Cytotoxic Potential of Bioactive Compounds from Aspergillus flavus, an Endophytic Fungus Isolated from Cynodon dactylon, against Breast Cancer: Experimental and Computational Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Cell Lines and Culture Media
2.3. Collection of Plant and Isolation of Endophytic Fungus
2.4. Molecular Identification of Endophytic Fungi
2.5. Evaluation of Anticancer Activity
2.5.1. MTT Assay
2.5.2. Detection of Cytotoxicity Using Dual Staining Assay
2.5.3. Apoptosis Analysis Using Flow Cytometry
2.5.4. Assessment of Mitochondrial Transmembrane Potential
2.5.5. Nuclear Integrity Measurement
2.5.6. Determination of Reactive Oxygen Species (ROS)
2.6. Gas Chromatography-Mass Spectrum (GC-MS) Analysis
2.7. In Silico Molecular Docking
2.8. Molecular Dynamics (MD) Simulation
2.9. Statistical Analysis
3. Results
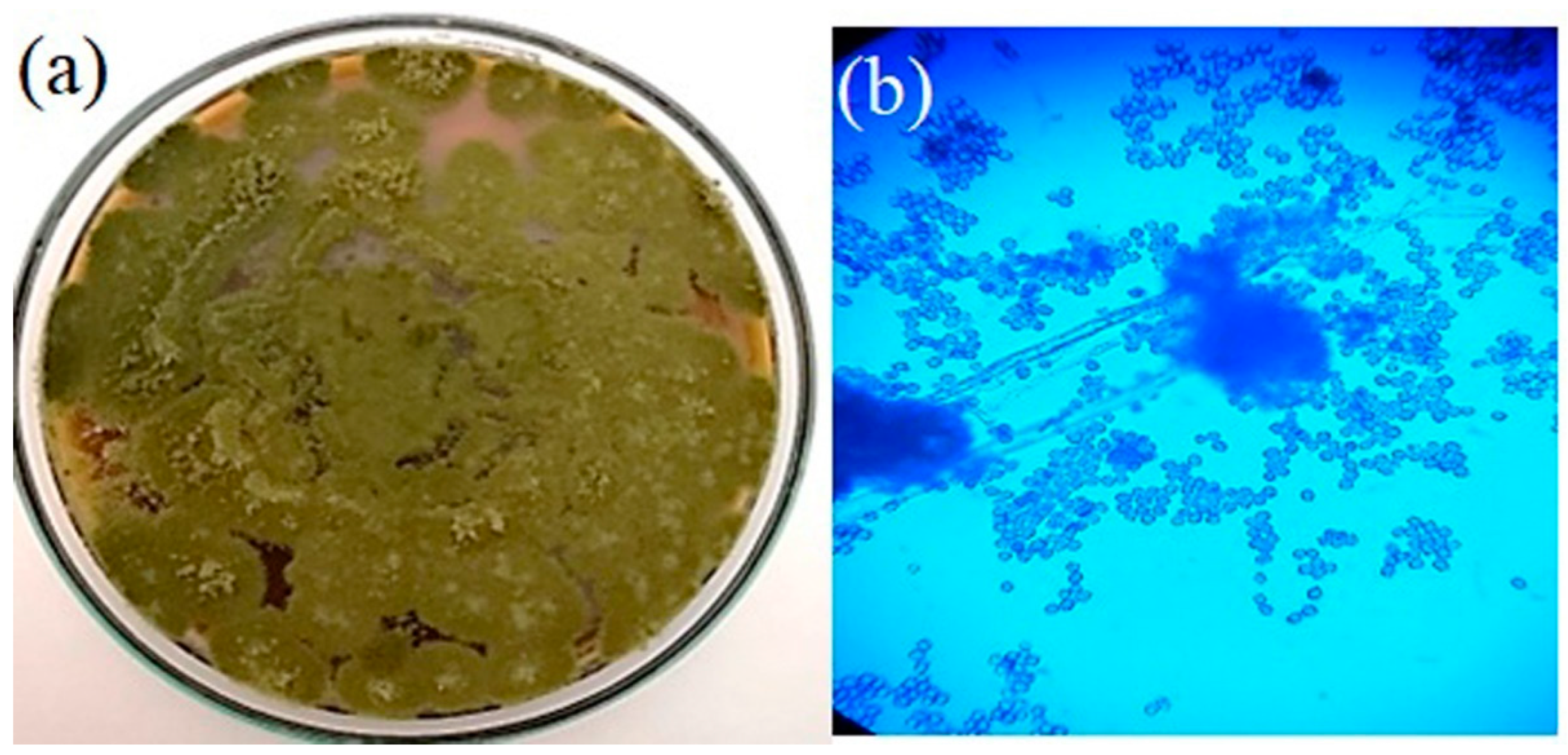
3.1. Isolation of Fungi and Microscopic Examination
3.2. Phylogenetic Analysis
3.3. Anticancer Activity
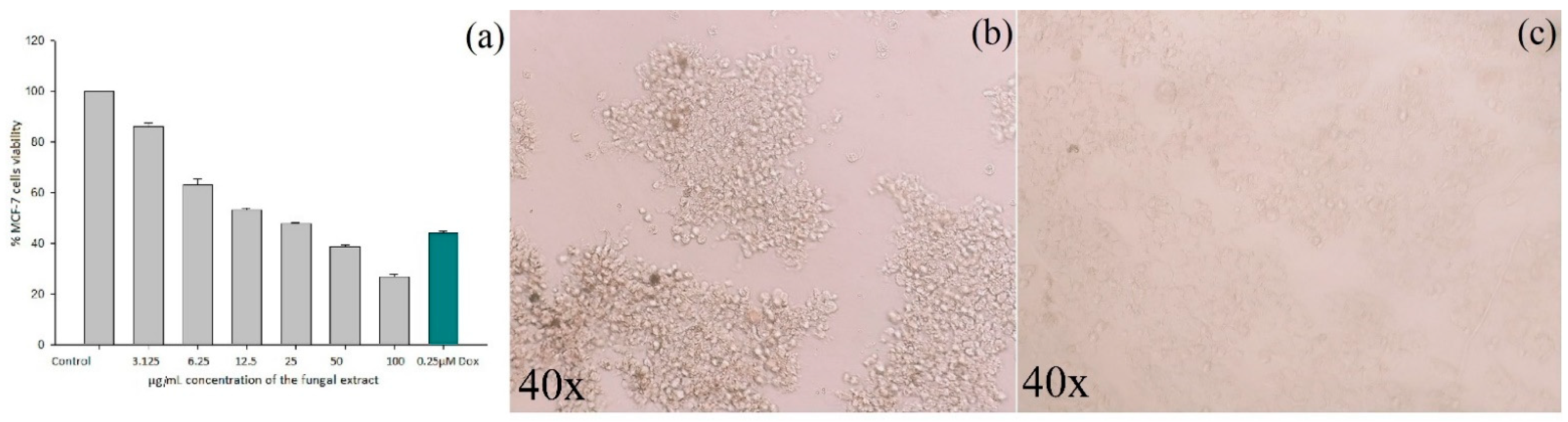
3.3.1. Cytotoxicity Assay
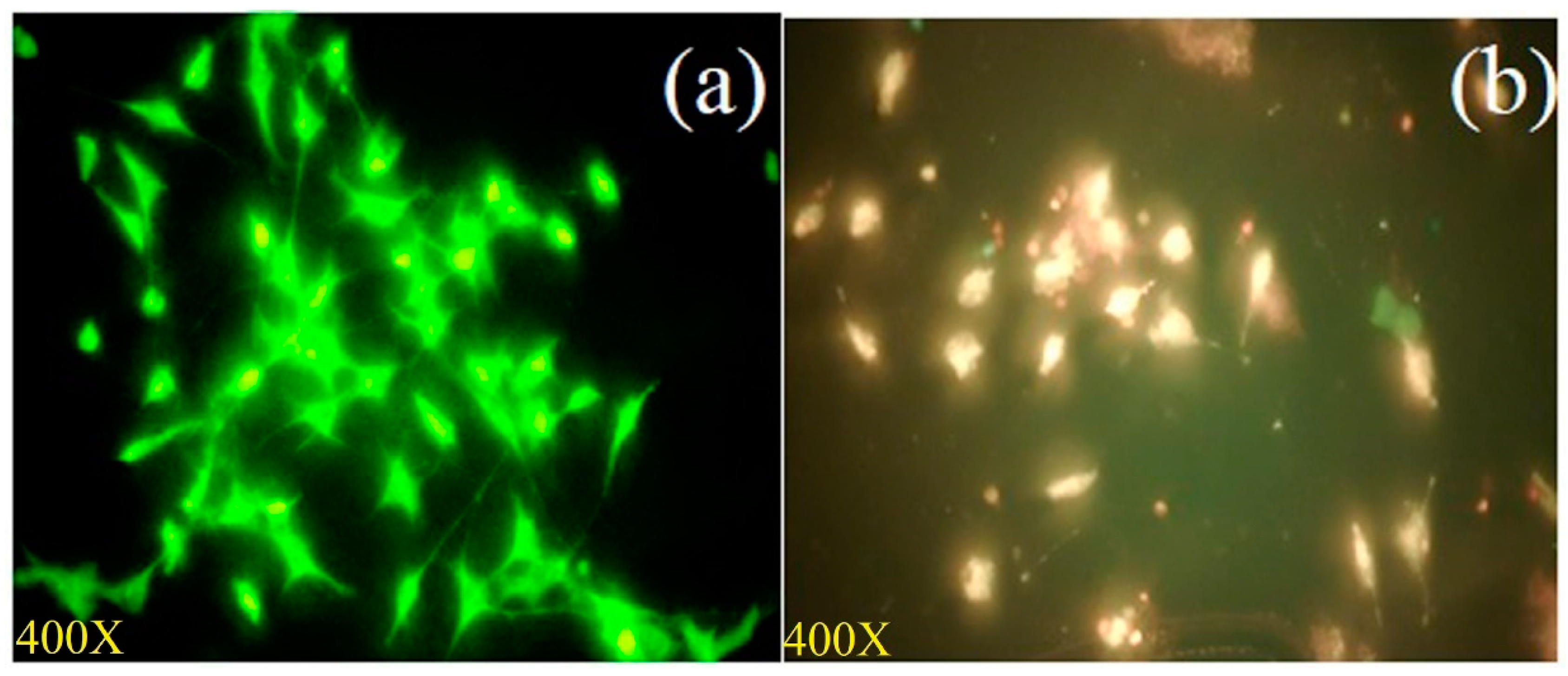
3.3.2. Analysis of Apoptosis Using Dual Staining
3.3.3. Assessment of Apoptosis by Flow Cytometer
3.3.4. Measurement of Mitochondrial Transmembrane Potential
3.3.5. Assessment of Nuclear Integrity by DAPI Staining
3.3.6. Assessment of Intracellular Reactive Oxygen Species
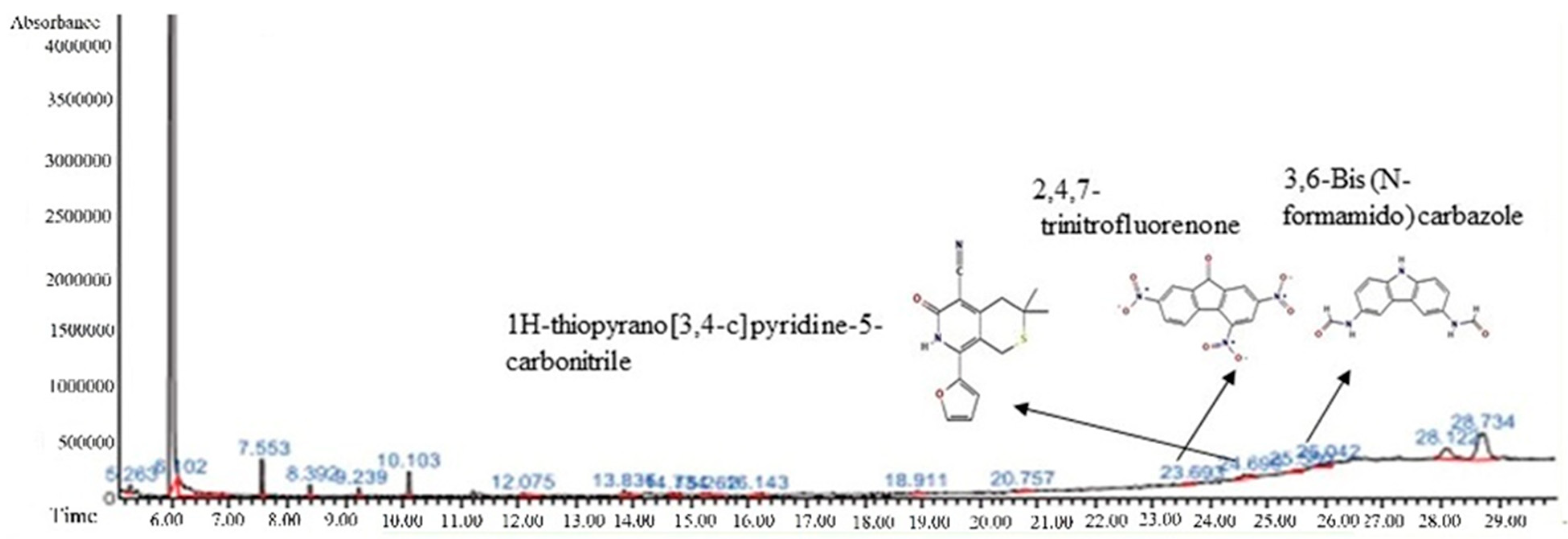
3.4. GC-MS Analysis
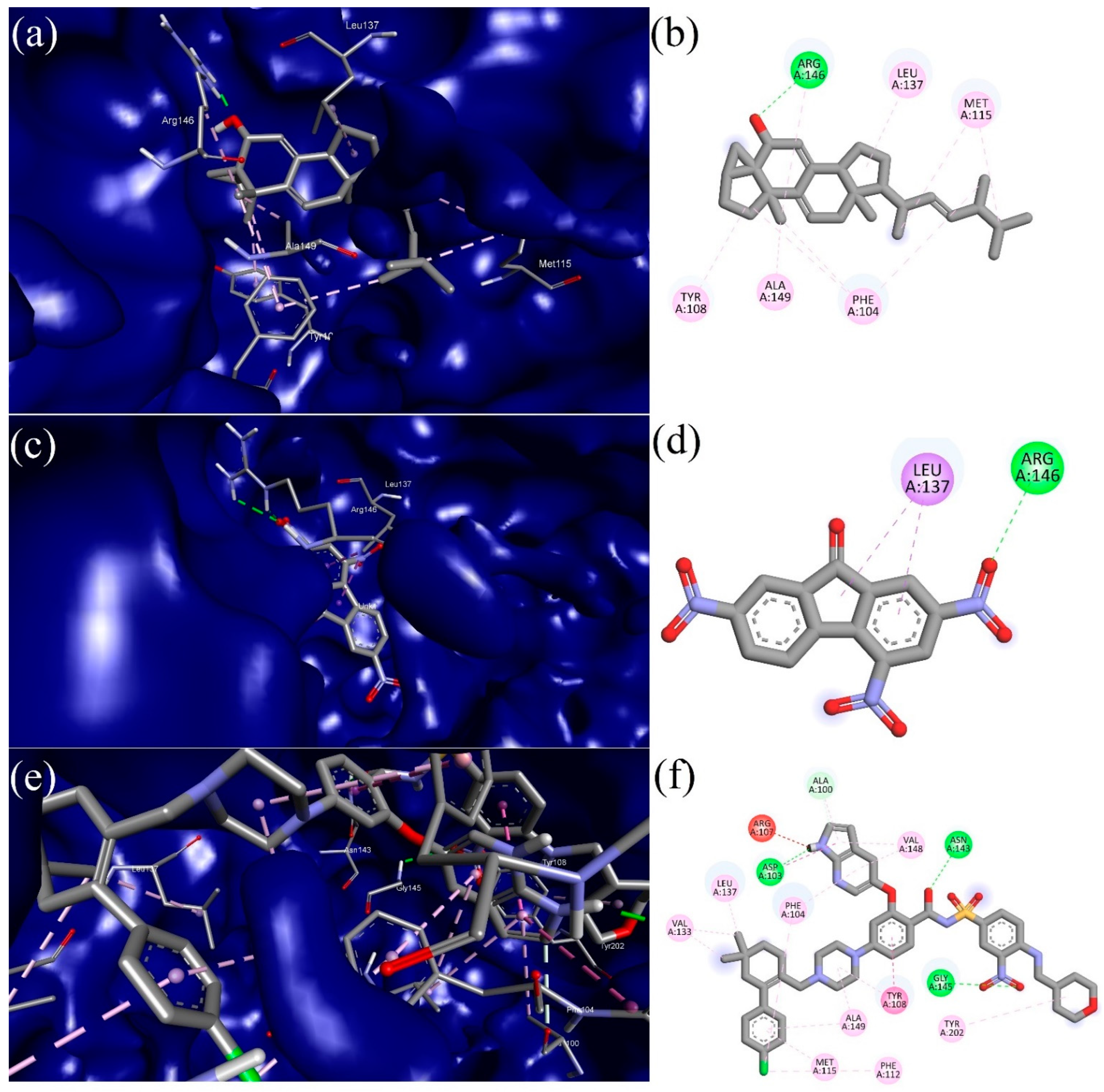
3.5. Molecular Docking (MD)
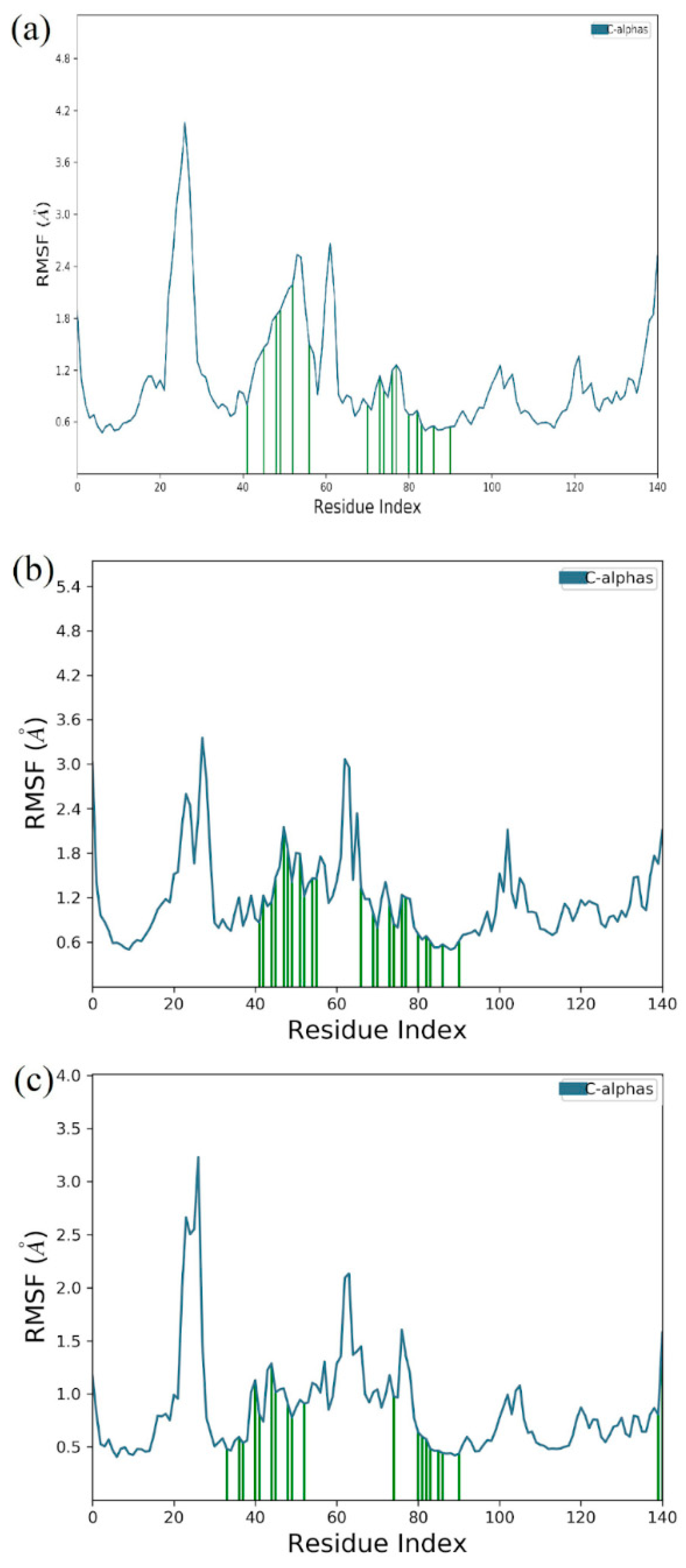
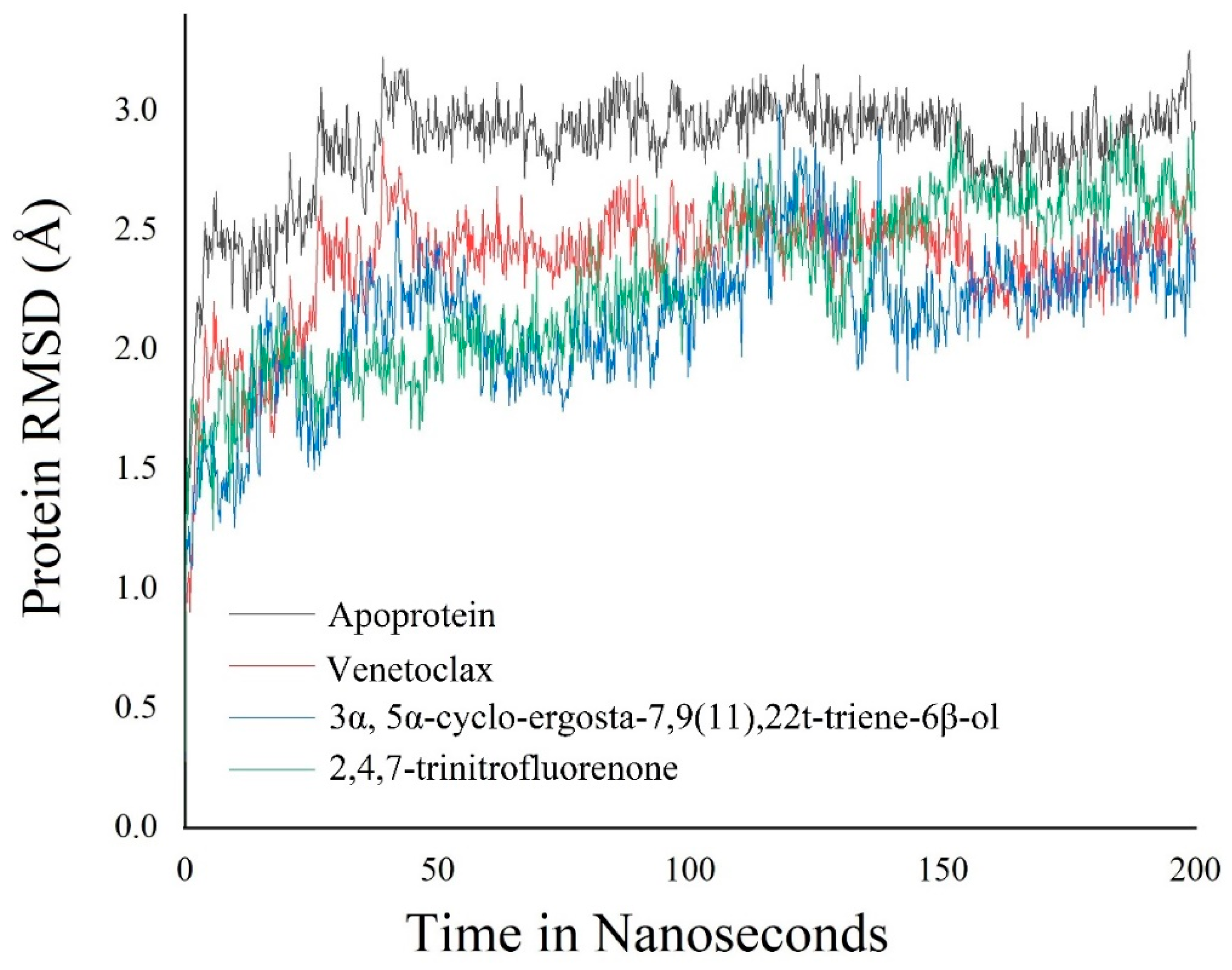
3.6. Molecular Dynamics Simulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wild, C.P. World Cancer Report 2014; Wild, C.P., Stewart, B.W., Eds.; World Health Organization: Geneva, Switzerland, 2014; pp. 482–494. [Google Scholar]
- Schiffman, J.D.; Breen, M. Comparative oncology: What dogs and other species can teach us about humans with cancer. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140231. [Google Scholar] [CrossRef] [PubMed]
- Eccles, S.A.; Aboagye, E.O.; Ali, S.; Anderson, A.S.; Armes, J.; Berditchevski, F.; Blaydes, J.P.; Brennan, K.; Brown, N.J.; Bryant, H.E.; et al. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res. 2013, 15, R92. [Google Scholar] [CrossRef]
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Shetty, S. International evaluation of an AI system for breast cancer screening. Nature 2020, 577, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Spezia, M.; Huang, S.; Yuan, C.; Zeng, Z.; Zhang, L.; Ji, X.; Liu, W.; Huang, B.; Luo, W.; et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018, 5, 77–106. [Google Scholar] [CrossRef]
- Williams, M.M.; Cook, R.S. Bcl-2 family proteins in breast development and cancer: Could Mcl-1 targeting overcome therapeutic resistance. Oncotarget 2015, 6, 3519–3530. [Google Scholar] [CrossRef]
- Chowdhury, A.; Kunjiappan, S.; Panneerselvam, T.; Somasundaram, B.; Bhattacharjee, C. Nanotechnology and nanocarrier-based approaches on treatment of degenerative diseases. Int. Nano Lett. 2017, 7, 91–122. [Google Scholar] [CrossRef]
- Runowicz, C.D.; Leach, C.R.; Henry, N.L.; Henry, K.S.; Mackey, H.T.; Cowens-Alvarado, R.L.; Cannady, R.S.; Pratt-Chapman, M.L.; Edge, S.B.; Jacobs, L.A.; et al. American cancer society/American society of clinical oncology breast cancer survivorship care guideline. CA A Cancer J. Clin. 2016, 66, 43–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Zhao, H.; Liang, B.; Du, Q. Clusterin confers resistance to TNF-alpha-induced apoptosis in breast cancer cells through NF-kappaB activation and Bcl-2 overexpression. J. Chemother. 2012, 24, 348–357. [Google Scholar] [CrossRef]
- Clark, A.M. Natural products as a resource for new drugs. Pharm. Res. 1996, 13, 1133–1141. [Google Scholar] [CrossRef]
- Lam, K.S. New aspects of natural products in drug discovery. Trends Microbiol. 2007, 15, 279–289. [Google Scholar] [CrossRef]
- Selvaraj, K.; Theivendren, P.; Pavadai, P.; Ravishankar, V.; Palanisamy, P.; Gopal, M.; Dharmalingam, S.R.; Sankaranarayanan, M. Impact of Physicochemical Parameters on Effective Extraction of Bioactive Compounds from Natural Sources: An Overview. Curr. Bioact. Compd. 2022, 18, 11–27. [Google Scholar] [CrossRef]
- Antonelli, A.; Smith, R.J.; Fry, C.; Simmonds, M.S.J.; Kersey, P.J.; Pritchard, H.W.; Ainsworth, A.M. State of the World’s Plants and Fungi; Royal Botanic Gardens (Kew): London, UK, 2020. [Google Scholar]
- Holliday, J.; Cleaver, M.P. Medicinal value of the caterpillar fungi species of the genus Cordyceps (Fr.) Link (Ascomycetes). A review. Int. J. Med. Mushrooms 2008, 10, 1–12. [Google Scholar] [CrossRef]
- Rappé, M.S.; Giovannoni, S.J. The uncultured microbial majority. Annu. Rev. Microbiol. 2003, 57, 369–394. [Google Scholar] [CrossRef] [PubMed]
- Cole, R.J.; Schweikert, M.A.; Jarvis, B.B. Handbook of Secondary Fungal Metabolites; Gulf Professional Publishing: Houston, TX, USA, 2003; Volume 3. [Google Scholar]
- Dewick, P.M. Medicinal Natural Products: A Biosynthetic Approach; John Wiley & Sons: Hoboken, NJ, USA, 2002; 552p. [Google Scholar]
- Turner, W.B. Fungal Metabolites; Academic Press: Cambridge, MA, USA, 1971; 446p. [Google Scholar]
- Kharwar, R.N.; Mishra, A.; Gond, S.K.; Stierle, A.; Stierle, D. Anticancer compounds derived from fungal endophytes: Their importance and future challenges. Nat. Prod. Rep. 2011, 28, 1208–1228. [Google Scholar] [CrossRef]
- Zhan, J.; Burns, A.M.; Liu, M.X.; Faeth, S.H.; Gunatilaka, A.L. Search for cell motility and angiogenesis inhibitors with potential anticancer activity: Beauvericin and other constituents of two endophytic strains of Fusarium oxysporum. J. Nat. Prod. 2007, 70, 227–232. [Google Scholar] [CrossRef]
- Yu, H.; Zhang, L.; Li, L.; Zheng, C.; Guo, L.; Li, W.; Sun, P.; Qin, L. Recent developments and future prospects of antimicrobial metabolites produced by endophytes. Microbiol. Res. 2010, 165, 437–449. [Google Scholar] [CrossRef]
- Khan, A.L.; Shahzad, R.; Al-Harrasi, A.; Lee, I.J. Endophytic microbes: A resource for producing extracellular enzymes. In Endophytes: Crop Productivity and Protection; Springer: Cham, Switzerland, 2017; pp. 95–110. [Google Scholar]
- Ratnaweera, P.B.; de Silva, E.D.; Williams, D.E.; Andersen, R.J. Antimicrobial activities of endophytic fungi obtained from the arid zone invasive plant Opuntia dillenii and the isolation of equisetin, from endophytic Fusarium sp. BMC Complement. Altern. Med. 2015, 15, 220. [Google Scholar] [CrossRef]
- Li, C.; Wang, F.; Wu, X.; Cao, S. A new 24-homo-30-nor-cycloartane triterpenoid from a Hawaiian endophytic fungal strain. Tetrahedron Lett. 2020, 61, 151508. [Google Scholar] [CrossRef]
- Kaliyaperumal, A.; Kumarakurubaran, S.; Saradha, D.M. Cynodon dactylon (L.) Pers.: An updated review of its phytochemistry and pharmacology. J. Med. Plants Res. 2013, 7, 3477–3483. [Google Scholar]
- Kowsalya, R.; Kaliaperumal, J.; Vaishnavi, M.; Namasivayam, E. Anticancer activity of Cynodon dactylon L. root extract against diethyl nitrosamine induced hepatic carcinoma. South Asian J. Cancer 2015, 4, 083–087. [Google Scholar] [CrossRef]
- Kleensang, A.; Vantangoli, M.M.; Odwin-DaCosta, S.; Andersen, M.E.; Boekelheide, K.; Bouhifd, M.; Fornace, A.J.; Li, H.-H.; Livi, C.B.; Madnick, S. Genetic variability in a frozen batch of MCF-7 cells invisible in routine authentication affecting cell function. Sci. Rep. 2016, 6, 28994. [Google Scholar] [CrossRef] [PubMed]
- Tapfuma, K.I.; Uche-Okereafor, N.; Sebola, T.E.; Hussan, R.; Mekuto, L.; Makatini, M.M.; Green, E.; Mavumengwana, V. Cytotoxic activity of crude extracts from Datura stramonium’s fungal endophytes against A549 lung carcinoma and UMG87 glioblastoma cell lines and LC-QTOF-MS/MS based metabolite profiling. BMC Complement. Altern. Med. 2019, 19, 330. [Google Scholar] [CrossRef] [PubMed]
- Sette, L.D.; Passarini, M.R.Z.; Delarmelina, C.; Salati, F.; Duarte, M.C.T. Molecular characterization and antimicrobial activity of endophytic fungi from coffee plants. World J. Microbiol. Biotechnol. 2006, 22, 1185–1195. [Google Scholar] [CrossRef]
- Kunjiappan, S.; Theivendran, P.; Baskararaj, S.; Sankaranarayanan, B.; Palanisamy, P.; Saravanan, G.; Arunachalam, S.; Sankaranarayanan, M.; Natarajan, J.; Somasundaram, B.; et al. Modeling a pH-sensitive Zein-co-acrylic acid hybrid hydrogels loaded 5-fluorouracil and rutin for enhanced anticancer efficacy by oral delivery. 3 Biotech 2019, 9, 185. [Google Scholar] [CrossRef]
- Kalimuthu, A.K.; Parasuraman, P.; Sivakumar, P.; Murugesan, S.; Arunachalam, S.; Pandian, S.R.K.; Ravishankar, V.; Ammunje, D.N.; Sampath, M.; Panneerselvam, T.; et al. In silico, in vitro screening of antioxidant and anticancer potentials of bioactive secondary metabolites from an endophytic fungus (Curvularia sp.) from Phyllanthus niruri L. Environ. Sci. Pollut. Res. 2022, 29, 48908–48925. [Google Scholar] [CrossRef]
- Mohan, U.P.; Sriram, B.; Panneerselvam, T.; Devaraj, S.; MubarakAli, D.; Parasuraman, P.; Palanisamy, P.; Premanand, A.; Arunachalam, S.; Kunjiappan, S. Utilization of plant-derived Myricetin molecule coupled with ultrasound for the synthesis of gold nanoparticles against breast cancer. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2020, 393, 1963–1976. [Google Scholar] [CrossRef]
- Kunjiappan, S.; Sankaranarayanan, M.; Kumar, B.K.; Pavadai, P.; Babkiewicz, E.; Maszczyk, P.; Glodkowska-Mrowka, E.; Arunachalam, S.; Pandian, S.R.K.; Ravishankar, V.; et al. Capsaicin-loaded solid lipid nanoparticles: Design, biodistribution, in silico modeling and in vitro cytotoxicity evaluation. Nanotechnology 2020, 32, 095101. [Google Scholar] [CrossRef]
- Kunjiappan, S.; Panneerselvam, T.; Govindaraj, S.; Kannan, S.; Parasuraman, P.; Arunachalam, S.; Sankaranarayanan, M.; Baskararaj, S.; Palanisamy, P.; Ammunje, D.N. Optimization and analysis of ultrasound-assisted extraction of bioactive polyphenols from Garcinia indica using RSM and ANFIS modeling and its anticancer activity. J. Iran. Chem. Soc. 2020, 17, 789–801. [Google Scholar] [CrossRef]
- Release, S. 3: Desmond molecular dynamics system, DE Shaw research, New York, NY, 2017. In Maestro-Desmond Interoperability Tools; Schrödinger: New York, NY, USA, 2017. [Google Scholar]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Basha, S.H.; Bethapudi, P.; Majji Rambabu, F. Anti-angiogenesis property by Quercetin compound targeting VEGFR2 elucidated in a computational approach. Eur. J. Biotechnol. Biosci. 2014, 2, 30–46. [Google Scholar]
- Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511–519. [Google Scholar] [CrossRef]
- Gunjan, M.; Naing, T.W.; Saini, R.S.; Ahmad, A.; Naidu, J.R.; Kumar, I. Marketing trends & future prospects of herbal medicine in the treatment of various disease. World J. Pharm. Res. 2015, 4, 132–155. [Google Scholar]
- Ijäs, H.; Shen, B.; Heuer-Jungemann, A.; Keller, A.; Kostiainen, M.A.; Liedl, T.; Ihalainen, J.A.; Linko, V. Unraveling the interaction between doxorubicin and DNA origami nanostructures for customizable chemotherapeutic drug release. Nucleic Acids Res. 2021, 49, 3048–3062. [Google Scholar] [CrossRef]
- Sadananda, T.; Govindappa, M.; Ramachandra, Y.; Chandrappa, C.; Umashankar, T. In Vitro Apoptotic Activity of Endophytic Fungal Lectin Isolated from Endophyte, Aspergillus flavus of Viscum album on Human Breast Adenocarcinoma Cell Line (MCF-7). Metabolomics 2016, 6, 162. [Google Scholar] [CrossRef]
- Pereira, S.; Castro, P. Diversity and characterization of culturable bacterial endophytes from Zea mays and their potential as plant growth-promoting agents in metal-degraded soils. Environ. Sci. Pollut. Res. 2014, 21, 14110–14123. [Google Scholar] [CrossRef]
- Siddiqui, M.; Rajkumar, S.V. The high cost of cancer drugs and what we can do about it. In Mayo Clinic Proceedings; No. 10; Elsevier: Amsterdam, The Netherlands, 2012; Volume 87, pp. 935–943. [Google Scholar]
- Pal, S.K.; Shukla, Y. Herbal medicine: Current status and the future. Asian Pac. J. Cancer Prev. 2003, 4, 281–288. [Google Scholar]
- Dakh, K.S.; Patekar, R.R.; Choudhary, H.B.; Momin, A.Z.; Undale, V.R.; Mahadik, P.; Desai, S.; Asalkar, M.; Shaikh, H. Herbal approach for tuberculosis management: A systematic review. World J. Adv. Res. Rev. 2022, 14, 637–647. [Google Scholar] [CrossRef]
- Tabish, S.A. Complementary and alternative healthcare: Is it evidence-based? Int. J. Health Sci. 2008, 2, V–IX. [Google Scholar]
- Amador, M.L.; Jimeno, J.; Paz-Ares, L.; Cortes-Funes, H.; Hidalgo, M. Progress in the development and acquisition of anticancer agents from marine sources. Ann. Oncol. 2003, 14, 1607–1615. [Google Scholar] [CrossRef]
- Rayan, A.; Raiyn, J.; Falah, M. Nature is the best source of anticancer drugs: Indexing natural products for their anticancer bioactivity. PLoS ONE 2017, 12, e0187925. [Google Scholar] [CrossRef]
- Das, K.; Tiwari, R.K.S.; Shrivastava, D.K. Techniques for evaluation of medicinal plant products as antimicrobial agent: Current methods and future trends. J. Med. Plants Res. 2010, 4, 104–111. [Google Scholar]
- Partida-Martínez, L.P.; Heil, M. The microbe-free plant: Fact or artifact? Front. Plant Sci. 2011, 2, 100. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S. Endophytic fungi: Novel sources of anticancer lead molecules. Appl. Microbiol. Biotechnol. 2012, 95, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Sopalun, K.; Laosripaiboon, W.; Wachirachaikarn, A.; Iamtham, S. Biological potential and chemical composition of bioactive compounds from endophytic fungi associated with thai mangrove plants. South Afr. J. Bot. 2021, 141, 66–76. [Google Scholar] [CrossRef]
- Firáková, S.; Šturdíková, M.; Múčková, M. Bioactive secondary metabolites produced by microorganisms associated with plants. Biologia 2007, 62, 251–257. [Google Scholar] [CrossRef]
- Nagori, B.P.; Solanki, R. Cynodon dactylon (L.) Pers.: A valuable medicinal plant. Res. J. Med. Plant 2011, 5, 508–514. [Google Scholar] [CrossRef]
- Selvaraj, K.; Chowdhury, R.; Bhattacharjee, C. Optimization of the solvent extraction of bioactive polyphenolic compounds from aquatic fern Azolla microphylla using response surface methodology. Int. Food Res. J. 2014, 21, 204–221. [Google Scholar]
- Ganesan, V.; Gurumani, V.; Kunjiappan, S.; Panneerselvam, T.; Somasundaram, B.; Kannan, S.; Chowdhury, A.; Saravanan, G.; Bhattacharjee, C. Optimization and analysis of microwave-assisted extraction of bioactive compounds from Mimosa pudica L. using RSM & ANFIS modeling. J. Food Meas. Charact. 2018, 12, 228–242. [Google Scholar]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2013, 1830, 3670–3695. [Google Scholar] [CrossRef]
- Majoumouo, M.S.; Tincho, M.B.; Toghueo, R.M.K.; Morris, T.; Hiss, D.C.; Boyom, F.F.; Mandal, C. Cytotoxicity potential of endophytic fungi extracts from Terminalia catappa against human cervical cancer cells. J. Toxicol. 2020, 2020, 8871152. [Google Scholar] [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Taggart, L.E.; McMahon, S.J.; Currell, F.J.; Prise, K.M.; Butterworth, K.T. The role of mitochondrial function in gold nanoparticle mediated radiosensitisation. Cancer Nanotechnol. 2014, 5, 5. [Google Scholar] [CrossRef]
- Redza-Dutordoir, M.; Averill-Bates, D.A. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2016, 1863, 2977–2992. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.; Yaglioglu, A.S.; Kishali, N.H.; Sahin, E.; Kara, Y. Evaluation of cytotoxic potentials of some isoindole-1, 3-dione derivatives on HeLa, C6 and A549 cancer cell lines. Med. Chem. 2020, 16, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wu, P.; Gao, J.; Zhang, L.; Ma, T.; Ma, B.; Yang, S.; Shao, G.; Yu, Y.; Huang, X.; et al. Heptadecanoic acid inhibits cell proliferation in PC9 nonsmallcell lung cancer cells with acquired gefitinib resistance. Oncol. Rep. 2019, 41, 3499–3507. [Google Scholar]
- Rhetso, T.; Shubharani, R.; Roopa, M.S.; Sivaram, V. Chemical constituents, antioxidant, and antimicrobial activity of Allium chinense G. Don. Future J. Pharm. Sci. 2020, 6, 102. [Google Scholar] [CrossRef]
- Ban, F.; Dalal, K.; Li, H.; LeBlanc, E.; Rennie, P.S.; Cherkasov, A. Best practices of computer-aided drug discovery: Lessons learned from the development of a preclinical candidate for prostate cancer with a new mechanism of action. J. Chem. Inf. Model. 2017, 57, 1018–1028. [Google Scholar] [CrossRef]
- Macalino, S.J.Y.; Gosu, V.; Hong, S.; Choi, S. Role of computer-aided drug design in modern drug discovery. Arch. Pharm. Res. 2015, 38, 1686–1701. [Google Scholar] [CrossRef]
- Baig, M.H.; Ahmad, K.; Rabbani, G.; Danishuddin, M.; Choi, I. Computer aided drug design and its application to the development of potential drugs for neurodegenerative disorders. Curr. Neuropharmacol. 2018, 16, 740–748. [Google Scholar] [CrossRef]
- Kønig, S.M.; Rissler, V.; Terkelsen, T.; Lambrughi, M.; Papaleo, E. Alterations of the interactome of Bcl-2 proteins in breast cancer at the transcriptional, mutational and structural level. PLoS Comput. Biol. 2019, 15, e1007485. [Google Scholar] [CrossRef] [PubMed]
- Kuwana, T.; Newmeyer, D.D. Bcl-2-family proteins and the role of mitochondria in apoptosis. Curr. Opin. Cell Biol. 2003, 15, 691–699. [Google Scholar] [CrossRef] [PubMed]

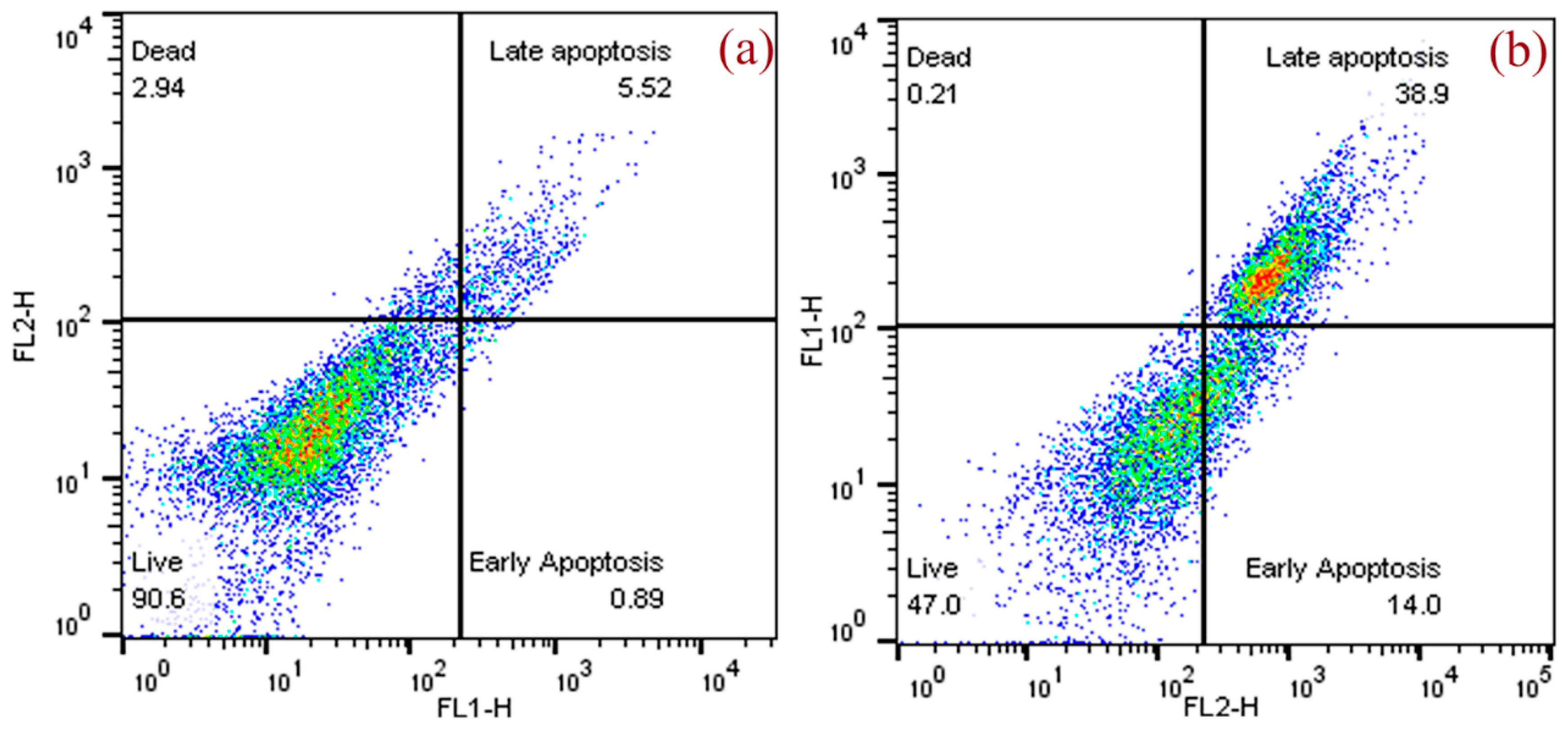

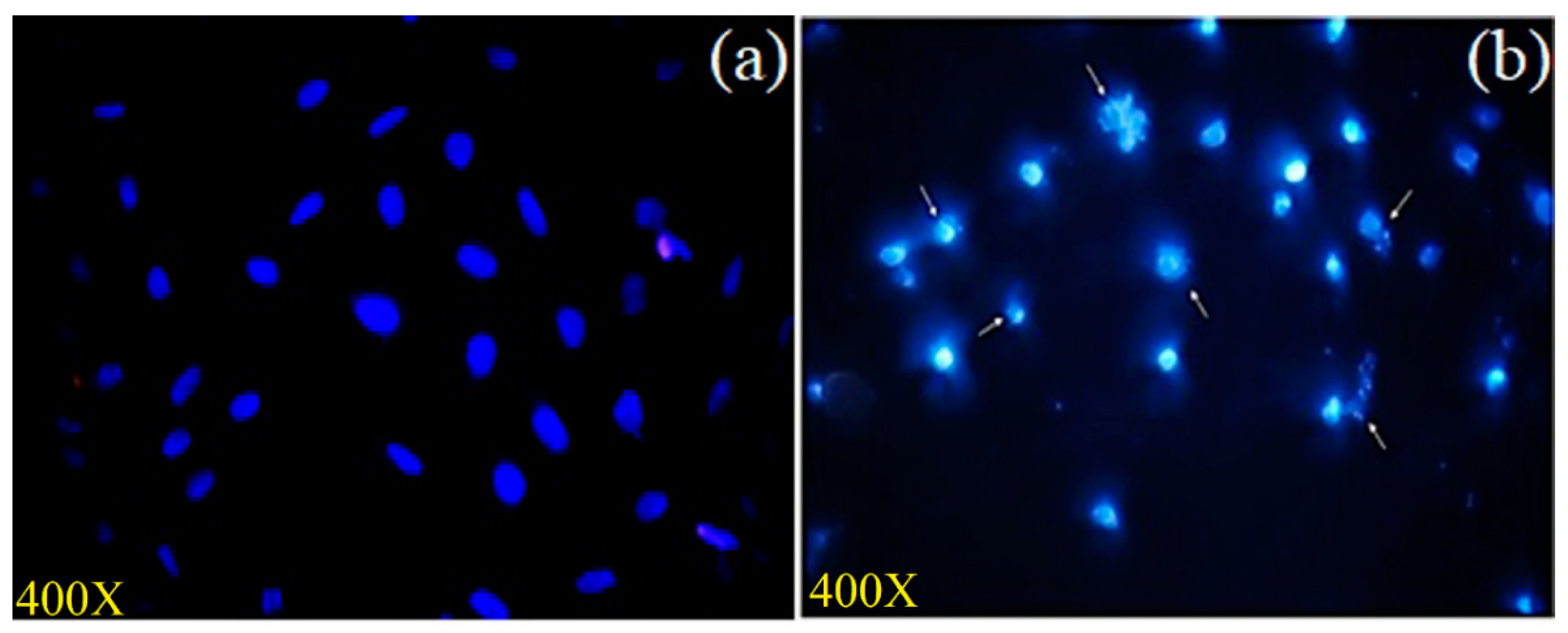
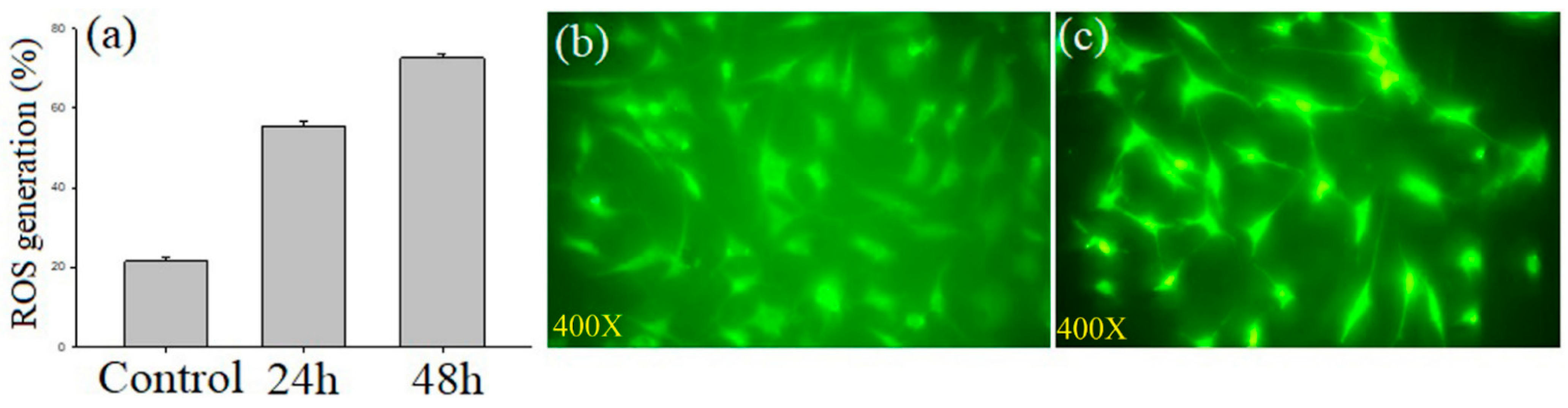
| MCF-7 Cells | Q4 Live (%) | Q3 Early Apoptosis (%) | Q2 Late Apoptosis (%) | Q1 Dead (%) |
|---|---|---|---|---|
| Control | 90.6 | 0.89 | 5.52 | 2.94 |
| EF extract | 47.0 | 14.0 | 38.9 | 0.21 |
| S. No | Retention Time | % Area of Peak | Phytochemical Compounds | Molecular Formula | Molecular Weight (in g/mol) | Structure |
|---|---|---|---|---|---|---|
| 1 | 23.693 | 0.25 | 2,4,7-trinitrofluorenone | C13H5N3O7 | 315.19 | 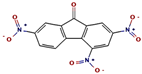 |
| 2 | 25.580 | 0.80 | 3,6-Bis (N-formamido) carbazole | C14H11N3O2 | 253.26 | 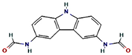 |
| 3 | 24.699 | 0.86 | 1H-thiopyrano[3,4-c] pyridine-5-carbonitrile | C15H14N2O2S | 286.4 | 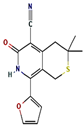 |
| 4 | 28.734 | 6.00 | 1H-isoindole-1,3(2H)-dione | C13H10N2O4 | 258.23 |  |
| 5 | 18.911 | 0.30 | Heptadecanoic acid | C17H34O2 | 270.5 |  |
| 6 | 18.911 | 0.30 | Methyl 2,8-dimethyltridecanoate | C16H32O2 | 256.42 | 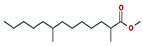 |
| 7 | 26.042 | 1.15 | 3α,5α-cyclo-ergosta-7,9(11),22t-triene-6β-ol | C28H42O | 394.6 |  |
| 8 | 20.757 | 0.33 | 2,3,4-trimethyllevoglucosan | C9H16O5 | 204.22 |  |
| 9 | 14.734 | 0.21 | Tritetracontane | C43H88 | 605.2 |  |
| S. No | CID | Compound | Binding Affinity (Kcal/mol) |
|---|---|---|---|
| 1 | 8521 | 2,4,7-trinitrofluorenone | −9.2 |
| 2 | 620086 | 3,6-Bis (N-formamido) carbazole | −6.2 |
| 3 | 6809 | 1H-isoindole-1,3(2H)-dione | −6.1 |
| 4 | 658451 | 1H-thiopyrano [3,4-c] pyridine-5-carbonitrile | −6.2 |
| 5 | 10465 | Heptadecanoic acid | −5.4 |
| 6 | 560473 | Methyl 2,8-dimethyltridecanoate | −5.5 |
| 7 | 5363271 | 3α, 5α-cyclo-ergosta-7,9(11),22t-triene-6β-ol | −9.5 |
| 8 | 91699158 | 2,3,4-trimethyllevoglucosan | −4.5 |
| 9 | 522398 | Tritetracontane | −5.3 |
| Standard drug | |||
| 10 | 49846579 | Venetoclax | −10.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalimuthu, A.K.; Pavadai, P.; Panneerselvam, T.; Babkiewicz, E.; Pijanowska, J.; Mrówka, P.; Rajagopal, G.; Deepak, V.; Sundar, K.; Maszczyk, P.; et al. Cytotoxic Potential of Bioactive Compounds from Aspergillus flavus, an Endophytic Fungus Isolated from Cynodon dactylon, against Breast Cancer: Experimental and Computational Approach. Molecules 2022, 27, 8814. https://doi.org/10.3390/molecules27248814
Kalimuthu AK, Pavadai P, Panneerselvam T, Babkiewicz E, Pijanowska J, Mrówka P, Rajagopal G, Deepak V, Sundar K, Maszczyk P, et al. Cytotoxic Potential of Bioactive Compounds from Aspergillus flavus, an Endophytic Fungus Isolated from Cynodon dactylon, against Breast Cancer: Experimental and Computational Approach. Molecules. 2022; 27(24):8814. https://doi.org/10.3390/molecules27248814
Chicago/Turabian StyleKalimuthu, Arjun Kumar, Parasuraman Pavadai, Theivendren Panneerselvam, Ewa Babkiewicz, Joanna Pijanowska, Piotr Mrówka, Gopalan Rajagopal, Venkataraman Deepak, Krishnan Sundar, Piotr Maszczyk, and et al. 2022. "Cytotoxic Potential of Bioactive Compounds from Aspergillus flavus, an Endophytic Fungus Isolated from Cynodon dactylon, against Breast Cancer: Experimental and Computational Approach" Molecules 27, no. 24: 8814. https://doi.org/10.3390/molecules27248814
APA StyleKalimuthu, A. K., Pavadai, P., Panneerselvam, T., Babkiewicz, E., Pijanowska, J., Mrówka, P., Rajagopal, G., Deepak, V., Sundar, K., Maszczyk, P., & Kunjiappan, S. (2022). Cytotoxic Potential of Bioactive Compounds from Aspergillus flavus, an Endophytic Fungus Isolated from Cynodon dactylon, against Breast Cancer: Experimental and Computational Approach. Molecules, 27(24), 8814. https://doi.org/10.3390/molecules27248814








