Comparative Phosphoproteomic Analysis of Sporulated Oocysts and Tachyzoites of Toxoplasma gondii Reveals Stage-Specific Patterns
Abstract
:1. Introduction
2. Results
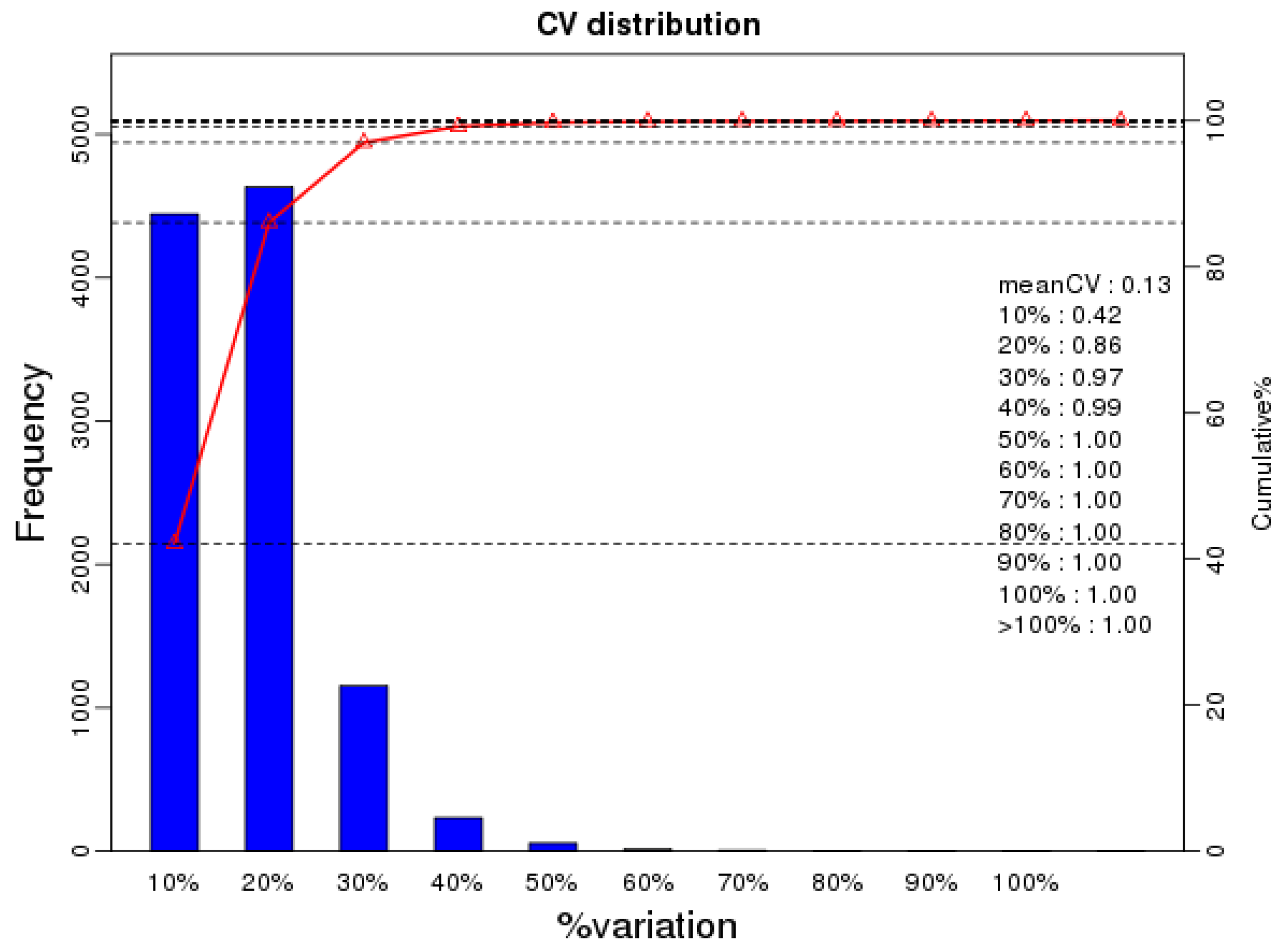
2.1. Phosphopeptide Profiling of Mature Oocysts and Tachyzoites of T. gondii
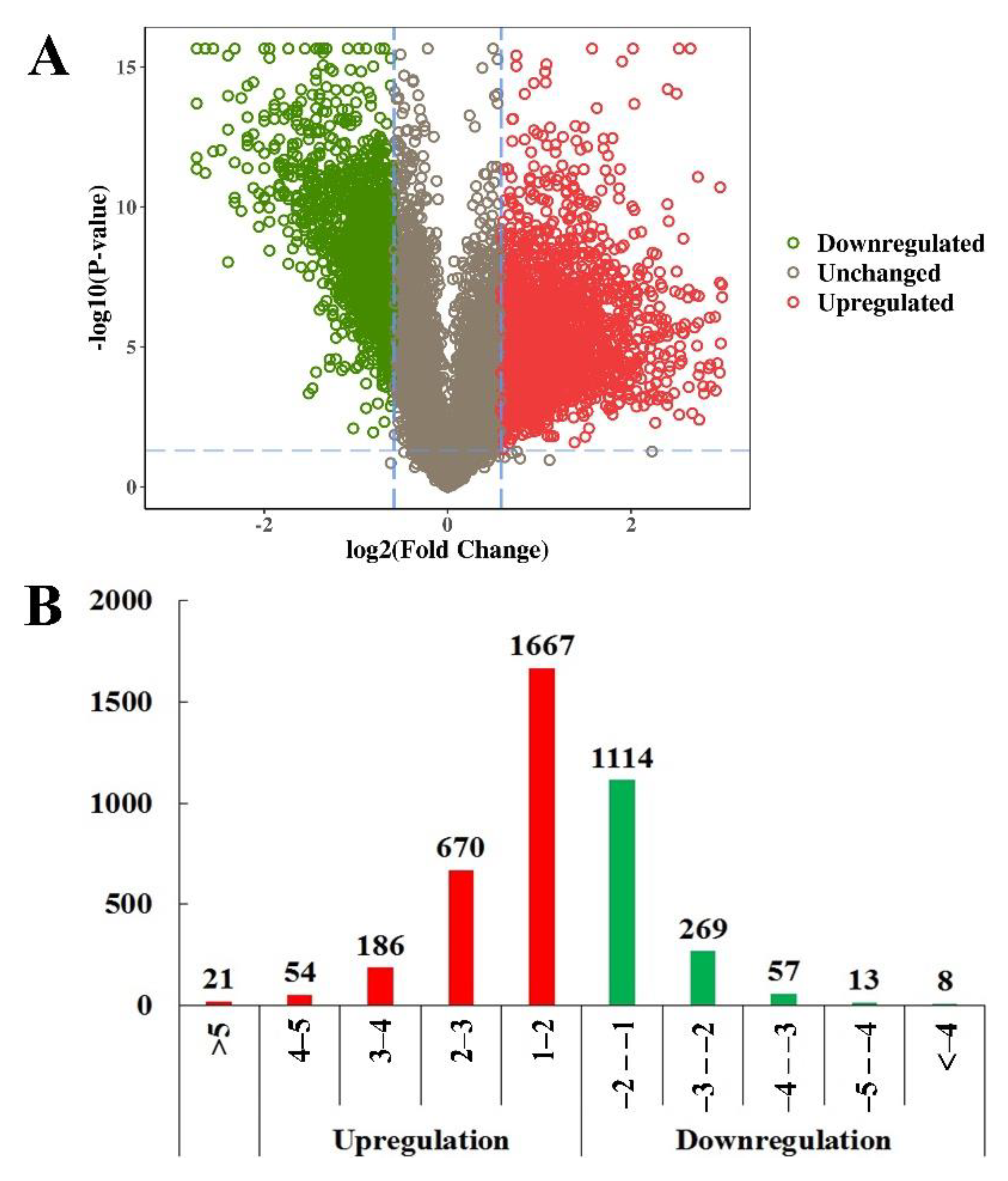
2.2. Identification of Differentially Expressed Phosphopeptides (DEPs)
2.3. Phosphorylation Motif Analysis for Quantitative Phosphopeptides
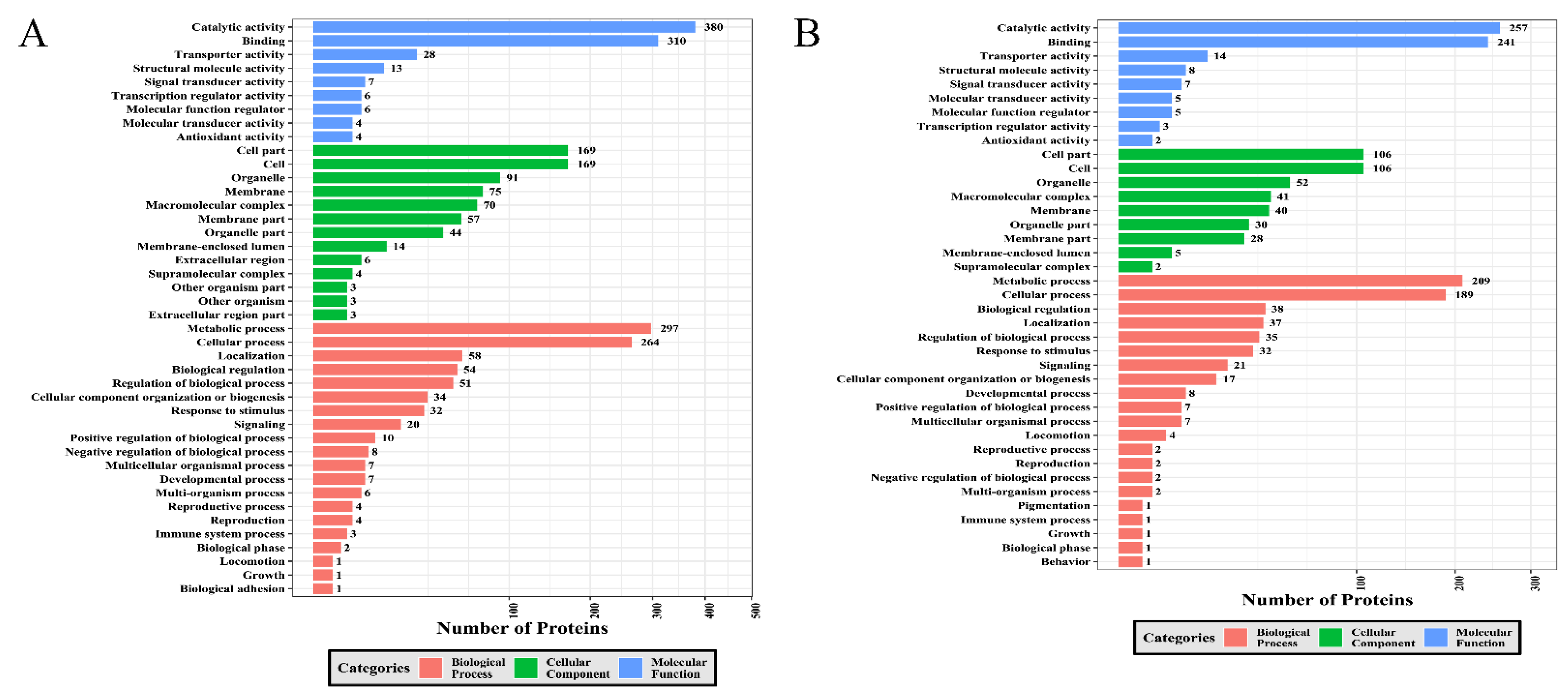
2.4. GO Category Analysis of DEPs
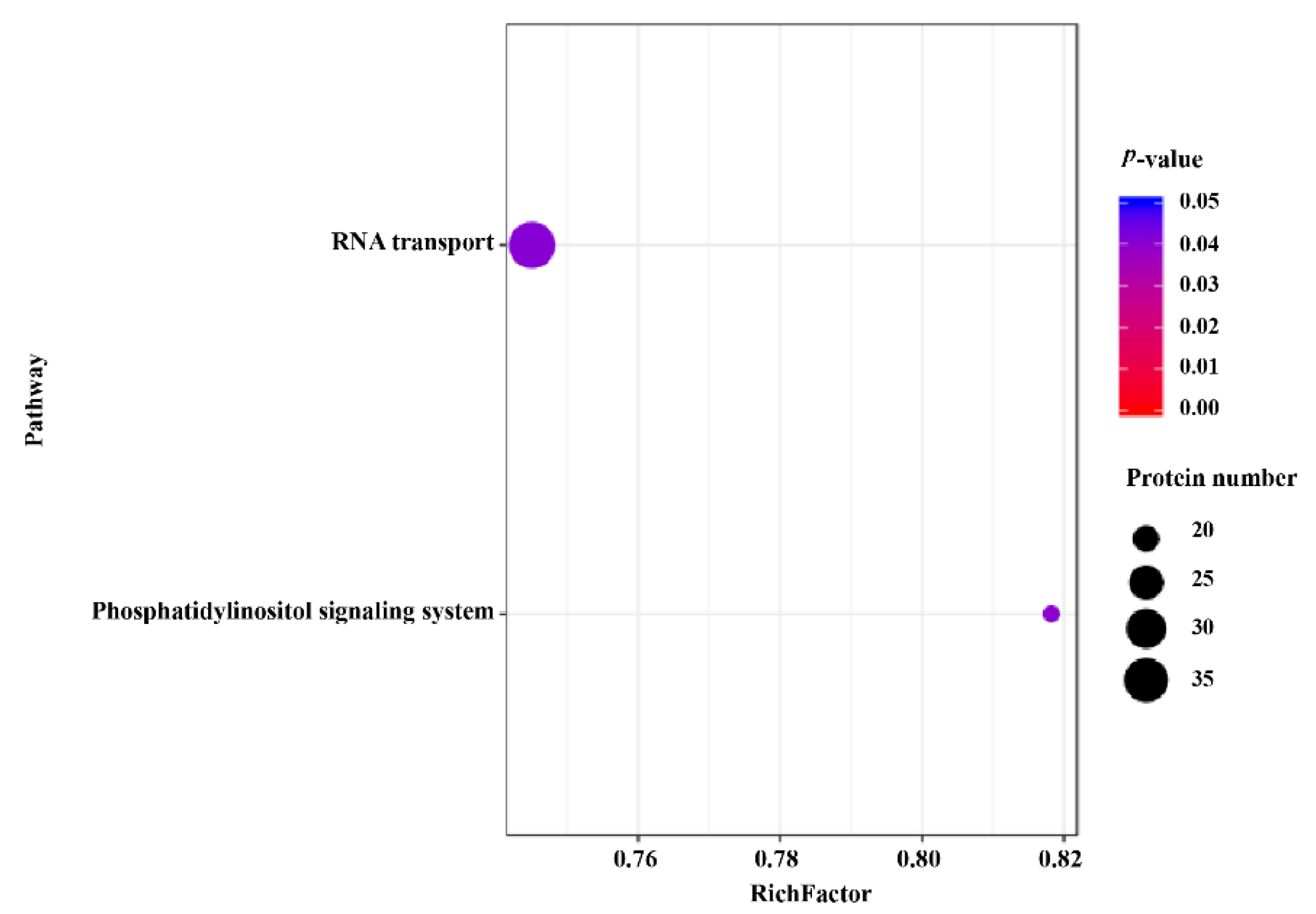
2.5. KEGG Pathway Analysis
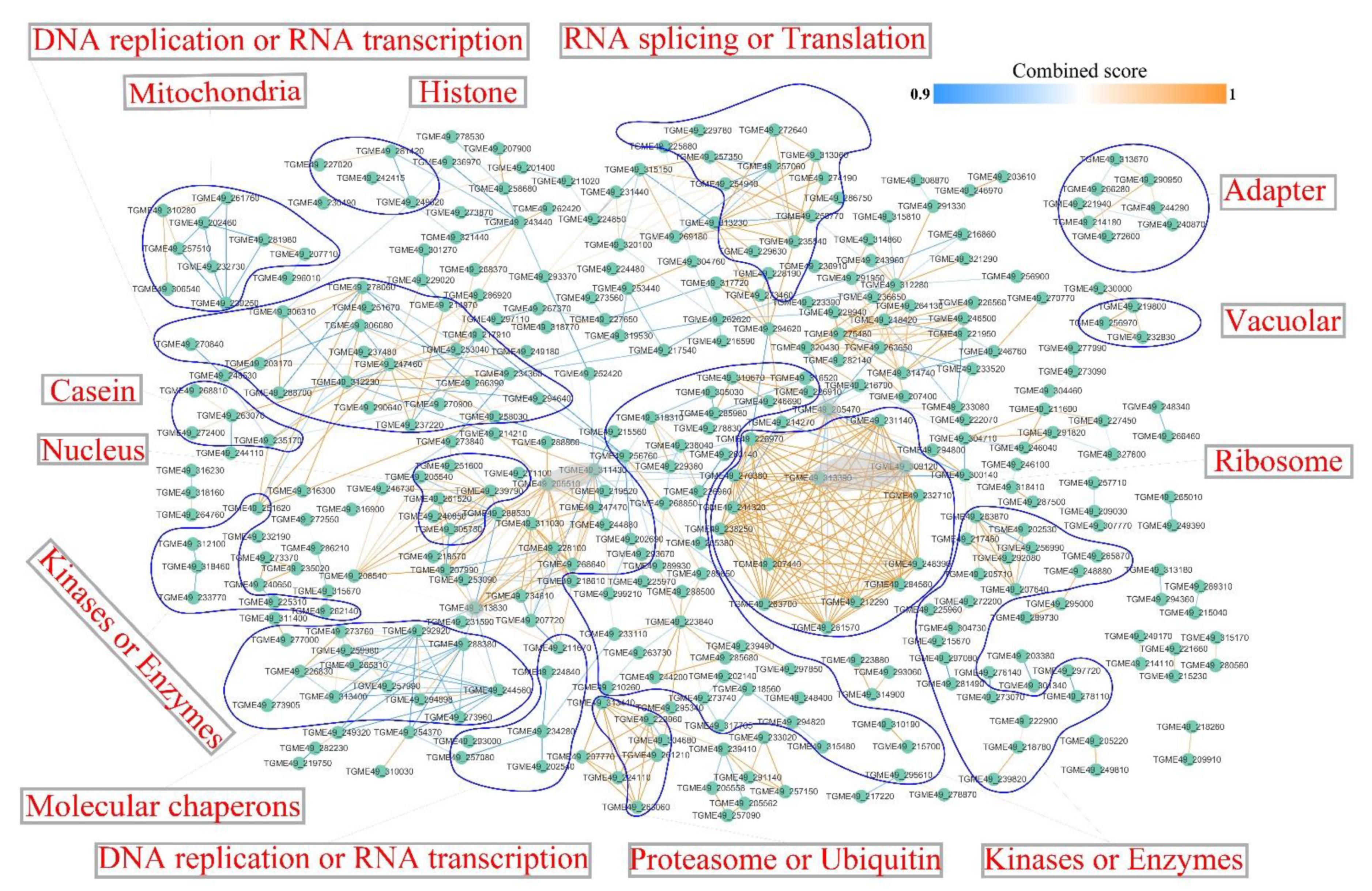
2.6. Interaction Network of Identified Phosphoproteins
2.7. Kinase Connected Network
2.8. Potential Phosphor-Dependent Interactions
3. Discussion
3.1. Motifs
3.2. GO and KEGG Analysis
3.3. PPI Network
3.4. Kinase–Protein Interactions
4. Material and Methods
4.1. Ethics Statements
4.2. Mice, Guinea Pigs, Cats, and Parasite Strains
4.3. Preparation of Tachyzoites and Sporulated Oocysts
4.3.1. Collection and Purification of Tachyzoites
4.3.2. Collection and Purification of Sporulated Oocysts
4.4. Extraction and Digestion of Protein
4.5. IBT Labeling
4.6. Enrichment of Phosphopeptides by Affinity Chromatography
4.7. LC-MS/MS
4.8. Data Analysis
4.9. Bioinformatics
4.10. Relevance Analysis of T. gondii Protein Kinase with Phosphorylated Peptides
4.11. Forecast of Protein Coactions Activated or Inactivated by Phosphorylation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| Ab1 | Abelson kinase |
| ACN | Acetonitrile |
| αSNAP | Alpha soluble N-ethylmaleimide-sensitive fusion protein attachment protein |
| ATM | Ataxia telangiectasia mutated kinase |
| CsCl | Cesium chloride |
| CDPK | Calcium dependent protein kinase |
| Cam-II | Calmodulin-dependent protein kinase II |
| CDC2 | Cell division cycle protein kinase p34 |
| CDK | Cyclin-dependent kinase |
| cis-SNARE | cis-soluble N-ethylmaleimide sensitive factor attachment protein receptor |
| CKI | Casein kinase I |
| CKII | Casein kinase II |
| CV | Coefficient of variation |
| MAPK | Cyclin-dependent kinase of mitogen-activated protein kinase |
| GRAs | Dense granule proteins |
| DEPs | Differentially expressed phosphopeptides |
| DTT | Dithiothreitol |
| EGFR | Epithelial growth factor receptor |
| FDR | False-discovery rate |
| FA | Formic acid |
| GO | Gene ontology |
| IKK | ikappab kinase |
| INSR | Insulin receptor |
| Jak | Janus kinase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MAPK | Mitogen-activated protein kinase |
| OG | Orthologous groups |
| OWP | Oocyst wall protein |
| PBS | Phosphate-buffered saline |
| PKA | Camp-dependent protein kinase |
| PKB | Protein kinase B |
| PKC | Protein kinase C |
| PKG | cGMP-dependent protein kinase |
| PTM | Posttranslational modification |
| ROPs | Rhoptry proteins |
| RP | ribosomal protein |
| STRING | Search tool for the retrieval of interacting genes/protein |
| SDZ | Sulfadiazine |
| SAGE | Serial analysis of gene expression |
| SPF | Specific pathogen free |
| Src | Sarcoma gene kinase |
| Syk | Cytoplasmic tyrosine kinases |
References
- Kochanowsky, J.A.; Koshy, A.A. Toxoplasma gondii. Curr. Biol. 2018, 28, R770–R771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sibley, L.D.; Khan, A.; Ajioka, J.W.; Rosenthal, B.M. Genetic diversity of Toxoplasma gondii in animals and humans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 2749–2761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Yolken, R.H. Strain hypothesis of Toxoplasma gondii infection on the outcome of human diseases. Acta Physiol. 2015, 21, 828–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howe, D.K.; Sibley, L.D. Toxoplasma gondii comprises three clonal lineages: Correlation of parasite genotype with human disease. J. Infect. Dis. 1995, 172, 1561–1566. [Google Scholar] [CrossRef]
- Khan, A.; Dubey, J.P.; Su, C.; Ajioka, J.W.; Rosenthal, B.M.; Sibley, L.D. Genetic analyses of atypical Toxoplasma gondii strains reveal a fourth clonal lineage in North America. Int. J. Parasitol. 2011, 41, 645–655. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Su, R.; Lu, Y.; Wang, M.; Liu, J.; Jian, F.; Yang, Y. Prevalence, risk factors, and genotypes of Toxoplasma gondii in food animals and humans (2000–2017) from China. Front. Microbiol. 2018, 9, 2108. [Google Scholar] [CrossRef]
- Wang, C.; Cheng, W.; Yu, Q.; Xing, T.; Chen, S.; Liu, L.; Yu, L.; Du, J.; Luo, Q.; Shen, J.; et al. Toxoplasma Chinese 1 strain of WH3Δrop16I/III/gra15II genetic background contributes to abnormal pregnant outcomes in murine model. Front. Immunol. 2018, 9, 1222. [Google Scholar] [CrossRef]
- Ferguson, D. Toxoplasma gondii and sex: Essential or optional extra? Trends Parasitol. 2002, 18, 351. [Google Scholar] [CrossRef]
- Dubey, J.P. Comparative infectivity of oocysts and bradyzoites of Toxoplasma gondii for intermediate (mice) and definitive (cats) hosts. Vet. Parasitol. 2006, 140, 69–75. [Google Scholar] [CrossRef]
- Freppel, W.; Ferguson, D.; Shapiro, K.; Dubey, J.P.; Puech, P.H.; Dumètre, A. Structure, composition, and roles of the Toxoplasma gondii oocyst and sporocyst walls. Cell Surf. 2018, 5, 100016. [Google Scholar] [CrossRef]
- Jones, J.L.; Dubey, J.P. Waterborne toxoplasmosis—Recent developments. Exp. Parasitol. 2010, 124, 10–25. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, K.E.; Lagunas-Solar, M.; Miller, M.A.; Barr, B.C.; Gardner, I.A.; Pina, C.; Melli, A.C.; Packham, A.E.; Zeng, N.; Truong, T.; et al. Physical inactivation of Toxoplasma gondii oocysts in water. Appl. Environ. Microbiol. 2007, 73, 5663–5666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wainwright, K.E.; Miller, M.A.; Barr, B.C.; Gardner, I.A.; Melli, A.C.; Essert, T.; Packham, A.E.; Truong, T.; Lagunas-Solar, M.; Conrad, P.A. Chemical inactivation of Toxoplasma gondii oocysts in water. J. Parasitol. 2007, 93, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Dumètre, A.; Dubey, J.P.; Ferguson, D. Effect of household bleach on the structure of the sporocyst wall of Toxoplasma gondii. Parasite 2021, 28, 68. [Google Scholar] [CrossRef] [PubMed]
- Possenti, A.; Fratini, F.; Fantozzi, L.; Pozio, E.; Dubey, J.P.; Ponzi, M.; Pizzi, E.; Spano, F. Global proteomic analysis of the oocyst/sporozoite of Toxoplasma gondii reveals commitment to a host-independent lifestyle. BMC Genom. 2013, 14, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumètre, A.; Dubey, J.P.; Ferguson, D.J.; Bongrand, P.; Azas, N.; Puech, P.H. Mechanics of the Toxoplasma gondii oocyst wall. Proc. Natl. Acad. Sci. USA 2013, 110, 11535–11540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rousseau, A.; Escotte-Binet, S.; La Carbona, S.; Dumètre, A.; Chagneau, S.; Favennec, L.; Kubina, S.; Dubey, J.P.; Majou, D.; Bigot-Clivot, A.; et al. Toxoplasma gondii oocyst infectivity assessed using a sporocyst-based cell culture assay combined with quantitative PCR for environmental applications. Appl. Environ. Microbiol. 2019, 85, e01189-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndao, O.; Puech, P.H.; Bérard, C.; Limozin, L.; Rabhi, S.; Azas, N.; Dubey, J.P.; Dumètre, A. Dynamics of Toxoplasma gondii Oocyst phagocytosis by macrophages. Front. Cell. Infect. Microbiol. 2020, 10, 207. [Google Scholar] [CrossRef]
- Fritz, H.M.; Bowyer, P.W.; Bogyo, M.; Conrad, P.A.; Boothroyd, J.C. Proteomic analysis of fractionated Toxoplasma oocysts reveals clues to their environmental resistance. PLoS ONE 2012, 7, e29955. [Google Scholar] [CrossRef] [Green Version]
- Blader, I.J.; Coleman, B.I.; Chen, C.T.; Gubbels, M.J. Lytic cycle of Toxoplasma gondii: 15 years later. Annu. Rev. Microbiol. 2015, 69, 463–485. [Google Scholar] [CrossRef] [Green Version]
- Radke, J.R.; Behnke, M.S.; Mackey, A.J.; Radke, J.B.; Roos, D.S.; White, M.W. The transcriptome of Toxoplasma gondii. BMC Biol. 2005, 3, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.F.; Han, X.L.; Li, F.X.; Yao, Y.Y.; Fang, J.P.; Liu, X.J.; Li, X.C.; Wu, K.; Liu, M.; Chen, X.G. Comparative studies of Toxoplasma gondii transcriptomes: Insights into stage conversion based on gene expression profiling and alternative splicing. Parasites Vectors 2018, 11, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.X.; Zhou, C.X.; Elsheikha, H.M.; He, S.; Zhou, D.H.; Zhu, X.Q. Proteomic differences between developmental stages of Toxoplasma gondii revealed by iTRAQ-based quantitative proteomics. Front. Microbiol. 2017, 8, 985. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Sundaram, S. An analysis of phosphorylation sites in protein kinases from Leishmania. Bioinformation 2016, 12, 249–253. [Google Scholar] [CrossRef] [Green Version]
- Yakubu, R.R.; Weiss, L.M.; Silmon de Monerri, N.C. Post-translational modifications as key regulators of apicomplexan biology: Insights from proteome-wide studies. Mol. Microbiol. 2018, 107, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Tsigankov, P.; Gherardini, P.F.; Helmer-Citterich, M.; Späth, G.F.; Zilberstein, D. Phosphoproteomic analysis of differentiating Leishmania parasites reveals a unique stage-specific phosphorylation motif. J. Proteome Res. 2013, 12, 3405–3412. [Google Scholar] [CrossRef]
- Treeck, M.; Sanders, J.L.; Elias, J.E.; Boothroyd, J.C. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell Host Microbe 2011, 10, 410–419. [Google Scholar] [CrossRef] [Green Version]
- Alam, M.M.; Solyakov, L.; Bottrill, A.R.; Flueck, C.; Siddiqui, F.A.; Singh, S.; Mistry, S.; Viskaduraki, M.; Lee, K.; Hopp, C.S.; et al. Phosphoproteomics reveals malaria parasite Protein Kinase G as a signalling hub regulating egress and invasion. Nat. Commun. 2015, 6, 7285. [Google Scholar] [CrossRef] [Green Version]
- Taylor, S.; Barragan, A.; Su, C.; Fux, B.; Fentress, S.J.; Tang, K.; Beatty, W.L.; Hajj, H.E.; Jerome, M.; Behnke, M.S.; et al. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science 2006, 314, 1776–1780. [Google Scholar] [CrossRef] [Green Version]
- Saeij, J.P.; Coller, S.; Boyle, J.P.; Jerome, M.E.; White, M.W.; Boothroyd, J.C. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature 2007, 445, 324–327. [Google Scholar] [CrossRef]
- Fentress, S.J.; Behnke, M.S.; Dunay, I.R.; Mashayekhi, M.; Rommereim, L.M.; Fox, B.A.; Bzik, D.J.; Taylor, G.A.; Turk, B.E.; Lichti, C.F.; et al. Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe 2010, 8, 484–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosowski, E.E.; Saeij, J.P. Toxoplasma gondii clonal strains all inhibit STAT1 transcriptional activity but polymorphic effectors differentially modulate IFNγ induced gene expression and STAT1 phosphorylation. PLoS ONE 2012, 7, e51448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, K.D.; Hu, K.; Whitmarsh, R.J.; Hassan, M.A.; Julien, L.; Lu, D.; Chen, L.; Hunter, C.A.; Saeij, J.P. Toxoplasma gondii rhoptry 16 kinase promotes host resistance to oral infection and intestinal inflammation only in the context of the dense granule protein GRA15. Infect. Immun. 2013, 81, 2156–2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellegrini, E.; Palencia, A.; Braun, L.; Kapp, U.; Bougdour, A.; Belrhali, H.; Bowler, M.W.; Hakimi, M.A. Structural basis for the subversion of MAP kinase signaling by an intrinsically disordered parasite secreted agonist. Structure 2017, 25, 16–26. [Google Scholar] [CrossRef] [Green Version]
- Braun, L.; Brenier-Pinchart, M.P.; Yogavel, M.; Curt-Varesano, A.; Curt-Bertini, R.L.; Hussain, T.; Kieffer-Jaquinod, S.; Coute, Y.; Pelloux, H.; Tardieux, I.; et al. A Toxoplasma dense granule protein, GRA24, modulates the early immune response to infection by promoting a direct and sustained host p38 MAPK activation. J. Exp. Med. 2013, 210, 2071–2086. [Google Scholar] [CrossRef]
- Wang, Z.X.; Zhou, C.X.; Calderón-Mantilla, G.; Petsalaki, E.; He, J.J.; Song, H.Y.; Elsheikha, H.M.; Zhu, X.Q. iTRAQ-based global phosphoproteomics reveals novel molecular differences between Toxoplasma gondii strains of different genotypes. Front. Cell. Infect. Microbiol. 2019, 9, 307. [Google Scholar] [CrossRef] [Green Version]
- Becker, C.H.; Bern, M. Recent developments in quantitative proteomics. Mutat. Res. 2011, 722, 171–182. [Google Scholar] [CrossRef] [Green Version]
- Borchert, N.; Krug, K.; Gnad, F.; Sinha, A.; Sommer, R.J.; Macek, B. Phosphoproteome of Pristionchus pacificus provides insights into architecture of signaling networks in nematode models. Mol. Cell. Proteom. 2012, 11, 1631–1639. [Google Scholar] [CrossRef] [Green Version]
- Amorim, J.C.; Batista, M.; da Cunha, E.S.; Lucena, A.; Lima, C.; Sousa, K.; Krieger, M.A.; Marchini, F.K. Quantitative proteome and phosphoproteome analyses highlight the adherent population during Trypanosoma cruzi metacyclogenesis. Sci. Rep. 2017, 7, 9899. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, D.; Gygi, S.P. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 2005, 23, 1391–1398. [Google Scholar] [CrossRef]
- Pi, E.; Qu, L.; Hu, J.; Huang, Y.; Qiu, L.; Lu, H.; Jiang, B.; Liu, C.; Peng, T.; Zhao, Y.; et al. Mechanisms of soybean roots’ tolerances to salinity revealed by proteomic and phosphoproteomic comparisons between two cultivars. Mol. Cell. Proteomics 2016, 15, 266–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez, M.A.; Contreras, I.; Hallé, M.; Tremblay, M.L.; McMaster, R.W.; Olivier, M. Leishmania GP63 alters host signaling through cleavage-activated protein tyrosine phosphatases. Sci. Signal. 2009, 2, ra58. [Google Scholar] [CrossRef]
- Cheng, W.; Liu, F.; Li, M.; Hu, X.; Chen, H.; Pappoe, F.; Luo, Q.; Wen, H.; Xing, T.; Xu, Y.; et al. Variation detection based on next-generation sequencing of type Chinese 1 strains of Toxoplasma gondii with different virulence from China. BMC Genom. 2015, 16, 888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shwab, E.K.; Saraf, P.; Zhu, X.Q.; Zhou, D.H.; McFerrin, B.M.; Ajzenberg, D.; Schares, G.; Hammond-Aryee, K.; van Helden, P.; Higgins, S.A.; et al. Human impact on the diversity and virulence of the ubiquitous zoonotic parasite Toxoplasma gondii. Proc. Natl. Acad. Sci. USA 2018, 115, E6956–E6963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Q.L.; Sun, L.X.; Elsheikha, H.M.; Cao, X.Z.; Nie, L.B.; Li, T.T.; Li, T.S.; Zhu, X.Q.; Wang, J.L. RHΔgra17Δnpt1 strain of Toxoplasma gondii elicits protective immunity against acute, chronic and congenital Toxoplasmosis in mice. Microorganisms 2020, 8, 352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, J.C.; Broncel, M.; Teague, H.; Russell, M.; McGovern, O.L.; Renshaw, M.; Frith, D.; Snijders, A.P.; Collinson, L.; Carruthers, V.B.; et al. Phosphorylation of Toxoplasma gondii secreted proteins during acute and chronic stages of infection. mSphere 2020, 5, e00792-20. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Xu, M.Z.; Pan, S.; Wang, H.; Peng, H.J.; Liu, Z.Z. iTRAQ-based phosphoproteomic analysis of Toxoplasma gondii tachyzoites provides insight into the role of phosphorylation for its invasion and egress. Front. Cell. Infect. Microbiol. 2020, 10, 586466. [Google Scholar] [CrossRef]
- Bansal, P.; Antil, N.; Kumar, M.; Yamaryo-Botté, Y.; Rawat, R.S.; Pinto, S.; Datta, K.K.; Katris, N.J.; Botté, C.Y.; Prasad, T.; et al. Protein kinase TgCDPK7 regulates vesicular trafficking and phospholipid synthesis in Toxoplasma gondii. PLoS Pathog. 2021, 17, e1009325. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.J.; Ferguson, D.J.; Whitehead, L.; Bradin, C.H.; Wu, H.J.; Tonkin, C.J. Phosphorylation of αSNAP is required for secretory organelle biogenesis in Toxoplasma gondii. Traffic 2016, 17, 102–116. [Google Scholar] [CrossRef] [Green Version]
- Selseleh, M.; Modarressi, M.; Shojaee, S.; Mohebali, M.; Eshraghian, M.; Selseleh, M.; Keshavarz, H. Brain tissue cysts in infected mice with RH-strain of Toxoplasma gondii and evaluation of BAG1 and SAG1 genes expression. Iran. J. Parasitol. 2013, 8, 40–46. [Google Scholar]
- Frenkel, J.K.; Dubey, J.P.; Hoff, R.L. Loss of stages after continuous passage of Toxoplasma gondii and Besnoitia jellisoni. J. Protozool. 1976, 23, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Boothroyd, J.C.; Grigg, M.E. Population biology of Toxoplasma gondii and its relevance to human infection: Do different strains cause different disease? Curr. Opin. Microbiol. 2002, 5, 438–442. [Google Scholar] [CrossRef]
- Asghari, A.; Majidiani, H.; Nemati, T.; Fatollahzadeh, M.; Shams, M.; Naserifar, R.; Kordi, B. Toxoplasma gondii tyrosine-rich oocyst wall protein: A closer look through an in silico prism. Biomed Res. Int. 2021, 2021, 1315618. [Google Scholar] [CrossRef] [PubMed]
- Hansson, M.; Nygren, P.A.; Ståhl, S. Design and production of recombinant subunit vaccines. Biotechnol. Appl. Biochem. 2000, 32, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Melo-Braga, M.N.; Verano-Braga, T.; León, I.R.; Antonacci, D.; Nogueira, F.C.; Thelen, J.J.; Larsen, M.R.; Palmisano, G. Modulation of protein phosphorylation, N-glycosylation and Lys-acetylation in grape (Vitis vinifera) mesocarp and exocarp owing to Lobesia botrana infection. Mol. Cell. Proteom. 2012, 11, 945–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Tang, H.; Abuzeid, A.; Tan, L.; Wang, A.; Wan, X.; Zhang, H.; Liu, Y.; Li, G. Protein phosphorylation networks in spargana of Spirometra erinaceieuropaei revealed by phosphoproteomic analysis. Parasites Vectors 2020, 13, 248. [Google Scholar] [CrossRef]
- Eaton, M.S.; Weiss, L.M.; Kim, K. Cyclic nucleotide kinases and tachyzoite-bradyzoite transition in Toxoplasma gondii. Int. J. Parasitol. 2006, 36, 107–114. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; Marq, J.B.; Bisio, H.; Jacot, D.; Mueller, C.; Yu, L.; Choudhary, J.; Brochet, M.; Soldati-Favre, D. Crosstalk between PKA and PKG controls pH-dependent host cell egress of Toxoplasma gondii. EMBO J. 2017, 36, 3250–3267. [Google Scholar] [CrossRef]
- Hirst, N.L.; Nebel, J.C.; Lawton, S.P.; Walker, A.J. Deep phosphoproteome analysis of Schistosoma mansoni leads development of a kinomic array that highlights sex-biased differences in adult worm protein phosphorylation. PLoS Negl. Trop. Dis. 2020, 14, e0008115. [Google Scholar] [CrossRef]
- Hu, X.; O’Shaughnessy, W.J.; Beraki, T.G.; Reese, M.L. Loss of the conserved alveolate kinase MAPK2 decouples Toxoplasma cell growth from cell division. mBio 2020, 11, e02517-20. [Google Scholar] [CrossRef]
- Honaker, Y.; Piwnica-Worms, H. Casein kinase 1 functions as both penultimate and ultimate kinase in regulating Cdc25A destruction. Oncogene 2010, 29, 3324–3334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dissous, C.; Khayath, N.; Vicogne, J.; Capron, M. Growth factor receptors in helminth parasites: Signalling and host-parasite relationships. FEBS Lett. 2006, 580, 2968–2975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerávolo, I.P.; Chaves, A.C.; Bonjardim, C.A.; Sibley, D.; Romanha, A.J.; Gazzinelli, R.T. Replication of Toxoplasma gondii, but not Trypanosoma cruzi, is regulated in human fibroblasts activated with gamma interferon: Requirement of a functional JAK/STAT pathway. Infect. Immun. 1999, 67, 2233–2240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meenderink, L.M.; Ryzhova, L.M.; Donato, D.M.; Gochberg, D.F.; Kaverina, I.; Hanks, S.K. P130Cas Src-binding and substrate domains have distinct roles in sustaining focal adhesion disassembly and promoting cell migration. PLoS ONE 2010, 5, e13412. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zeng, L.; Wang, J.; Chau, J.F.; Lai, K.P.; Jia, D.; Poonepalli, A.; Hande, M.P.; Liu, H.; He, G.; et al. A positive role for c-Abl in Atm and Atr activation in DNA damage response. Cell Death Differ. 2011, 18, 5–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wengelnik, K.; Daher, W.; Lebrun, M. Phosphoinositides and their functions in apicomplexan parasites. Int. J. Parasitol. 2018, 48, 493–504. [Google Scholar] [CrossRef]
- Daher, W.; Morlon-Guyot, J.; Sheiner, L.; Lentini, G.; Berry, L.; Tawk, L.; Dubremetz, J.F.; Wengelnik, K.; Striepen, B.; Lebrun, M. Lipid kinases are essential for apicoplast homeostasis in Toxoplasma gondii. Cell. Microbiol. 2015, 17, 559–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cernikova, L.; Faso, C.; Hehl, A.B. Roles of phosphoinositides and their binding proteins in parasitic protozoa. Trends Parasitol. 2019, 35, 996–1008. [Google Scholar] [CrossRef]
- Sidik, S.M.; Huet, D.; Ganesan, S.M.; Huynh, M.H.; Wang, T.; Nasamu, A.S.; Thiru, P.; Saeij, J.; Carruthers, V.B.; Niles, J.C.; et al. A genome-wide crispr screen in Toxoplasma identifies essential apicomplexan genes. Cell 2016, 166, 1423–1435.e12. [Google Scholar] [CrossRef] [Green Version]
- Ren, B.; Kong, P.; Hedar, F.; Brouwers, J.F.; Gupta, N. Phosphatidylinositol synthesis, its selective salvage, and inter-regulation of anionic phospholipids in Toxoplasma gondii. Commun. Biol. 2020, 3, 750. [Google Scholar] [CrossRef]
- Tomavo, S.; Schwarz, R.T.; Dubremetz, J.F. Evidence for glycosyl- phosphatidylinositol anchoring of Toxoplasma gondii major surface antigens. Mol. Cell. Biol. 1989, 9, 4576–4580. [Google Scholar] [CrossRef] [PubMed]
- Séron, K.; Dzierszinski, F.; Tomavo, S. Molecular cloning, functional complementation in Saccharomyces cerevisiae and enzymatic properties of phosphatidylinositol synthase from the protozoan parasite Toxoplasma gondii. Eur. J. Biochem. 2000, 267, 6571–6579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nardelli, S.C.; Che, F.Y.; Silmon de Monerri, N.C.; Xiao, H.; Nieves, E.; Madrid-Aliste, C.; Angel, S.O.; Sullivan, W.J., Jr.; Angeletti, R.H.; Kim, K.; et al. The histone code of Toxoplasma gondii comprises conserved and unique posttranslational modifications. mBio 2013, 4, e00922-13. [Google Scholar] [CrossRef] [Green Version]
- Wei, F.; Wang, W.; Liu, Q. Protein kinases of Toxoplasma gondii: Functions and drug targets. Parasitol. Res. 2013, 112, 2121–2129. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, S.; Wang, W.; Gu, Y.; Liu, H.; Wei, F.; Liu, Q. Targeted disruption of CK1α in Toxoplasma gondii increases acute virulence in mice. Eur. J. Protistol. 2016, 56, 90–101. [Google Scholar] [CrossRef] [PubMed]
- White, M.W.; Radke, J.R.; Radke, J.B. Toxoplasma development—Turn the switch on or off? Cell. Microbiol. 2014, 16, 466–472. [Google Scholar] [CrossRef]
- Shaw, M.K.; He, C.Y.; Roos, D.S.; Tilney, L.G. Proteasome inhibitors block intracellular growth and replication of Toxoplasma gondii. Parasitology 2000, 121 Pt 1, 35–47. [Google Scholar] [CrossRef]
- Paugam, A.; Creuzet, C.; Dupouy-Camet, J.; Roisin, P. In vitro effects of gliotoxin, a natural proteasome inhibitor, on the infectivity and proteolytic activity of Toxoplasma gondii. Parasitol. Res. 2002, 88, 785–787. [Google Scholar] [CrossRef]
- Silmon de Monerri, N.C.; Yakubu, R.R.; Chen, A.L.; Bradley, P.J.; Nieves, E.; Weiss, L.M.; Kim, K. The Ubiquitin proteome of Toxoplasma gondii reveals roles for protein ubiquitination in cell-cycle transitions. Cell Host Microbe 2015, 18, 621–633. [Google Scholar] [CrossRef] [Green Version]
- Melatti, C.; Pieperhoff, M.; Lemgruber, L.; Pohl, E.; Sheiner, L.; Meissner, M. A unique dynamin-related protein is essential for mitochondrial fission in Toxoplasma gondii. PLoS Pathog. 2019, 15, e1007512. [Google Scholar] [CrossRef]
- De Miguel, N.; Braun, N.; Bepperling, A.; Kriehuber, T.; Kastenmüller, A.; Buchner, J.; Angel, S.O.; Haslbeck, M. Structural and functional diversity in the family of small heat shock proteins from the parasite Toxoplasma gondii. Biochim. Biophys. Acta 2009, 1793, 1738–1748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shonhai, A.; Maier, A.G.; Przyborski, J.M.; Blatch, G.L. Intracellular protozoan parasites of humans: The role of molecular chaperones in development and pathogenesis. Protein Pept. Lett. 2011, 18, 143–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toursel, C.; Dzierszinski, F.; Bernigaud, A.; Mortuaire, M.; Tomavo, S. Molecular cloning, organellar targeting and developmental expression of mitochondrial chaperone HSP60 in Toxoplasma gondii. Mol. Biochem. Parasitol. 2000, 111, 319–332. [Google Scholar] [CrossRef]
- Mitra, P.; Deshmukh, A.S.; Choudhury, C. Molecular chaperone function of stress inducible Hsp70 is critical for intracellular multiplication of Toxoplasma gondii. Biochim. Biophys. Acta Mol. Cell. Res. 2021, 1868, 118898. [Google Scholar] [CrossRef] [PubMed]
- Angel, S.O.; Figueras, M.J.; Alomar, M.L.; Echeverria, P.C.; Deng, B. Toxoplasma gondii Hsp90: Potential roles in essential cellular processes of the parasite. Parasitology 2014, 141, 1138–1147. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Cadahía, B.; Drobic, B.; Davie, J.R. H3 phosphorylation: Dual role in mitosis and interphase. Biochem. Cell. Biol. 2009, 87, 695–709. [Google Scholar] [CrossRef]
- Lo, W.S.; Duggan, L.; Emre, N.C.; Belotserkovskya, R.; Lane, W.S.; Shiekhattar, R.; Berger, S.L. Snf1—A histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 2001, 293, 1142–1146. [Google Scholar] [CrossRef]
- Saksouk, N.; Bhatti, M.M.; Kieffer, S.; Smith, A.T.; Musset, K.; Garin, J.; Sullivan, W.J., Jr.; Cesbron-Delauw, M.F.; Hakimi, M.A. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol. Cell. Biol. 2005, 25, 10301–10314. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, W.J., Jr.; Hakimi, M.A. Histone mediated gene activation in Toxoplasma gondii. Mol. Biochem. Parasitol. 2006, 148, 109–116. [Google Scholar] [CrossRef]
- Drewry, L.L.; Sibley, L.D. Toxoplasma actin is required for efficient host cell invasion. mBio 2015, 6, e00557. [Google Scholar] [CrossRef] [Green Version]
- Staggs, S.E.; See, M.J.; Dubey, J.P.; Villegas, E.N. Obtaining highly purified Toxoplasma gondii oocysts by a discontinuous cesium chloride gradient. J. Vis. Exp. 2009, 33, 1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Betts, M.J.; Lu, Q.; Jiang, Y.; Drusko, A.; Wichmann, O.; Utz, M.; Valtierra-Gutiérrez, I.A.; Schlesner, M.; Jaeger, N.; Jones, D.T.; et al. Mechismo: Predicting the mechanistic impact of mutations and modifications on molecular interactions. Nucleic Acids Res. 2015, 43, e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]


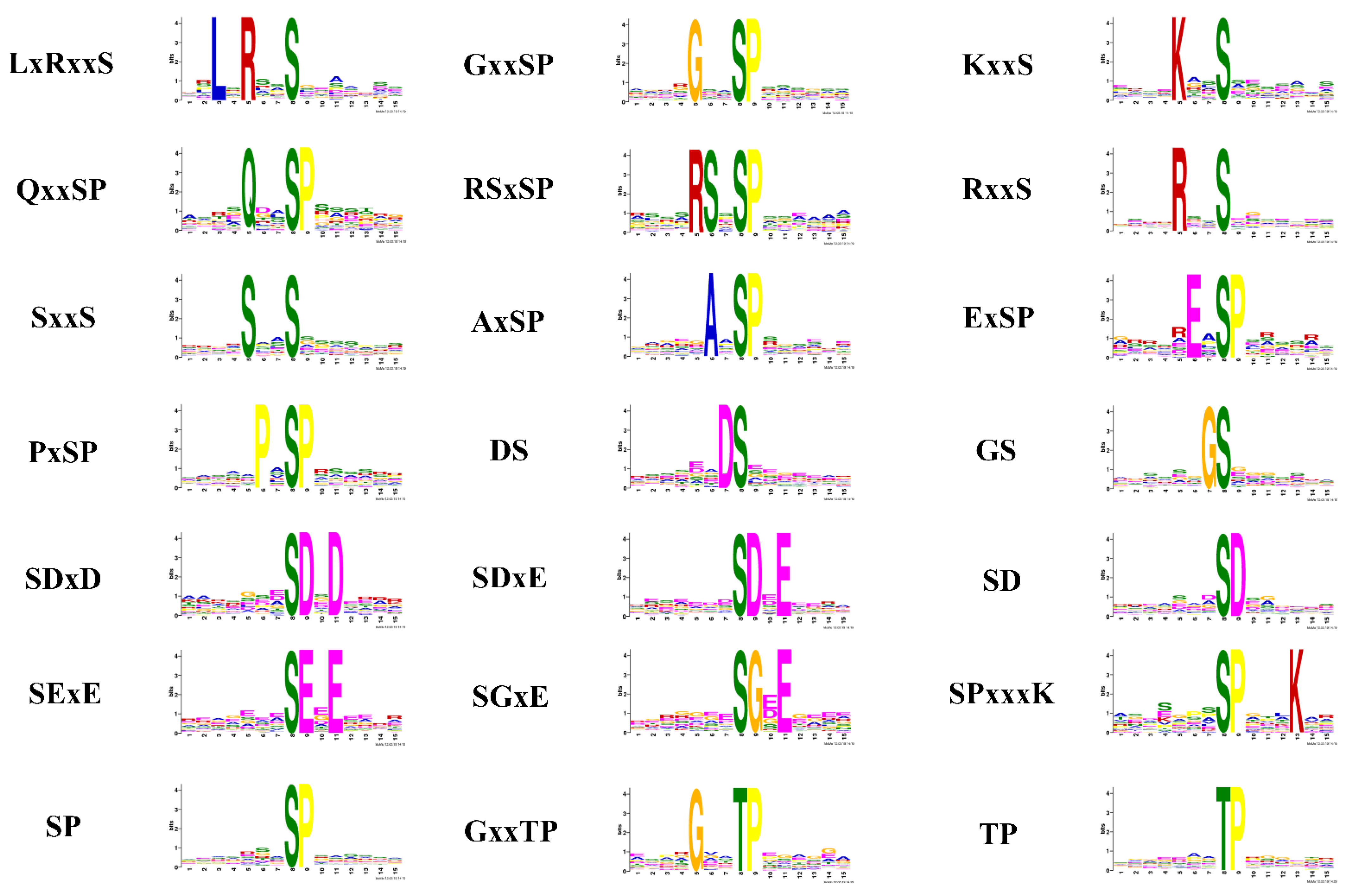
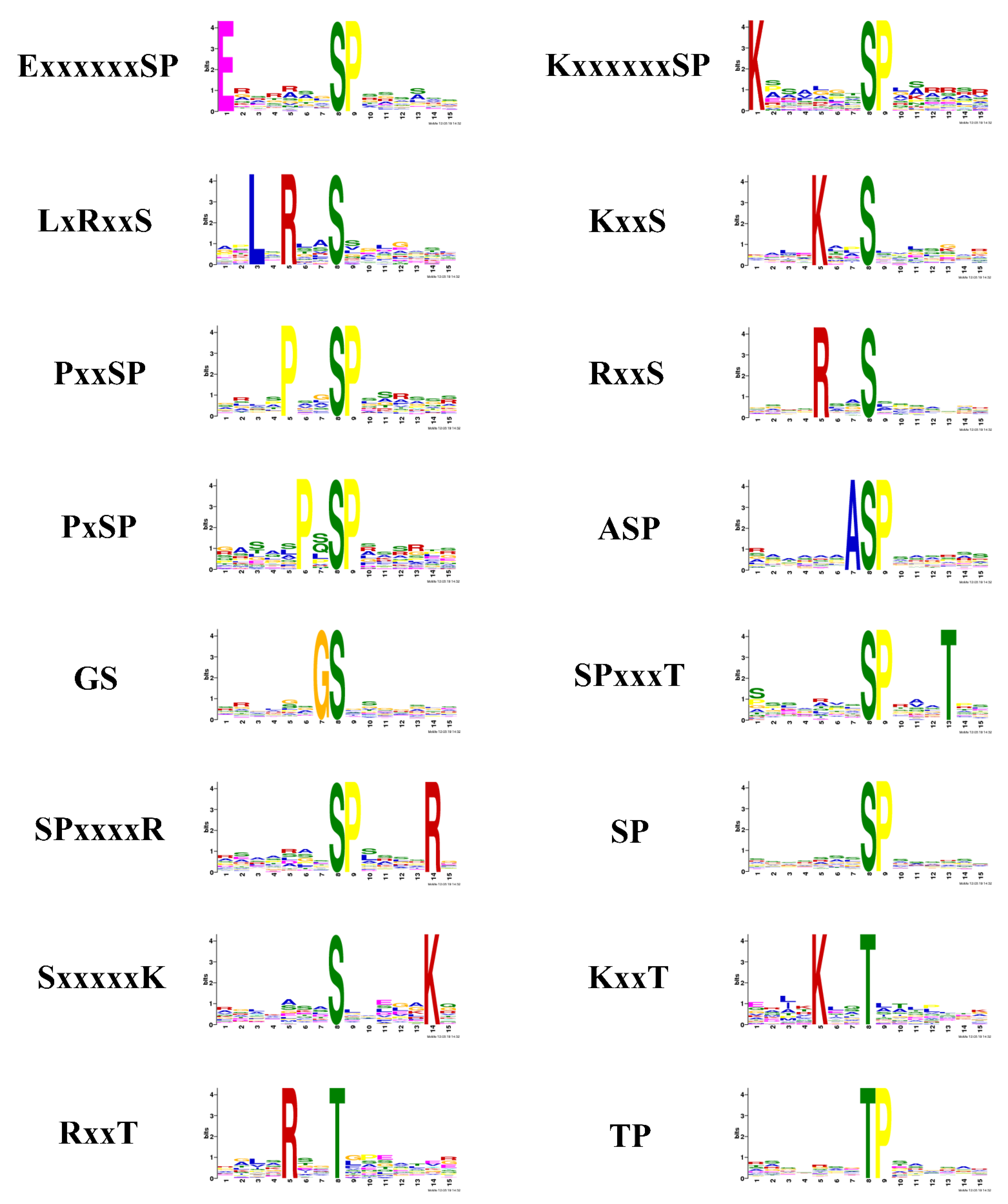

| Motif | Kinase Classes |
|---|---|
| P..SP | ATM, CDC2, CDK, CKI, CKII, IKK, MAPK, PKA, PKC, and PKG |
| E……SP | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, INSR, MAPK, PKA, PKB, PKC, PKG, and Syk |
| SP…T | Ab1, ATM, CaM-II, CDC2, CDK, CKI, IKK, INSR, MAPK, PKA, PKB, PKC, and PKG |
| ASP | Ab1, ATM, CDC2, CDK, CKI, IKK, INSR, MAPK, PKA, PKB, PKC, PKG |
| K……SP | ATM, CDC2, CDK, CKI, CKII, IKK, MAPK, PKA, and PKG |
| SP….R | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, Jak, MAPK, PKA, PKB, PKC, and PKG |
| P.SP | ATM, CaM-II, CDC2, CDK, CKI, IKK, MAPK, PKA, PKC, and PKG |
| SP | Ab1, ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, INSR, MAPK, Src, Syk, PKA, PKB, PKC, and PKG |
| L.R..S | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, MAPK, PKA, PKB, PKC, and PKG |
| R..S | ATM, CaM-II, CDC2, CKI, CKII, IKK, MAPK, PKA, PKB, PKC, and PKG |
| GS | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, INSR, Jak, MAPK, PKA, PKC, PKG, Src, and Syk |
| K..S | ATM, CDC2, CDK, CKI, IKK, MAPK, PKA, PKC, PKG, and Syk |
| S…..K | ATM, CaM-II, CDK, CKI, CKII, IKK, INSR, Jak, MAPK, PKB, PKC, PKG, and Syk |
| TP | ATM, CaM-II, CDC2, CDK, CKI, CKII, EGFR, IKK, INSR, Jak, MAPK, PKA, PKB, PKC, PKG, and Syk |
| R..T | Ab1, CaM-II, CDC2, EGFR, IKK, MAPK, PKA, PKC, PKG, and Src |
| K..T | CDK, IKK, MAPK, PKA, PKC, and PKG |
| Motif | Kinase Classes |
|---|---|
| RS.SP | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, MAPK, PKA, PKB, PKC, and PKG |
| SP…K | ATM, PKA, CDC2, CDK, IKK, MAPK, PKC, and PKG |
| A.SP | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, MAPK, PKA, PKB, PKC, PKG, and Syk |
| P.SP | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, MAPK, PKA, PKB, PKC, and PKG |
| Q..SP | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, MAPK, PKA, PKB, PKC, and PKG |
| G..SP | Ab1, ATM, CDC2, CDK, CKI, CKII, IKK, INSR, Jak, MAPK, PKB, PKC, PKG, and Syk |
| E.SP | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, INSR, MAPK, PKA, PKB, PKC, and PKG |
| SP | Ab1, ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, INSR, MAPK, PKA, PKB, PKC, PKG, Src, and Syk |
| SD.E | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, MAPK, PKA, PKB, PKC, and PKG |
| L.R..S | ATM, CaM-II, CDC2,CKI, CKII, IKK, INSR, MAPK, PKA, PKB, PKC, and PKG |
| R..S | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, INSR, MAPK, PKA, PKB, PKC, PKG, Src, and Syk |
| SS.D | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, MAPK, PKA, PKC, and PKG |
| SG.E | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, INSR, MAPK, PKA, PKB, PKC, PKG, and Syk |
| SE.E | ATM, CaM-II, CDC2, CDK, CKI, CKII, EGFR, IKK, INSR, MAPK, PKA, PKB, PKC, PKG, and Syk |
| SD | Ab1, ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, INSR, MAPK, PKA, PKB, PKC, PKG, and Syk |
| GS | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, INSR, MAPK, PKA, PKB, PKC, PKG, Src, and Syk |
| K..S | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, INSR, MAPK, PKA, PKB, PKC, and PKG |
| DS | ATM, CaM-II, CDC2, CDK, CKI, CKII, EGFR, IKK, INSR, Jak, MAPK, PKA, PKC, PKC, and PKG |
| S..S | ATM, CaM-II, CDC2, CDK, CKI, CKII, IKK, MAPK, PKA, PKB, PKC, and PKG |
| G..TP | ATM, CDC2, CDK, CKII, IKK, MAPK, PKC, and PKG, |
| TP | ATM, CDC2, CDK, CKI, CKII, IKK, MAPK, PKA, PKB, and PKG, |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.-X.; Che, L.; Hu, R.-S.; Sun, X.-L. Comparative Phosphoproteomic Analysis of Sporulated Oocysts and Tachyzoites of Toxoplasma gondii Reveals Stage-Specific Patterns. Molecules 2022, 27, 1022. https://doi.org/10.3390/molecules27031022
Wang Z-X, Che L, Hu R-S, Sun X-L. Comparative Phosphoproteomic Analysis of Sporulated Oocysts and Tachyzoites of Toxoplasma gondii Reveals Stage-Specific Patterns. Molecules. 2022; 27(3):1022. https://doi.org/10.3390/molecules27031022
Chicago/Turabian StyleWang, Ze-Xiang, Liang Che, Rui-Si Hu, and Xiao-Lin Sun. 2022. "Comparative Phosphoproteomic Analysis of Sporulated Oocysts and Tachyzoites of Toxoplasma gondii Reveals Stage-Specific Patterns" Molecules 27, no. 3: 1022. https://doi.org/10.3390/molecules27031022







