS; CELL_PROJECTION_ORGANIZATION; CMA & PROTEIN_CATABOLIC_PROCESS; PROTEIN_FOLDING; PROTEIN_STABILIZATION;PROTEIN_CONTAINING_COMPLEX_ORGANIZATION; MITOCHONDRION_ORGANIZATION; TELOMERE_ORGANIZATION; PROGRAMMED_CELL_DEATH |
| EMT [21] | |||||
| HSPA1A | Heat shock 70 kDa protein 1A variant | potential biomarker for BC, overexpressed in BC, promotes progression, inhibits apoptosis, extracellularly-activates proinflammatory immunity [22] | PT | COMPLEMENT | REGULATION_OF_CELL_DIFFERENTIATION; MITOTIC_SPINDLE_ORGANIZATION; MITOTIC_CELL_CYCLE_PROCESS; CYTOSKELETON_ORGANIZATION; PROTEIN_FOLDING/CMPF; PROTEIN_REFOLDING; PROTEIN_STABILIZATION; LYSOSOMAL_TRANSPORT; PROTEIN_CATABOLIC_PROCESS; RNA-CATABOLIC_PROCESS; MITOCHONDRION_ORGANIZATION; REGULATION_OF_DNA_TEMPLATED_TRANSCRIPTION_IN_RESPONSE_TO_STRESS PROGRAMMED_CELL_DEATH |
| EMT [23] | |||||
| RAN | RAS-related nuclear protein/GTP-binding nuclear protein RAN | BC progression, associated with histological grade of tumor, nerve invasion and metastasis, vascular metastasis and Ki-67 [24] | PT | MYC_TARGETS_V1 | INTRACELLULAR_PROTEIN_TRANSPORT; MITOTIC_SPINDLE; MITOTIC_CELL_CYCLE_PROCESS; CHROMOSOME_SEGREGATION; RIBOSOME_BIOGENESIS |
| E2F_TARGETS | |||||
| RPS14 | 40S ribosomal protein S14 | overexpressed in ER+ BC, enhances cell proliferation, cell cycle, metastasis, anti-apoptotic affect, stimulates interferon signaling pathways [25] | PT | UPR | CYTOPLASMIC_TRANSLATION; RIBOSOME_BIOGENESIS/RIBOSOME_ASSEMBLY |
| RPL6 | Human 60s ribosomal protein L6 | up-regulated in multidrug-resistant gastric cancer cells, overexpression is anti-apoptotic, accelerates cell growth and colony forming ability [26] | PT | MYC_TARGETS_V1 | GOBP_CYTOPLASMIC_TRANSLATION; RIBOSOME_BIOGENESIS/RIBOSOME_ASSEMBLY; Peptide chain elongation (genecards.org) |
| IFITM2 | Interferon- induced transmembrane protein 2 | tumor progression and lymphatic metastasis in ccRCC [27] | PT | IFN-α_RESPONSE | DEFENSE_RESPONSE; RESPONSE_TO_INTERFERON_ALPHA; RESPONSE_TO_INTERFERON_BETA; RESPONSE_TO_INTERFERON_GAMMA |
| IFN-γ_RESPONSE | |||||
| TPM3 | Tropomyosin alpha-3 chain | overexpressed in BC, promotes cancer cell migration [28], proliferation, invasion, EMT [29] | PT | EMT [30] | CYTOSKELETON_ORGANIZATION |
| TUBA4A | Tubulin alpha-4a chain | oncogenic role, drug resistance [31], cell movement and development [32], microtentacles formation and metastatic dissemination [33] | PT | MITOTIC_SPINDLE | MITOTIC_CELL_CYCLE; CYTOSKELETON_ORGANIZATION |
| MTORC1_SIGNALING | |||||
| UV_RESPONSE_UP | |||||
| TUBB2A | Tubulin beta-2A chain | overexpressed in invasive BC cell lines, predictive biomarker for distant metastasis in BC, cell proliferation, movement, adhesion [34] | PT | UPR | MITOTIC_CELL_CYCLE; CYTOSKELETON_ORGANIZATION; CELL_MIGRATION |
| TNFA_SIGNALING_VIA_NFKB | |||||
| LAMP2 | Lysosome-associated membrane protein type 2 | overexpressed in BC tissue and BC cell lines, promotes proliferation [35], protein degradation and turnover [36]; co-overexpressed with HSPA8, promotes cancer cell survival during OS [35] | PT | COMPLEMENT | LYSOSOMAL_TRANSPORT; CMA & PROTEIN_CATABOLIC_PROCESS |
| PROTEIN_SECRETION | |||||
| COAGULATION | |||||
| PRKCSH | Protein kinase C substrate 80K-H/Hepatocystin [37]/Glucosidase 2 subunit beta | promotes tumorigenesis, overexpressed in tumors, correlated with the progression of lymph node metastasis in BC; induction of tumor-promoting factors and tumor resistance to ER stress [37] | PT | UPR, ERAD pathway [37] | CARBOHYDRATE_DERIVATIVE_METABOLIC_PROCESS; Metabolism of proteins; protein N-glycosylation processing phase (genecards.org); |
| PPIA/CYPA | Peptidyl-prolyl isomerase A/cyclophilin A | overexpressed in BC, cell survival [17], growth, malignant transformation, metastasis, drug resistance [38], anti-apoptosis [39]; promotes EMT in NSCLC cells [40] | PT | GLYCOLYSIS | CELL_ADHESION; CELL_MIGRATION; PROTEIN_FOLDING; PROTEIN_MODIFICATION_BY_SMALL_PROTEIN_CONJUGATION_OR_REMOVAL; CELL_DEATH_IN_RESPONSE_TO_OXIDATIVE_STRESS |
| MYC_TARGETS_V1 | |||||
| EMT [40] | |||||
| ENO2 | Neuron–specific enolase | promotes cell proliferation, glycolysis [41]; overexpressed in lung cancer [42] and glycolytic subtype of TNBC [43] | PT | GLYCOLYSIS | CARBOHYDRATE_METABOLIC_PROCESS |
| HYPOXIA | |||||
| APOPTOSIS | |||||
| EMT | |||||
| FAM | |||||
| UV_RESPONSE_UP | |||||
| SLC25A5/AAC2/ANT2 | Solute carrier family 25 member 5/mitochondrial ADP/ATP carrier-2/adenine nucleotide translocase 2 | overexpressed in cancer cells, including BC, induces cellsurvival in hypoxic condition, depletion inhibits tumor cell growth and proliferation, stimulates apoptosis, and facilitates chemotherapy-induced apoptosis [44,45] | PT | OXPHOS | MITOCHONDRION_ORGANIZATION; AUTOPHAGY_OF_MITOCHONDRION; CHROMOSOME_SEGREGATION; NUCLEOTIDE_TRANSMEMBRANE_TRANSPORT; PROGRAMMED_CELL_DEATH |
| EEF1A1 | Eukaryotic translation elongation factor-1 alpha-1 | overexpressed in tumors, including BC, controls cell proliferation and cell death [46], promotes heat shock response, protecting cancer cells from proteotoxic stress, sustains cancer cell survival [47], oncogenesis, pro-apoptotic/anti-apoptotic activity | PT | - | CMA & PROTEIN_CATABOLIC_PROCESS; TRANSLATIONAL_ELONGATION; Cytoskeleton modulation |
| CAND1 | Cullin-associated and neddylation dissociated protein 1 | overexpressed in PCa, promotes cell viability, proliferation, anti-apoptotic role [48]; mediates invasion and metastasis in ER+ BC through activation of estrogen and androgen signaling pathways [49] | PT | - | REGULATION_OF_DNA_TEMPLATED_TRANSCRIPTION_INITIATION; Centriole duplication control [48] |
| CREBZF | CREB/ATF bZIP transcription factor | putative tumor-suppressive activity, participates in modulation of p53 [50], reduces MCF7 cell proliferation, migration, and invasion, its knockdown facilitating BC development [51] | AT | - | GOBP_CHROMATIN_ORGANIZATION |
| Downregulated proteins and pathways | |||||
| FASN | Fatty acid synthase | overexpressed in cancer cells, enhances cancer malignant progression [52], tumor cell migration, metastasis [53]; inhibition reduces cell proliferation, suppresses migration and invasion and induces apoptosis [54] | AT | FAM | LIPID_BIOSYNTHETIC_PROCESS; FATTY_ACID_BIOSYNTHETIC_PROCESS; FATTY_ACID_METABOLIC_PROCESS |
| CHOLESTEROL_HOMEOSTASIS | |||||
| ESTROGEN_RESPONSE_EARLY | |||||
| TPI1 | Triosephosphate isomerase | upregulated in multiple cancers, promotes tumor development and progression of BC in tissue and cell lines, promotes glycolysis, proliferation, metastasis, activates PI3K/Akt/mTOR, regulates EMT [55] | AT | GLYCOLYSIS | CARBOHYDRATE_METABOLIC_PROCESS |
| MTORC1_SIGNALING | |||||
| HYPOXIA | |||||
| PRDX1 | Peroxiredoxin-1 | regulates cell growth, differentiation, apoptosis, overexpressed in BC tissues and cell lines, controversial role, it could act as tumor suppressor or as a suppressor of tumor cell death [56]; knockout inhibits in vivo growth of mammary tumors derived from MCF7 cells and reduces survival of MCF7 cells under stress condition [57]; tumor suppressor in BC, its deletion promotes tumor growth in mice [58]; loss of PRDX1 results in development of cancer-associated fibroblasts (CAFs) in BC [59] | AT [57] | PEROXIZOME | CELL_REDOX_HOMEOSTASIS |
| ROS | |||||
| PDIA4 | Protein disulfide isomerase A4 | upregulated in BC, inhibition promotes reduction of OC cells growth and proliferation, induces apoptosis in MM cells [60] | AT | - | PROTEIN_FOLDING |
| SOD1 | Superoxide dismutase 1 (Cu-Zn) | downregulation promotes apoptosis and oncogene-induced senescence [61] | AT | ROS | CYTOSKELETON_ORGANIZATION; CELL_DEATH_IN_RESPONSE_TO_OS; LIPID_METABOLIC_PROCESS; INFLAMMATORY_RESPONSE; PROGRAMMED_CELL_DEATH |
| GLYCOLYSIS | |||||
| PEROXIZOME | |||||
| PROTEIN_SECRETION | |||||
| ENO1 | Alpha-enolase | overexpressed in BC, involved in cell growth, hypoxia tolerance, autoimmune activities, glycolysis pathway [62] | AT | GLYCOLYSIS | CARBOHYDRATE_METABOLIC_PROCESS |
| HYPOXIA | |||||
| MTORC1_SIGNALING | |||||
| TPD52L2 | Tumor protein D52-like 2 | overexpressed in BC, OC and PCa; its knockdown suppressed cell colony-forming potency, cell growth, and induces apoptosis and ER stressof oxaliplatin-resistant gastric carcinoma cells [63] | AT | ANDROGEN_RESPONSE | CARBOHYDRATE_METABOLIC_PROCESS |
| ACTN4 | Actinin alpha 4 | BC tumorigenesis, cell movement, proliferation, metastasis; depletion results in reduced proliferation, migration and metastasis, decreases estrogen-mediated cancer cell proliferation in MCF7 [64] | AT | APICAL_JUNCTION | CELL_MORPHOGENESIS; CYTOSKELETON_ORGANIZATION; TRANSMEMBRANE_TRANSPORT |
| MITOTICSPINDLE | |||||
| IQGAP2 | RAS GTP-ase-activating-like protein | tumor suppressor in most cancers, downregulation promotes proliferation and EMT, inhibits apoptosis, stimulates metastatic abilities of BC cells and lymphovascular invasion [65] | PT | ANDROGEN_RESPONSE | CYTOSKELETON_ORGANIZATION; ACTIN_CYTOSKELETON_REORGANIZATION; ACTIN_FILAMENT_BASED_PROCESS |
| CALM1 | Calcium-calmodulin N-terminal domain | knockdown inhibits proliferation, invasion, migration, induces cell cycle arrest and increases apoptosis in ESCC [66] | AT | COMPLEMENT | CYTOSOLIC_CALCIUM_ION_TRANSPORT; TRANSMEMBRANE_TRANSPORT; CELL_CYCLE_PROCESS; MITOTIC_CELL_CYCLE; CYTOKINESIS |
| AHSG | Fetuin-A/Alpha2- Heremans Schmid (HS) glycoprotein | synthetized, modified and secreted by tumor cells, downregulated, reduces growth, motility, adhesion and attachment of tumor cells [67] | AT | - | INFLAMMATORY_RESPONSE; VESICLE_MEDIATED_TRANSPORT; cell attachment [67] |
| EEF1A1 | Eukaryotic translation elongation factor-1 alpha-1 | overexpressed in tumors, including BC, controls cell proliferation and cell death [46], promotes heat shock response, protecting cancer cells from proteotoxic stress, sustains cancer cell survival [47], oncogenesis, pro-apoptotic/anti-apoptotic activity | AT | - | CMA &PROTEIN_CATABOLIC_PROCESS; TRANSLATIONAL_ELONGATION |
| PCBP1/ hn-RNP-E1 (HNRNP E1) | polyC-RNA-binding protein 1/heterogeneous nuclear riboproteinE1 | tumor suppressor, downregulated in human cancers promotes proliferation, migration and invasion of LUAD [68] | PT | - | REGULATION_OF_DNA-TEMPLATED TRANSCRIPTION |
| Downregulated JTB condition | |||||
| Upregulated proteins and pathways | |||||
| PSME1/ PA28α | Proteasome activator complex subunit 1 isoform 4 | tumor-associated protein/putative tumor biomarker/upregulated in hESCC [69], PC [70], SKCM [71], MM, when promotes cell growth and proliferation [72], anti-apoptotic [73]; downregulated in HCC [74] | PT | IFN-α_RESPONSE | PROTEIN_CATABOLIC_PROCESS; MITOTIC_CELL_CYCLE |
| IFN-γ_RESPONSE | |||||
| ENO2 | Neurone –specific enolase | promotes cell proliferation, glycolysis [41]; overexpressed in lung cancer [42] and glycolytic subtype of TNBC [43] | PT | GLYCOLYSIS | CARBOHYDRATE_METABOLIC_PROCESS |
| HYPOXIA | |||||
| APOPTOSIS | |||||
| FAM | |||||
| EMT | |||||
| UV_RESPONSE_UP | |||||
| PGK1 | Phosphoglycerate kinase 1 | overexpression associated with poor prognosis in BC, progression, metastases, potential survival biomarker and invasion promoter, regulates HIF-1α-mediated EMT [75] | PT | GLYCOLYSIS | CARBOHYDRATE_METABOLIC_PROCESS |
| MTORC1-SIGNALING | |||||
| HYPOXIA | |||||
| POTEKP/ACTBL3 | POTE ankyrin domain family member K/beta-actin-like protein 3 | involved in HCC [76], upregulated in HGSC [77]; lung cancer exosome-specific protein [78] | PT | - | CYTOSKELETON_ORGANIZATION |
| TUBA1A | Tubulin alpha-1a | upregulated in BC tissue [79], involved in cell division and cell movement; overexpression was correlated with poor overall survival and a more aggressive phenotype in GC [80] | PT | - | MICROTUBULE_BASED_PROCESS; CELL_DIVISION; CELL_JUNCTION_ORGANIZATION; CYTOSKELETON_ORGANIZATION; CYTOSKELETON_DEPENDENT_INTRACELLULAR_TRANSPORT |
| TUBB | Beta-tubulin | upregulated in BC tissue [79] | PT | E2F_TARGETS | MICROTUBULE_BASED_PROCESS; CELL_DIVISION; CELL_CYCLE; CYTOSKELETON_ORGANIZATION; CYTOSKELETON_DEPENDENT_INTRACELLULAR_TRANSPORT; CELL_JUNCTION_ORGANIZATION |
| HSPE1/ CH10 | Heat shock protein family E (HSP10) member/10kDa HSP | tumorigenesis [81], cancer cell survival, regulation of cell death [17] | PT | MYC_TARGETS_V1 | PROTEIN_FOLDING/CMPF; PROGRAMMED_CELL_DEATH |
| MTORC1-SIGNALING | |||||
| HSPD1 | Heat shock protein family D (HSP60) member 1 | cancer cell survival, regulation of cell death, proliferation [17], represses E-cadherin, promotes cell invasion, migration, poor prognosis [18] | PT | MYC_TARGETS_V1 | BIOLOGICAL_ADHESION; PROTEIN_FOLDING; PROTEIN_REFOLDING; PROTEIN_MATURATION; PROTEIN_CONTAINING_COMPLEX_ORGANIZATION; PROTEIN_STABILIZATION; PROTEIN_INTRACELLULAR_TRANSPORT; TRANSMEMBRANE_TRANSPORT; PROTEIN_TRANSMEMBRANE_TRANSPORT_INTO_INTRACELLULAR_ORGANELLE; MITOCHONDRION_ORGANIZATION; DNA_RECOMBINATION; IMMUNE_RESPONSE_TO_TUMOR_CELL; PROGRAMMED_CELL_DEATH |
| MTORC1_ SIGNALING | |||||
| EMT [18] | |||||
| HSPA1A | Heat shock 70 kDa protein 1A variant | potential biomarker for BC, overexpressed in BC, promotes progression, inhibits apoptosis, extracellularly-activates proinflammatory immunity [22] | PT | COMPLEMENT | REGULATION_OF_CELL_DIFFERENTIATION; MITOTIC_SPINDLE_ORGANIZATION; MITOTIC_CELL_CYCLE_PROCESS; CYTOSKELETON_ORGANIZATION; PROTEIN_FOLDING/CMPF; PROTEIN_REFOLDING; PROTEIN_STABILIZATION; LYSOSOMAL_TRANSPORT; PROTEIN_CATABOLIC_PROCESS; RNA-CATABOLIC_PROCESS; MITOCHONDRION_ORGANIZATION; REGULATION_OF_DNA_TEMPLATED_TRANSCRIPTION_IN_RESPONSE_TO_STRESS PROGRAMMED_CELL_DEATH |
| TMSB10 | Thymosin beta 10 | overexpressed in many cancers: RCC, pancreatic, lung, and thyroid carcinoma, promotes migration, invasion, and EMT [82]; positively associated with high-grade aggressive BC, significantly elevated in BC cells and tissues, proliferation, invasion migration of BC cells by activation of Akt/FOXO signaling, valuable serum biomarker for diagnosis and potential therapeutic target in BC [83] | PT | EMT [83] | CYTOSKELETON_ORGANIZATION; CELL_MIGRATION |
| PARK7/DJ-1 | Parkinsonism associated deglycase DJ-1 | oncogene upregulated in various cancers, involved in tumor initiation, progression, proliferation, metastasis, recurrence, resistance to chemotherapy [84], overexpression increases cell survival; highly expressed in cytoplasm of invasive BC cells [85] | PT | - | RAS_PROTEIN_SIGNAL_TRANSDUCTION; INTRACELLULAR_PROTEIN_TRANSPORT; PROTEIN_MODIFICATION_BY_SMALL_PROTEIN_REMOVAL; PROTEIN_CATABOLIC_PROCES; PROTEIN_REPAIR; CELLULAR_AMINO_ACID_BIOSYNTHETIC_PROCESS; NUCLEOBASE_CONTAINING_SMALL_MOLECULE_METABOLIC_PROCESS; DNA_REPAIR; TRANSMEMBRANE_TRANSPORT; REGULATION_OF_SIGNALING_RECEPTOR_ACTIVITY; REGULATION_OF_TRANSCRIPTION_REGULATORY_REGION_DNA_BINDING; GENERATION_OF_PRECURSOR_METABOLITES_AND_ENERGY; MITOCHONDRION_ORGANIZATION; INFLAMMATORY_RESPONSE; PROGRAMMED_CELL_DEATH; CELL_DEATH_IN_RESPONSE_TO_HYDROGEN_PEROXIDE; DETOXIFICATION |
| PRDX2 | Peroxiredoxin 2 | overexpressed in various cancers [86], highly upregulated in BC [57]; dual effect in carcinogenesis, in BC induces selective growth of metastatic cancer cells in lung by protecting them against OS [87]; reduces OS, cell damage and apoptosis [88] | PT | PEROXIZOME | CELL_REDOX_HOMEOSTASIS |
| ROS | |||||
| GAPDH1 | Glyceraldehyde-3-phosphate dehydrogenase 1 | overexpressed in many cancers [89], in association with BC cell proliferation and tumor aggressiveness [90]; several PTMs have pro-apoptotic role [73]; promotes cancer growth and metastasis by affecting EMT [91] | PT | GYCOLYSIS | CARBOHYDRATE_METABOLIC_PROCESS; nuclear tRNA export, DNA replication and repair, endocytosis, exocytosis, cytoskeletal organization, iron metabolism, cell death [89], membrane fusion, vesicle secretion, transcription co-activation, cell cycle regulation, mRNA stabilization [92]; DEFENSE_RESPONSE |
| HYPOXIA | |||||
| EMT [91] | |||||
| ACTC1 | Actin alpha cardiac muscle 1 | upregulated in BC and other malignancies [93]; promotes resistance to apoptosis, cell survival, controls cell migration [94] | PT | KRAS_SIGNALING_DN | Cell differentiation, anatomical structure development, cell cytoskeleton organization, programmed cell death |
| APICAL_JUNCTION | |||||
| EMT [95] | |||||
| PPIA/CYPA | Peptidylprolyl isomerase A | overexpressed in BC, cell survival [17], growth, malignant transformation, metastasis, drug resistance [38], anti-apoptosis [39] | PT | GLYCOLYSIS | CELL_ADHESION; CELL_MIGRATION; PROTEIN_FOLDING; PROTEIN_MODIFICATION_BY_SMALL_PROTEIN_CONJUGATION_OR_REMOVAL; CELL_DEATH_IN_RESPONSE_TO_OS |
| MYC_TARGETS_V1 | |||||
| Downregulated proteins and pathways | |||||
| ACTG1 | Actin gamma 1 | overexpressed in skin cancer and HCC, promotes growth, migration, proliferation, inhibits mitochondrial apoptotic pathway, increases aerobic glycolysis, role in microtubule integrity; depletion in BC cells resulted in centrosome amplification, formation of multipolar spindles, defects in chromosome segregation, leading to mitotic abnormalities [93] | AT | APICAL_JUNCTION | ACTOMYOSIN_STRUCTURE_ORGANIZATION; CYTOSKELETON_ORGANIZATION; CELL_MIGRATION; CELL_JUNCTION_ORGANIZATION; BIOLOGICAL_ADHESION |
| CHOLESTEROL_HOMEOSTASIS | |||||
| TPI1 | Triosephosphate isomerase | upregulated in multiple cancers, promotes tumor development and progression of BC in tissue and cell lines, promotes glycolysis, proliferation, metastasis, activates PI3K/Akt/mTOR, regulates EMT [55] | AT | GLYCOLYSIS | CARBOHYDRATE_METABOLIC_PROCESS |
| MTORC1_SIGNALING | |||||
| HYPOXIA | |||||
| HSPA1A | Heat shock 70kDa protein 1A variant | potential biomarker for BC, overexpressed in BC, promotes progression, inhibits apoptosis, extracellularly-activates proinflammatory immunity [22] | AT | COMPLEMENT | REGULATION_OF_CELL_DIFFERENTIATION; MITOTIC_SPINDLE_ORGANIZATION; MITOTIC_CELL_CYCLE_PROCESS/CELL_CYCLE_PROCESS; CYTOSKELETON_ORGANIZATION; PROTEIN_FOLDING/CMPF; PROTEIN_STABILIZATION; PROTEIN_CONTAINING_COMPLEX_ ORGANIZATION; LYSOSOMAL_TRANSPORT; PROTEIN_CATABOLIC_PROCESS; RNA-CATABOLIC_PROCESS; MITOCHONDRION_ORGANIZATION; REGULATION_OF_DNA_TEMPLATED_TRANSCRIPTION_IN_RESPONSE_TO_STRESS PROGRAMMED_CELL_DEATH |
| HSPB1 | Heat shock 27 kDa protein 1 | overexpressed in BC, downregulation was correlated with PTEN increase (tumor suppressor) that negatively regulates PI3K/AKT | AT | APOPTOSIS | PROTEIN_FOLDING/CMPF; CELL_ADHESION; CELL_MIGRATION; CYTOSKELETON_DEPENDENT_INTRACELLULAR_TRANSPORT; CELL_DEATH_IN_RESPONSE_TO_OXIDATIVE_STRESS; PROGRAMMED_CELL_DEATH |
| HSPA8 | Heat shock 70 kDa protein 8 | depletion suppresses cancer cells growth, induces apoptosis, and cell cycle arrest [96] | AT | G2M_CHECKPOINT | PROTEIN_FOLDING/CMPF; PROTEIN_REFOLDING;PROTEIN_CONTAINING_COMPLEX_ORGANIZATION; LYSOSOMAL_TRANSPORT; CMA & PROTEIN_CATABOLIC_PROCESS; CELL_JUNCTION_ORGANIZATION; CYTOSKELETON_DEPENDENT_INTRACELLULAR_TRANSPORT |
| FASN | Fatty acid synthase | inhibition reduces cell proliferation, suppresses migration and invasion and induces apoptosis [54] | AT | FAM | LIPID_BIOSYNTHETIC_PROCESS; FATTY_ACID_BIOSYNTHETIC_PROCESS; FATTY_ACID_METABOLIC_PROCESS |
| CHOLESTEROL HOMEOSTASIS | |||||
| ESTROGEN_RESPONSE_EARLY | |||||
| EEF1A1 | Eukaryotic translation elongation factor-1 alpha-1 | overexpressed in tumors, including BC, controls cell proliferation and cell death [46], promotes heat shock response, protecting cancer cells from proteotoxic stress, sustains cancer cell survival [47], oncogenesis, pro-apoptotic/anti-apoptotic activity | AT | - | CMA & PROTEIN_CATABOLIC_PROCESS; TRANSLATIONAL_ELONGATION |
| SOD1 | Superoxide dismutase 1 (Cu-Zn) | downregulation promotes apoptosis and oncogene-induced senescence [61] | AT | ROS | CYTOSKELETON_ORGANIZATION; CELL_DEATH_IN_RESPONSE_TO_OXIDATIVE_STRESS; LIPID_METABOLIC_PROCESS; INFLAMMATORY_RESPONSE; PROGRAMMED_CELL_DEATH |
| GLYCOLYSIS | |||||
| PEROXIZOME | |||||
| PROTEIN_SECRETION | |||||
| APOPTOSIS | |||||
| MKI67 | Proliferation marker protein Ki-67 | overexpressed in cancer cells [97]; downregulated, reduces migration, invasion, tumor progression; knockout induces transcriptome remodeling, alters EMT and suppresses stem cell characteristics [98] | AT | G2M_CHECKPOINT | CHROMATIN_ORGANIZATION; CHROMOSOME_ORGANIZATION; CHROMOSOME_SEGREGATION; MITOTIC_NUCLEAR_DIVISION |
| CALM1 | Calcium-calmodulin N-terminal domain | knockdown inhibits proliferation, invasion, migration, induces cell cycle arrest and increases apoptosis in ESCC [66] | AT | COMPLEMENT | CYTOSOLIC_CALCIUM_ION_TRANSPORT; TRANSMEMBRANE_TRANSPORT; CELL_CYCLE_PROCESS; MITOTIC_CELL_CYCLE; CYTOKINESIS |
| IFITM2 | Interferon- induced transmembrane protein 2 | downregulation inhibits migration and invasion in ccRCC [27]; knocking out IFITM2 enhanced activation of the endogenous IFN-α pathway that may alter the immune and stromal cells in TME enhancing the invasive abilities of cancer cells [99] | AT/ PT | IFN-α_RESPONSE | DEFENSE_RESPONSE; RESPONSE_TO_INTERFERON_ALPHA; RESPONSE_TO_INTERFERON_BETA; RESPONSE_TO_INTERFERON_GAMMA |
| IFN-γ_RESPONSE | |||||
| RPS5 | 40S ribosomal protein S5 | when overexpressed, negatively regulates the expression of p53 and plays an anti-apoptotic role in cancer cells [100] | AT | MYC_TARGETS_V1 | CYTOPLASMIC_TRANSLATION; RIBOSOME_BIOGENESIS/RIBOSOME_ASSEMBLY |
| CTNNB1 | Catenin beta 1 | Nuclear CTNNB1 plays a key role in most cancers as an oncogene; downregulation inhibited cell proliferation, migration, and invasion (EMT) and induced apoptosis in RCC [101] | AT | WNT_BETA_CATENIN_SIGNALING | CELL_MORPHOGENESIS; BIOLOGICAL_ADHESION; CELL_CELL_JUNCTION_ASSEMBLY/CELL_JUNCTION_ORGANIZATION; REGULATION_OF_EPITHELIAL_TO_MESENCHYMAL_TRANSITION; CELL_MIGRATION; REGULATION_OF_DNA_TEMPLATED_TRANSCRIPTION_ELONGATION; RESPONSE_TO_ESTRADIOL; TELOMERE_ORGANIZATION; PROGRAMMED_CELL_DEATH |
| CHOLESTEROL_HOMEOSTASIS | |||||
| APOPTOSIS | |||||
| TGF_BETA_SIGNALING | |||||
| EMT [102] | |||||
| ISG15 | Interferon-stimulated 15/Ubiquitin-like protein ISG15 | putative oncogene, aberrantly expressed in human cancers, protumor/antitumor functions, overexpressed in highly metastatic MDA-MB-231 BC cell line, enhances proliferation or invasiveness [103], cell cycle progression, cell motility and tumor growth [104], overexpressed in BC tissue [105] | AT | IFN-α_RESPONSE | PROTEIN_CONTAINING_COMPLEX_ORGANIZATION; PROTEIN_MODIFICATION_BY_SMALL_PROTEIN_CONJUGATION; PROTEIN_MODIFICATION_BY_SMALL_PROTEIN_CONJUGATION_OR_REMOVAL; DEFENSE_RESPONSE, immune system modulation [105] |
| IFN-γ_RESPONSE | |||||
| ANXA2 | Annexin A | depletion resulted in ROS elevation upon OS and activation of ROS-mediated cellular damage/death, elevated protein oxidation, decreased tumor growth [106] | AT | HYPOXIA | PROTEIN_MATURATION; cellular redox regulation [106] MEMBRANE_ORGANIZATION; VESICLE_MEDIATED_TRANSPORT |
| MYC_TARGETS_V1 | MTORC1_SIGNALING | MITOTIC_SPINDLE | FAM | CHOLESTEROL_HOMEOSTASIS | COMPLEMENT | COAGULATION | UPR | TNFA_SIGNALING_VIA_NFKB | E2F_TARGETS | IF ALPHA | IF-GAMMA | EMT | PROTEIN_SECRETION | ERAD | GLYCOLYSIS | HYPOXIA | APOPTOSIS | OXPHOS | APICAL_JUNCTION | ESTOGEN_RESPONSE_EARLY | ANDROGEN_RESPONSE | PEROXIZOME | ROS | UV_RESPONSE_UP | WNT_BETA_CATENIN_SIGNALING | TGF_BETA_SIGNALING | KRAS_SIGNALING_DN | G2M_CHECKPOINT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UP in UP | |||||||||||||||||||||||||||||
| HSPD1 | [18] | ||||||||||||||||||||||||||||
| HSP90AA1 | [107] | [21] | |||||||||||||||||||||||||||
| HSPA1A | [23] | ||||||||||||||||||||||||||||
| RAN | |||||||||||||||||||||||||||||
| RPL6 | |||||||||||||||||||||||||||||
| PPIA/CYPA | [40] | ||||||||||||||||||||||||||||
| ENO2 | |||||||||||||||||||||||||||||
| TUBA4A | |||||||||||||||||||||||||||||
| TUBB2A | |||||||||||||||||||||||||||||
| RPS14 | |||||||||||||||||||||||||||||
| PRKCSH | |||||||||||||||||||||||||||||
| IFITM2 | |||||||||||||||||||||||||||||
| LAMP2 | |||||||||||||||||||||||||||||
| TPM3 | [30] | ||||||||||||||||||||||||||||
| SLC25A5 | [45] | ||||||||||||||||||||||||||||
| D in UP | |||||||||||||||||||||||||||||
| TPI1 | |||||||||||||||||||||||||||||
| FASN | |||||||||||||||||||||||||||||
| CALM1 | |||||||||||||||||||||||||||||
| ACTN4 | |||||||||||||||||||||||||||||
| TPD52L2 | |||||||||||||||||||||||||||||
| SOD1 | |||||||||||||||||||||||||||||
| ENO1 | |||||||||||||||||||||||||||||
| YWHAQ | |||||||||||||||||||||||||||||
| PRDX1 | [59] | ||||||||||||||||||||||||||||
| IQGAP2 | [65] | [65] | |||||||||||||||||||||||||||
| UP in D | |||||||||||||||||||||||||||||
| HSPD1 | [18] | ||||||||||||||||||||||||||||
| HSPE1 | |||||||||||||||||||||||||||||
| HSPA1A | [23] | ||||||||||||||||||||||||||||
| PPIA/CYPA | [40] | ||||||||||||||||||||||||||||
| PGK1 | [108] | ||||||||||||||||||||||||||||
| ENO2 | |||||||||||||||||||||||||||||
| PSME1 | |||||||||||||||||||||||||||||
| GAPDH1 | [91] | ||||||||||||||||||||||||||||
| PRDX2 | |||||||||||||||||||||||||||||
| TMSB10 | [82] | ||||||||||||||||||||||||||||
| TUBA1A | [80] | ||||||||||||||||||||||||||||
| TUBB | |||||||||||||||||||||||||||||
| ACTC1 | [95] | ||||||||||||||||||||||||||||
| PARK7 | [84] | [84] | |||||||||||||||||||||||||||
| D in D | |||||||||||||||||||||||||||||
| CTNNB1 | [109] | ||||||||||||||||||||||||||||
| TPI1 | |||||||||||||||||||||||||||||
| FASN | |||||||||||||||||||||||||||||
| SOD1 | |||||||||||||||||||||||||||||
| CALM1 | |||||||||||||||||||||||||||||
| ACTG1 | |||||||||||||||||||||||||||||
| HSPA1A | [23] | ||||||||||||||||||||||||||||
| HSPB1 | |||||||||||||||||||||||||||||
| HSPA8 | |||||||||||||||||||||||||||||
| ANXA2 | [106] | ||||||||||||||||||||||||||||
| ISG15 | |||||||||||||||||||||||||||||
| IFITM2 | |||||||||||||||||||||||||||||
| MKI67 | |||||||||||||||||||||||||||||
| RPS5 | |||||||||||||||||||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jayathirtha, M.; Whitham, D.; Alwine, S.; Donnelly, M.; Neagu, A.-N.; Darie, C.C. Investigating the Function of Human Jumping Translocation Breakpoint Protein (hJTB) and Its Interacting Partners through In-Solution Proteomics of MCF7 Cells. Molecules 2022, 27, 8301. https://doi.org/10.3390/molecules27238301
Jayathirtha M, Whitham D, Alwine S, Donnelly M, Neagu A-N, Darie CC. Investigating the Function of Human Jumping Translocation Breakpoint Protein (hJTB) and Its Interacting Partners through In-Solution Proteomics of MCF7 Cells. Molecules. 2022; 27(23):8301. https://doi.org/10.3390/molecules27238301
Chicago/Turabian StyleJayathirtha, Madhuri, Danielle Whitham, Shelby Alwine, Mary Donnelly, Anca-Narcisa Neagu, and Costel C. Darie. 2022. "Investigating the Function of Human Jumping Translocation Breakpoint Protein (hJTB) and Its Interacting Partners through In-Solution Proteomics of MCF7 Cells" Molecules 27, no. 23: 8301. https://doi.org/10.3390/molecules27238301
APA StyleJayathirtha, M., Whitham, D., Alwine, S., Donnelly, M., Neagu, A.-N., & Darie, C. C. (2022). Investigating the Function of Human Jumping Translocation Breakpoint Protein (hJTB) and Its Interacting Partners through In-Solution Proteomics of MCF7 Cells. Molecules, 27(23), 8301. https://doi.org/10.3390/molecules27238301








