Evolution of DIMBOA-Glc O-Methyltransferases from Flavonoid O-Methyltransferases in the Grasses
Abstract
:1. Introduction
2. Results
2.1. Close Homologs of the Flavonoid 5-OMT ZmFOMT2 Are Restricted to the Panicoideae Subfamily
2.2. ZmFOMT2 Homologs Have DIMBOA-Glc 4-OMT or Flavonoid 5-OMT Activity
2.3. The DIMBOA-Glc 4-OMTs ZmBX10-12 and ZnBX10 also Possess Flavonoid 3-OMT Activity with Flavonols
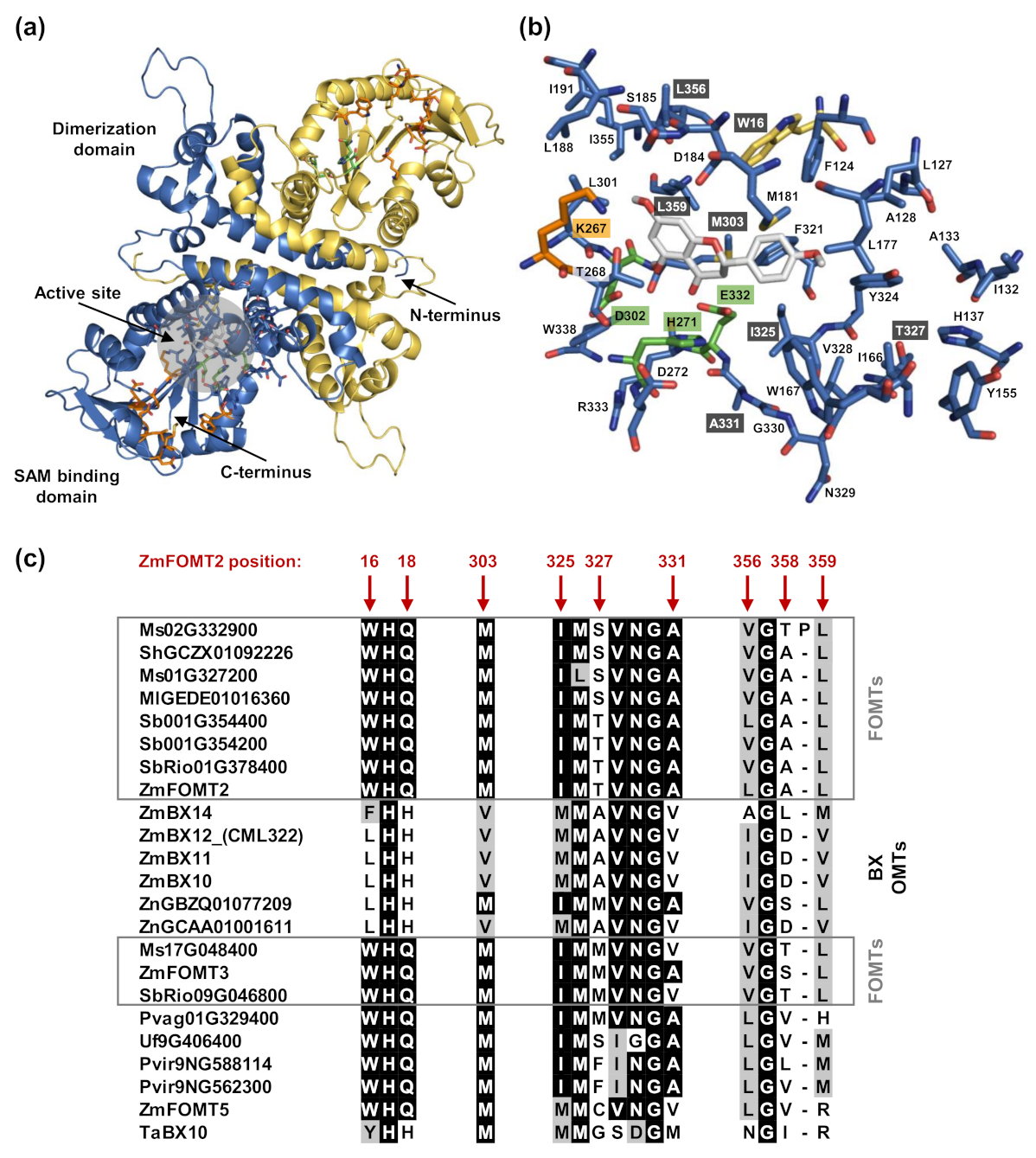
2.4. Identification of Active Site Residues Determining the Substrate Specificities and Activities of Flavonoid 5-OMTs and DIMBOA-Glc 4-OMTs
3. Discussion
4. Materials and Methods
4.1. Plants and Growth Conditions
4.2. RNA and cDNA Preparation
4.3. Gene Synthesis
4.4. Site-Directed Mutagenesis
4.5. Cloning and Heterologous Expression of OMT Genes in E. coli
4.6. In Vitro Enzyme Assays
4.7. Liquid Chromatography–Mass Spectrometry (LC–MS) Analysis of BXs and Flavonoids in Enzyme Assays
4.7.1. Targeted LC–MS/MS Analysis
4.7.2. Untargeted LC–MS Analysis with Accurate Mass Determination
4.8. Sequence Alignment and Phylogenetic Analysis
4.9. Homology Modeling and Molecular Docking
4.10. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Ibrahim, R.K.; Deluca, V.; Khouri, H.; Latchinian, L.; Brisson, L.; Charest, P.M. Enzymology and Compartmentation of Polymethylated Flavonol Glucosides in Chrysosplenium americanum. Phytochemistry 1987, 26, 1237–1245. [Google Scholar] [CrossRef]
- Ibrahim, R.K.; Bruneau, A.; Bantignies, B. Plant O-methyltransferases: Molecular analysis, common signature and classification. Plant Mol. Biol. 1998, 36, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Paiva, N.L. Stress-Induced Phenylpropanoid Metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Hammerschmidt, R. Phytoalexins: What Have we Learned After 60 Years? Annu. Rev. Phytopathol. 1999, 37, 285–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, C.P.; Chiang, V.L. Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferases. Plant Mol. Biol. 1998, 37, 663–674. [Google Scholar] [CrossRef]
- Lam, K.C.; Ibrahim, R.K.; Behdad, B.; Dayanandan, S. Structure, function, and evolution of plant O-methyltransferases. Genome 2007, 50, 1001–1013. [Google Scholar] [CrossRef] [Green Version]
- Noel, J.P.; Dixon, R.A.; Pichersky, E.; Zubieta, C.; Ferrer, J.L. Structural, functional, and evolutionary basis for methylation of plant small molecules. In Recent Advances in Phytochemistry; Romeo, J.T., Ed.; Elsevier: Amsterdam, The Netherlands, 2003; Volume 37, pp. 37–58. [Google Scholar]
- Qu, L.J.; Li, S.A.; Xing, S.F. Methylation of phytohormones by the SABATH methyltransferases. Chin. Sci. Bull. 2010, 55, 2211–2218. [Google Scholar] [CrossRef]
- Kim, B.G.; Sung, S.H.; Chong, Y.; Lim, Y.; Ahn, J.H. Plant Flavonoid O-Methyltransferases: Substrate Specificity and Application. J. Plant Biol. 2010, 53, 321–329. [Google Scholar] [CrossRef]
- Gauthier, A.; Gulick, P.J.; Ibrahim, R.K. Characterization of Two cDNA Clones Which Encode O-Methyltransferases for the Methylation of both Flavonoid and Phenylpropanoid Compounds. Arch. Biochem. Biophys. 1998, 351, 243–249. [Google Scholar] [CrossRef]
- Frick, S.; Kutchan, T.M. Molecular cloning and functional expression of O-methyltransferases common to isoquinoline alkaloid and phenylpropanoid biosynthesis. Plant J. 1999, 17, 329–339. [Google Scholar] [CrossRef]
- Deavours, B.E.; Liu, C.J.; Naoumkina, M.A.; Tang, Y.H.; Farag, M.A.; Sumner, L.W.; Noel, J.P.; Dixon, R.A. Functional analysis of members of the isoflavone and isoflavanone O-methyltransferase enzyme families from the model legume Medicago truncatula. Plant Mol. Biol. 2006, 62, 715–733. [Google Scholar] [CrossRef] [Green Version]
- Itoh, N.; Iwata, C.; Toda, H. Molecular cloning and characterization of a flavonoid-O-methyltransferase with broad substrate specificity and regioselectivity from Citrus depressa. BMC Plant Biol. 2016, 16, 180. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Wang, Y.; Chen, Y.; Xu, S.; Gong, Q.; Zhao, C.; Cao, J.; Sun, C. Characterization of a Flavonoid 3′/5′/7-O-Methyltransferase from Citrus reticulata and Evaluation of the In Vitro Cytotoxicity of Its Methylated Products. Molecules 2020, 25, 858. [Google Scholar] [CrossRef] [Green Version]
- Gang, D.R.; Lavid, N.; Zubieta, C.; Chen, F.; Beuerle, T.; Lewinsohn, E.; Noel, J.P.; Pichersky, E. Characterization of Phenylpropene O-Methyltransferases from Sweet Basil: Facile Change of Substrate Specificity and Convergent Evolution within a Plant O-Methyltransferase Family. Plant Cell 2002, 14, 505–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.M.; Lee, E.; Kanapathy-Sinnaiaha, F.; Park, Y.; Kornblatt, J.A.; Lim, Y.; Ibrahim, R.K. Structure-function relationships of wheat flavone O-methyltransferase: Homology modeling and site-directed mutagenesis. BMC Plant Biol. 2010, 10, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berim, A.; Gang, D.R. Methoxylated flavones: Occurrence, importance, biosynthesis. Phytochem. Rev. 2016, 15, 363–390. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The Origin and Evolution of Plant Flavonoid Metabolism. Front. Plant Sci. 2019, 10, 943. [Google Scholar] [CrossRef] [Green Version]
- Tohge, T.; de Souza, L.P.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028. [Google Scholar] [CrossRef] [PubMed]
- Kodama, O.; Miyakawa, J.; Akatsuka, T.; Kiyosawa, S. Sakuranetin, a Flavanone Phytoalexin from Ultraviolet-Irradiated Rice Leaves. Phytochemistry 1992, 31, 3807–3809. [Google Scholar] [CrossRef]
- Rakwal, R.; Tamogami, S.; Kodama, O. Role of Jasmonic Acid as a Signaling Molecule in Copper Chloride-elicited Rice Phytoalexin Production. Biosci. Biotech. Bioch. 1996, 60, 1046–1048. [Google Scholar] [CrossRef]
- Hasegawa, M.; Mitsuhara, I.; Seo, S.; Okada, K.; Yamane, H.; Iwai, T.; Ohashi, Y. Analysis on Blast Fungus-Responsive Characters of a Flavonoid Phytoalexin Sakuranetin; Accumulation in Infected Rice Leaves, Antifungal Activity and Detoxification by Fungus. Molecules 2014, 19, 11404–11418. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Kitano, T.; Yoshimoto, R.; Takata, R.; Ube, N.; Ueno, K.; Ueno, M.; Yabuta, Y.; Teraishi, M.; Holland, C.K.; et al. Natural variation in the expression and catalytic activity of a naringenin 7-O-methyltransferase influences antifungal defenses in diverse rice cultivars. Plant J. 2020, 101, 1103–1117. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Lin, F.Q.; Hasegawa, M.; Okada, K.; Nojiri, H.; Yamane, H. Purification and Identification of Naringenin 7-O-Methyltransferase, a Key Enzyme in Biosynthesis of Flavonoid Phytoalexin Sakuranetin in Rice. J. Biol. Chem. 2012, 287, 19315–19325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo, S.C.; Weiergang, I.; Bonham, C.; Hipskind, J.; Wood, K.; Nicholson, R.L. Phytoalexin accumulation in sorghum: Identification of a methyl ether of luteolinidin. Physiol. Mol. Plant Pathol. 1996, 49, 21–31. [Google Scholar] [CrossRef]
- Balmer, D.; de Papajewski, D.V.; Planchamp, C.; Glauser, G.; Mauch-Mani, B. Induced resistance in maize is based on organ-specific defence responses. Plant J. 2013, 74, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Ube, N.; Katsuyama, Y.; Kariya, K.; Tebayashi, S.; Sue, M.; Tohnooka, T.; Ueno, K.; Taketa, S.; Ishihara, A. Identification of methoxylchalcones produced in response to CuCl2 treatment and pathogen infection in barley. Phytochemistry 2021, 184, 112650. [Google Scholar] [CrossRef] [PubMed]
- Förster, C.; Handrick, V.; Ding, Y.; Nakamura, Y.; Paetz, C.; Schneider, B.; Castro-Falcón, G.; Hughes, C.C.; Luck, K.; Poosapati, S.; et al. Biosynthesis and antifungal activity of fungus-induced O-methylated flavonoids in maize. Plant Physiol. 2022, 188, 167–190. [Google Scholar] [CrossRef] [PubMed]
- Frey, M.; Schullehner, K.; Dick, R.; Fiesselmann, A.; Gierl, A. Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 2009, 70, 1645–1651. [Google Scholar] [CrossRef]
- Kokubo, Y.; Nishizaka, M.; Ube, N.; Yabuta, Y.; Tebayashi, S.; Ueno, K.; Taketa, S.; Ishihara, A. Distribution of the tryptophan pathway-derived defensive secondary metabolites gramine and benzoxazinones in Poaceae. Biosci. Biotech. Bioch. 2017, 81, 431–440. [Google Scholar] [CrossRef] [Green Version]
- Meihls, L.N.; Handrick, V.; Glauser, G.; Barbier, H.; Kaur, H.; Haribal, M.M.; Lipka, A.E.; Gershenzon, J.; Buckler, E.S.; Erb, M.; et al. Natural Variation in Maize Aphid Resistance Is Associated with 2,4-Dihydroxy-7-Methoxy-1,4-Benzoxazin-3-One Glucoside Methyltransferase Activity. Plant Cell 2013, 25, 2341–2355. [Google Scholar] [CrossRef] [Green Version]
- Handrick, V.; Robert, C.A.M.; Ahern, K.R.; Zhou, S.Q.; Machado, R.A.R.; Maag, D.; Glauser, G.; Fernandez-Penny, F.E.; Chandran, J.N.; Rodgers-Melnik, E.; et al. Biosynthesis of 8-O-Methylated Benzoxazinoid Defense Compounds in Maize. Plant Cell 2016, 28, 1682–1700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glauser, G.; Marti, G.; Villard, N.; Doyen, G.A.; Wolfender, J.L.; Turlings, T.C.J.; Erb, M. Induction and detoxification of maize 1,4-benzoxazin-3-ones by insect herbivores. Plant J. 2011, 68, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Maag, D.; Köhler, A.; Robert, C.A.M.; Frey, M.; Wolfender, J.L.; Turlings, T.C.J.; Glauser, G.; Erb, M. Highly localized and persistent induction of Bx1-dependent herbivore resistance factors in maize. Plant J. 2016, 88, 976–991. [Google Scholar] [CrossRef] [PubMed]
- Cambier, V.; Hance, T.; De Hoffmann, E. Effects of 1,4-Benzoxazin-3-One Derivatives from Maize on Survival and Fecundity of Metopolophium dirhodum (Walker) on Artificial Diet. J. Chem. Ecol. 2001, 27, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, A.; Ishihara, A.; Hasegawa, M.; Kodama, O.; Iwamura, H. Induced accumulation of 2-hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one glucoside (HDMBOA-Glc) in maize leaves. Phytochemistry 2001, 56, 669–675. [Google Scholar] [CrossRef]
- Oikawa, A.; Ishihara, A.; Tanaka, C.; Mori, N.; Tsuda, M.; Iwamura, H. Accumulation of HDMBOA-Glc is induced by biotic stresses prior to the release of MBOA in maize leaves. Phytochemistry 2004, 65, 2995–3001. [Google Scholar] [CrossRef]
- Oikawa, A.; Ishihara, A.; Iwamura, H. Induction of HDMBOA-Glc accumulation and DIMBOA-Glc 4-O-methyltransferase by jasmonic acid in poaceous plants. Phytochemistry 2002, 61, 331–337. [Google Scholar] [CrossRef]
- Li, B.; Förster, C.; Robert, C.A.M.; Züst, T.; Hu, L.; Machado, R.A.R.; Berset, J.-D.; Handrick, V.; Knauer, T.; Hensel, G.; et al. Convergent evolution of a metabolic switch between aphid and caterpillar resistance in cereals. Sci. Adv. 2018, 4, eaat6797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makowska, B.; Bakera, B.; Rakoczy-Trojanowska, M. The genetic background of benzoxazinoid biosynthesis in cereals. Acta Physiol. Plant. 2015, 37, 176. [Google Scholar] [CrossRef] [Green Version]
- Soreng, R.J.; Peterson, P.M.; Romaschenko, K.; Davidse, G.; Teisher, J.K.; Clark, L.G.; Barbera, P.; Gillespie, L.J.; Zuloaga, F.O. A worldwide phylogenetic classification of the Poaceae (Gramineae) II: An update and a comparison of two 2015 classifications. J. Syst. Evol. 2017, 55, 259–290. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.J.; Deavours, B.E.; Richard, S.B.; Ferrer, J.L.; Blount, J.W.; Huhman, D.; Dixon, R.A.; Noel, J.P. Structural Basis for Dual Functionality of Isoflavonoid O-Methyltransferases in the Evolution of Plant Defense Responses. Plant Cell 2006, 18, 3656–3669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joe, E.J.; Kim, B.G.; An, B.C.; Chong, Y.; Ahn, J.H. Engineering of Flavonoid O-Methyltransferase for a Novel Regioselectivity. Mol. Cells 2010, 30, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Vianney, Y.M.; Weisz, K.; Grathwol, C.W.; Link, A.; Bornscheuer, U.T.; Pavlidis, I.V. Influence of Substrate Binding Residues on the Substrate Scope and Regioselectivity of a Plant O-Methyltransferase against Flavonoids. ChemCatChem 2020, 12, 3721–3727. [Google Scholar] [CrossRef]
- Greenhagen, B.T.; O’Maille, P.E.; Noel, J.P.; Chappell, J. Identifying and manipulating structural determinates linking catalytic specificities in terpene synthases. Proc. Natl. Acad. Sci. USA 2006, 103, 9826–9831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köllner, T.G.; Degenhardt, J.; Gershenzon, J. The Product Specificities of Maize Terpene Synthases TPS4 and TPS10 Are Determined Both by Active Site Amino Acids and Residues Adjacent to the Active Site. Plants 2020, 9, 552. [Google Scholar] [CrossRef]
- Liu, H.T.; Naismith, J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008, 8, 91. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. Software News and Update AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Förster, C.; Gershenzon, J.; Köllner, T.G. Evolution of DIMBOA-Glc O-Methyltransferases from Flavonoid O-Methyltransferases in the Grasses. Molecules 2022, 27, 1007. https://doi.org/10.3390/molecules27031007
Förster C, Gershenzon J, Köllner TG. Evolution of DIMBOA-Glc O-Methyltransferases from Flavonoid O-Methyltransferases in the Grasses. Molecules. 2022; 27(3):1007. https://doi.org/10.3390/molecules27031007
Chicago/Turabian StyleFörster, Christiane, Jonathan Gershenzon, and Tobias G. Köllner. 2022. "Evolution of DIMBOA-Glc O-Methyltransferases from Flavonoid O-Methyltransferases in the Grasses" Molecules 27, no. 3: 1007. https://doi.org/10.3390/molecules27031007
APA StyleFörster, C., Gershenzon, J., & Köllner, T. G. (2022). Evolution of DIMBOA-Glc O-Methyltransferases from Flavonoid O-Methyltransferases in the Grasses. Molecules, 27(3), 1007. https://doi.org/10.3390/molecules27031007





