Quantitative Assessment of Serine-8 Phosphorylated β-Amyloid Using MALDI-TOF Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
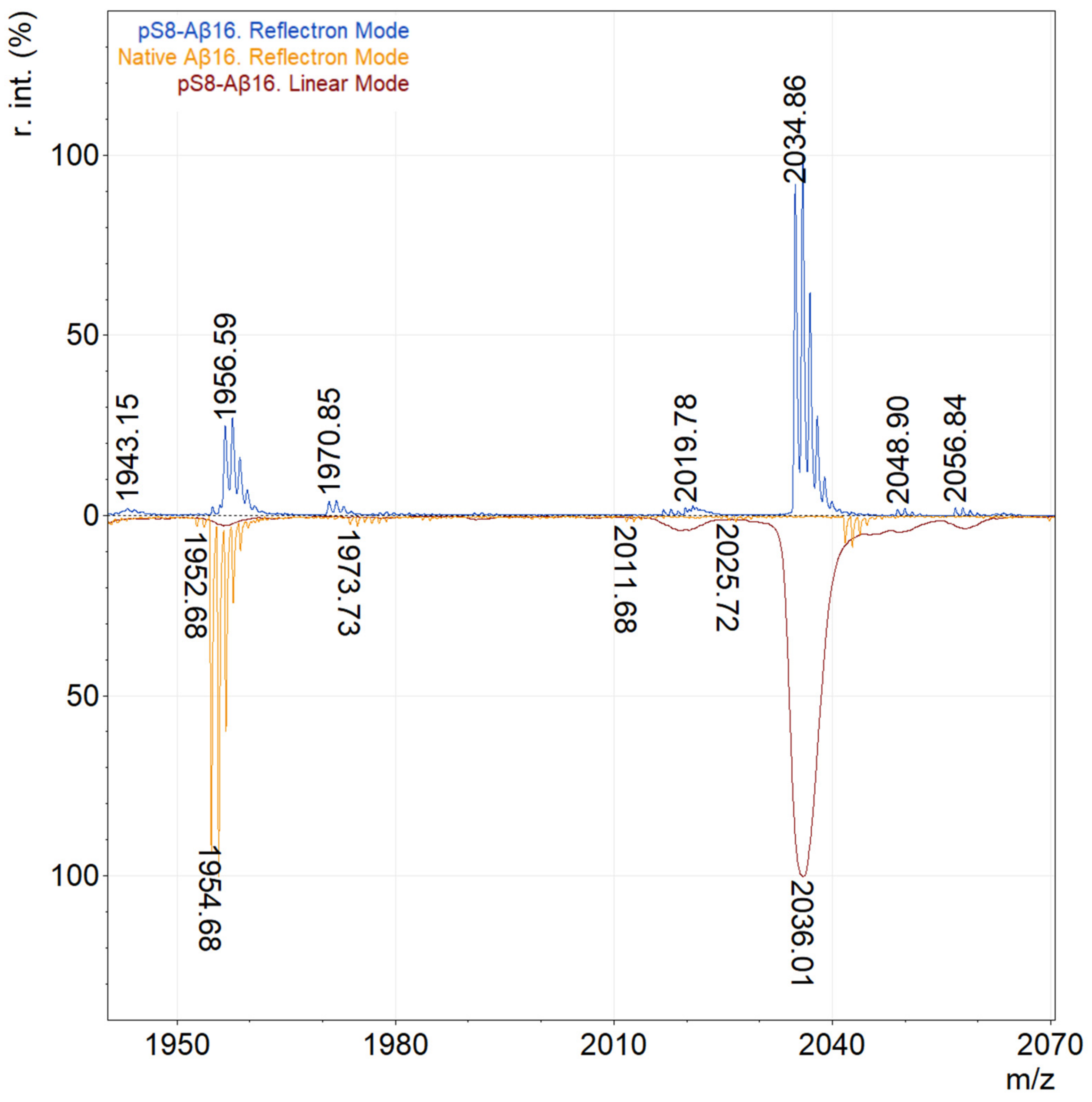
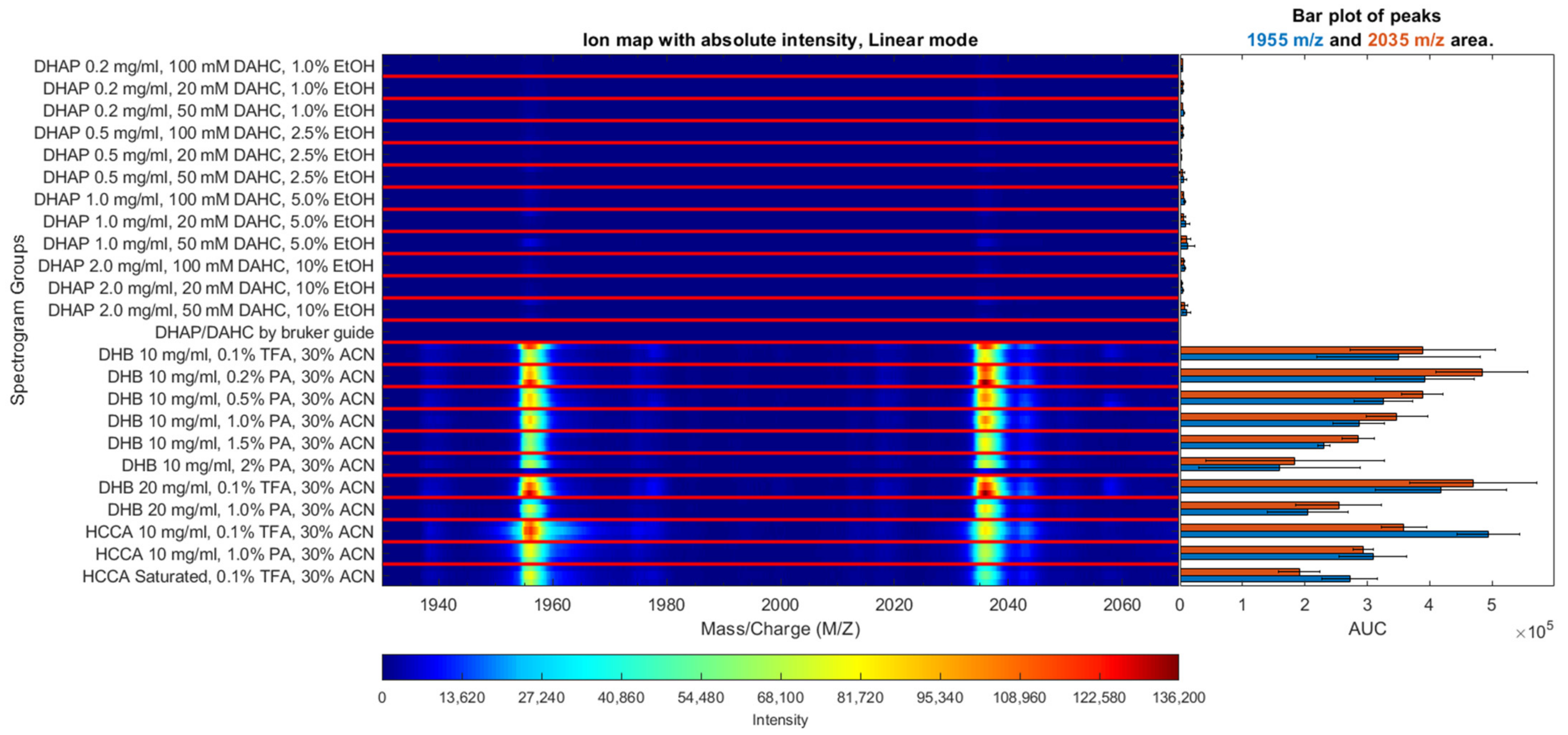
2.1. Optimization of Experimental Conditions for MALDI-TOF Analysis
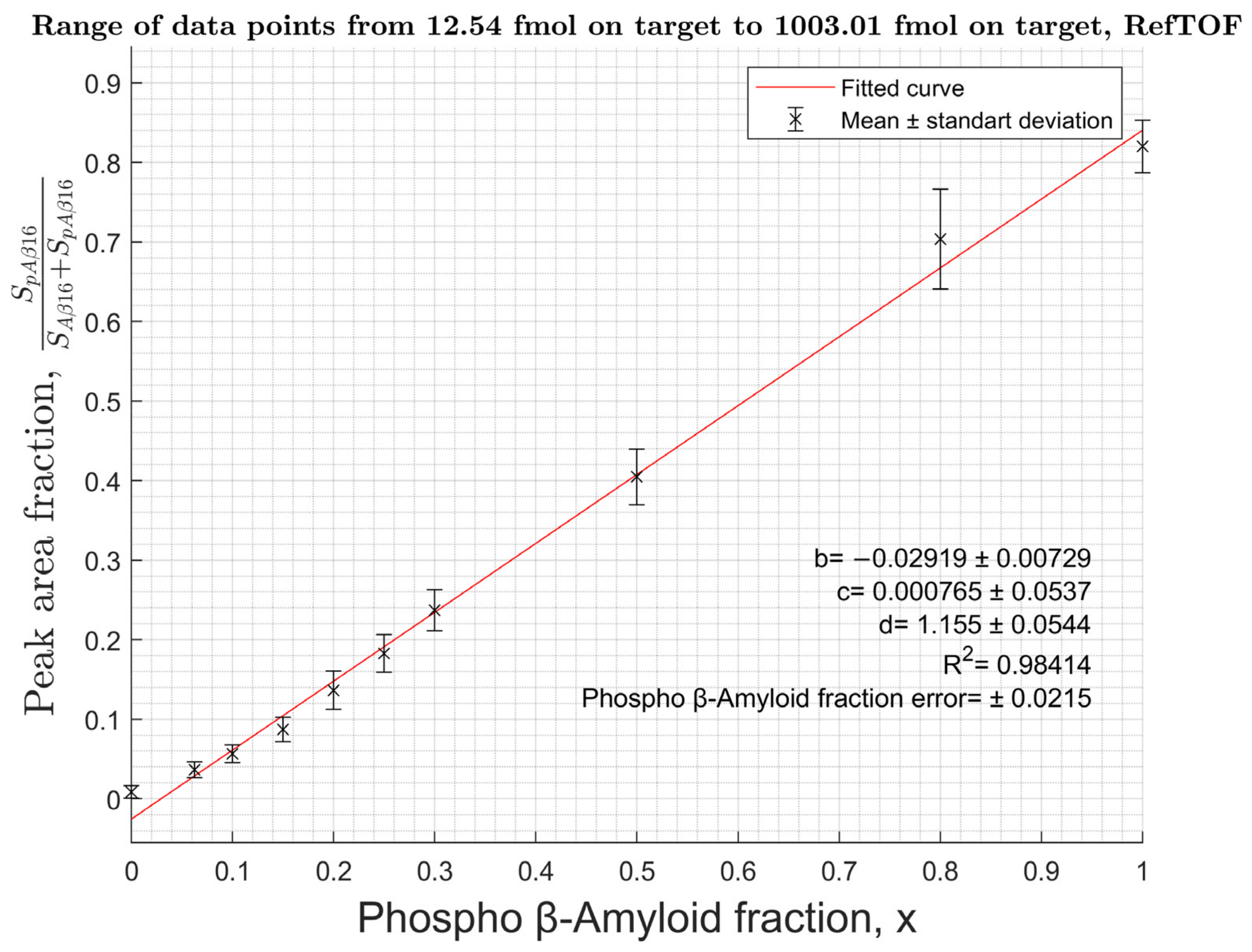
2.2. Calibration Curves
3. Materials and Methods
3.1. Materials
3.2. Selection of the Optimal Matrix for Aβ Ionization
3.3. Construction of Calibration Curves to Determine the Proportion of pS8-Aβ
3.4. Model Sample Preparation and Immunoprecipitation
3.5. Hydrolysis
3.6. Mass Spectra Measurements
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaugler, J.; James, B.; Johnson, T.; Reimer, J.; Weuve, J. 2021 Alzheimer’s Disease Facts and Figures. Alzheimer’s Dement. 2021, 17, 327–406. [Google Scholar] [CrossRef]
- Prince, M.; Bryce, R.; Albanese, E.; Wimo, A.; Ribeiro, W.; Ferri, C.P. The Global Prevalence of Dementia: A Systematic Review and Metaanalysis. Alzheimer’s Dement. 2013, 9, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.F.; Xu, T.H.; Yan, Y.; Zhou, Y.R.; Jiang, Y.; Melcher, K.; Xu, H.E. Amyloid Beta: Structure, Biology and Structure-Based Therapeutic Development. Acta Pharmacol. Sin. 2017, 38, 1205–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakharova, N.V.; Kononikhin, A.S.; Indeykina, M.I.; Bugrova, A.E.; Strelnikova, P.; Pekov, S.; Kozin, S.A.; Popov, I.A.; Mitkevich, V.; Makarov, A.A.; et al. Mass Spectrometric Studies of the Variety of Beta-Amyloid Proteoforms in Alzheimer’s Disease. Mass Spectrom. Rev. 2022, e21775. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Rezaei-Ghaleh, N.; Terwel, D.; Thal, D.R.; Richard, M.; Hoch, M.; Mc Donald, J.M.; Wüllner, U.; Glebov, K.; Heneka, M.T.; et al. Extracellular Phosphorylation of the Amyloid β 2-Peptide Promotes Formation of Toxic Aggregates during the Pathogenesis of Alzheimer’s Disease. EMBO J. 2011, 30, 2255–2265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Wirths, O.; Theil, S.; Gerth, J.; Bayer, T.A.; Walter, J. Early Intraneuronal Accumulation and Increased Aggregation of Phosphorylated Abeta in a Mouse Model of Alzheimer’s Disease. Acta Neuropathol. 2013, 125, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Rijal Upadhaya, A.; Kosterin, I.; Kumar, S.; Von Arnim, C.A.F.; Yamaguchi, H.; Fändrich, M.; Walter, J.; Thal, D.R. Biochemical Stages of Amyloid-β Peptide Aggregation and Accumulation in the Human Brain and Their Association with Symptomatic and Pathologically Preclinical Alzheimer’s Disease. Brain 2014, 137, 887–903. [Google Scholar] [CrossRef] [PubMed]
- Popov, I.A.; Indeikina, M.I.; Pekov, S.I.; Starodubtseva, N.L.; Kononikhin, A.S.; Nikolaeva, M.I.; Kukaev, E.N.; Kostyukevich, Y.I.; Kozin, S.A.; Makarov, A.A.; et al. Estimation of Phosphorylation Level of Amyloid-Beta Isolated from Human Blood Plasma: Ultrahigh-Resolution Mass Spectrometry. Mol. Biol. 2014, 48, 607–614. [Google Scholar] [CrossRef]
- Yin, Z.; Wang, S.; Shen, B.; Deng, C.; Tu, Q.; Jin, Y.; Shen, L.; Jiao, B.; Xiang, J. Coimmunocapture and Electrochemical Quantitation of Total and Phosphorylated Amyloid-β 40 Monomers. Anal. Chem. 2019, 91, 3539–3545. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Kaneko, N.; Villemagne, V.L.; Kato, T.; Doecke, J.; Doré, V.; Fowler, C.; Li, Q.X.; Martins, R.; Rowe, C.; et al. High Performance Plasma Amyloid-β Biomarkers for Alzheimer’s Disease. Nature 2018, 554, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Pekov, S.; Indeykina, M.; Popov, I.; Kononikhin, A.; Bocharov, K.; Kozin, S.A.; Makarov, A.A.; Nikolaev, E. Application of MALDI-TOF/TOF-MS for Relative Quantitation of α- and β-Asp7 Isoforms of Amyloid-β Peptide. Eur. J. Mass Spectrom. 2018, 24, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, D.G.; Indeykina, M.I.; Pekov, S.I.; Bugrova, A.E.; Kechko, O.I.; Iusupov, A.E.; Kononikhin, A.S.; Makarov, A.A.; Nikolaev, E.N.; Popov, I.A. Relative Quantitation of Beta-Amyloid Peptide Isomers with Simultaneous Isomerization of Multiple Aspartic Acid Residues by Matrix Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2020, 31, 164–168. [Google Scholar] [CrossRef] [PubMed]
- De Ranieri, E. When a velocitron meets a reflectron. Nat. Methods 2015, 12, 8. [Google Scholar] [CrossRef]
- Kaufmann, R.; Spengler, B.; Lützenkirchen, F. Mass Spectrometric Sequencing of Linear Peptides by Product-ion Analysis in a Reflectron Time-of-flight Mass Spectrometer Using Matrix-assisted Laser Desorption Ionization. Rapid Commun. Mass Spectrom. 1993, 7, 902–910. [Google Scholar] [CrossRef] [PubMed]
- Annan, R.S.; Carr, S.A. Phosphopeptide Analysis by Matrix-Assisted Laser Desorption Time-of-Flight Mass Spectrometry. Anal. Chem. 1996, 68, 3413–3421. [Google Scholar] [CrossRef] [PubMed]
- Suckau, D.; Resemann, A.; Schuerenberg, M.; Hufnagel, P.; Franzen, J.; Holle, A. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 2003, 376, 952–965. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, D.G.; Indeykina, M.I.; Pekov, S.I.; Iusupov, A.E.; Bugrova, A.E.; Kononikhin, A.S.; Nikolaev, E.N.; Popov, I.A. Probabilistic Model Applied to Ion Abundances in Product-Ion Spectra: Quantitative Analysis of Aspartic Acid Isomerization in Peptides. Anal. Bioanal. Chem. 2019, 411, 7783–7789. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Hou, J.; Xie, Z.; Xue, P.; Cui, Z.; Chen, X.; Li, J.; Cai, T.; Wu, P. Enhanced MALDI-TOF MS Analysis of Phosphopeptides Using an Optimized DHAP/DAHC Matrix. J. Biomed. Biotechnol. 2010, 2010, 759690. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuzin, A.A.; Stupnikova, G.S.; Strelnikova, P.A.; Danichkina, K.V.; Indeykina, M.I.; Pekov, S.I.; Popov, I.A. Quantitative Assessment of Serine-8 Phosphorylated β-Amyloid Using MALDI-TOF Mass Spectrometry. Molecules 2022, 27, 8406. https://doi.org/10.3390/molecules27238406
Kuzin AA, Stupnikova GS, Strelnikova PA, Danichkina KV, Indeykina MI, Pekov SI, Popov IA. Quantitative Assessment of Serine-8 Phosphorylated β-Amyloid Using MALDI-TOF Mass Spectrometry. Molecules. 2022; 27(23):8406. https://doi.org/10.3390/molecules27238406
Chicago/Turabian StyleKuzin, Andrey A., Galina S. Stupnikova, Polina A. Strelnikova, Ksenia V. Danichkina, Maria I. Indeykina, Stanislav I. Pekov, and Igor A. Popov. 2022. "Quantitative Assessment of Serine-8 Phosphorylated β-Amyloid Using MALDI-TOF Mass Spectrometry" Molecules 27, no. 23: 8406. https://doi.org/10.3390/molecules27238406
APA StyleKuzin, A. A., Stupnikova, G. S., Strelnikova, P. A., Danichkina, K. V., Indeykina, M. I., Pekov, S. I., & Popov, I. A. (2022). Quantitative Assessment of Serine-8 Phosphorylated β-Amyloid Using MALDI-TOF Mass Spectrometry. Molecules, 27(23), 8406. https://doi.org/10.3390/molecules27238406






