From the Physicochemical Characteristic of Novel Hesperetin Hydrazone to Its In Vitro Antimicrobial Aspects
Abstract
:1. Introduction
2. Materials and Methods
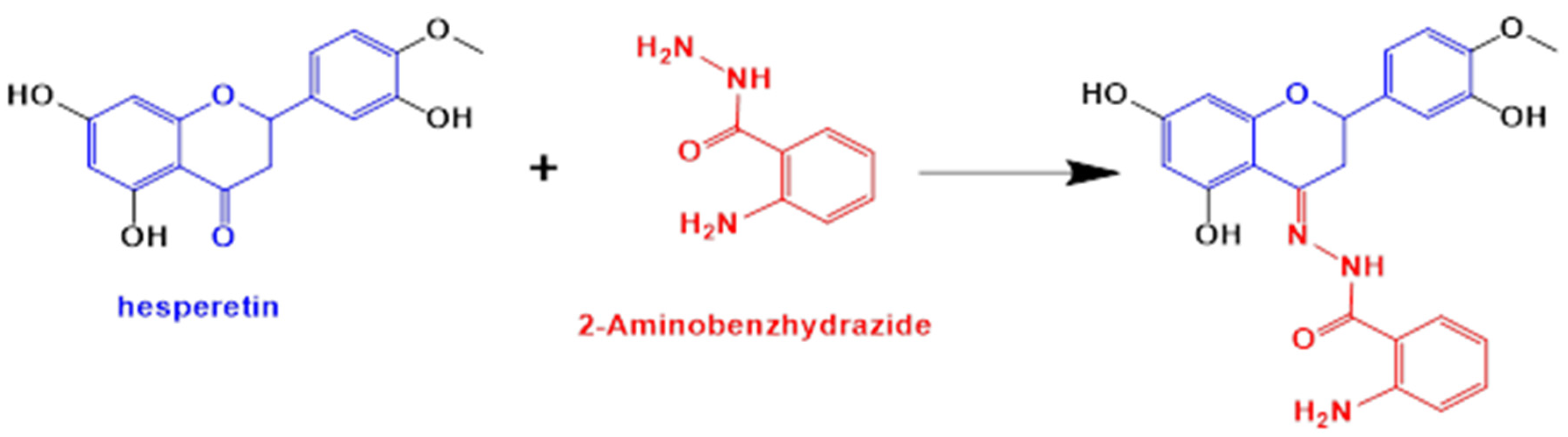
2.1. Materials in the Synthesis of Novel Hesperetin Derivatives
2.2. Apparatus
2.2.1. Characterization of HABH
2.2.2. Characterization of Copper(II) Complex with HABH in Solid State
2.2.3. Stoichiometry Determination of the Copper(II) Complex with HABH in Solution
Potentiometry
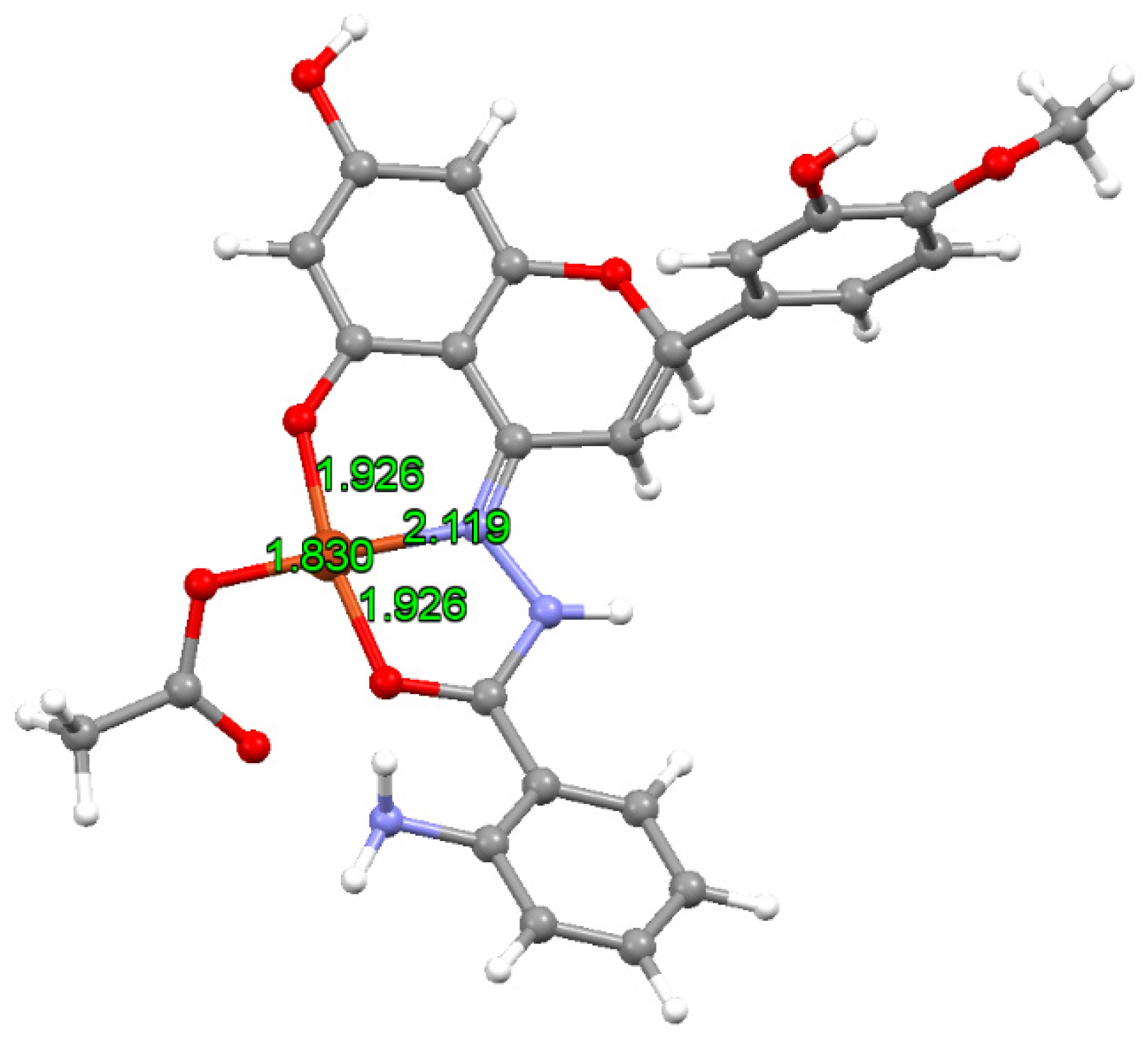
Spectroscopy and Computational Details
2.3. DNA Studies
2.3.1. Electronic Absorption Titration
2.3.2. Competitive Binding Fluorescence Measurement
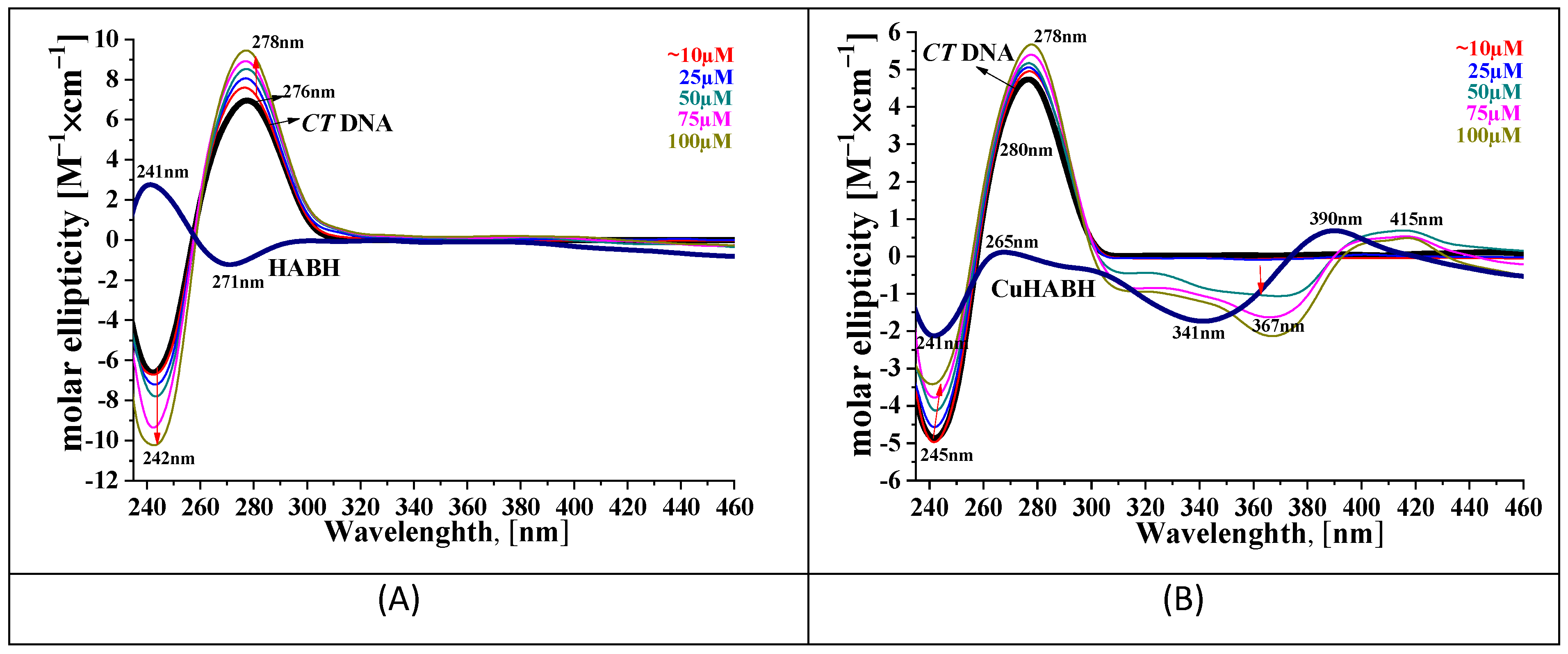
2.3.3. Circular Dichroism Spectroscopy Analysis
2.4. Antibacterial Activity
2.5. Antibiofilm Activity
3. Results and Discussion
3.1. Characterization of the Complex CuHABH
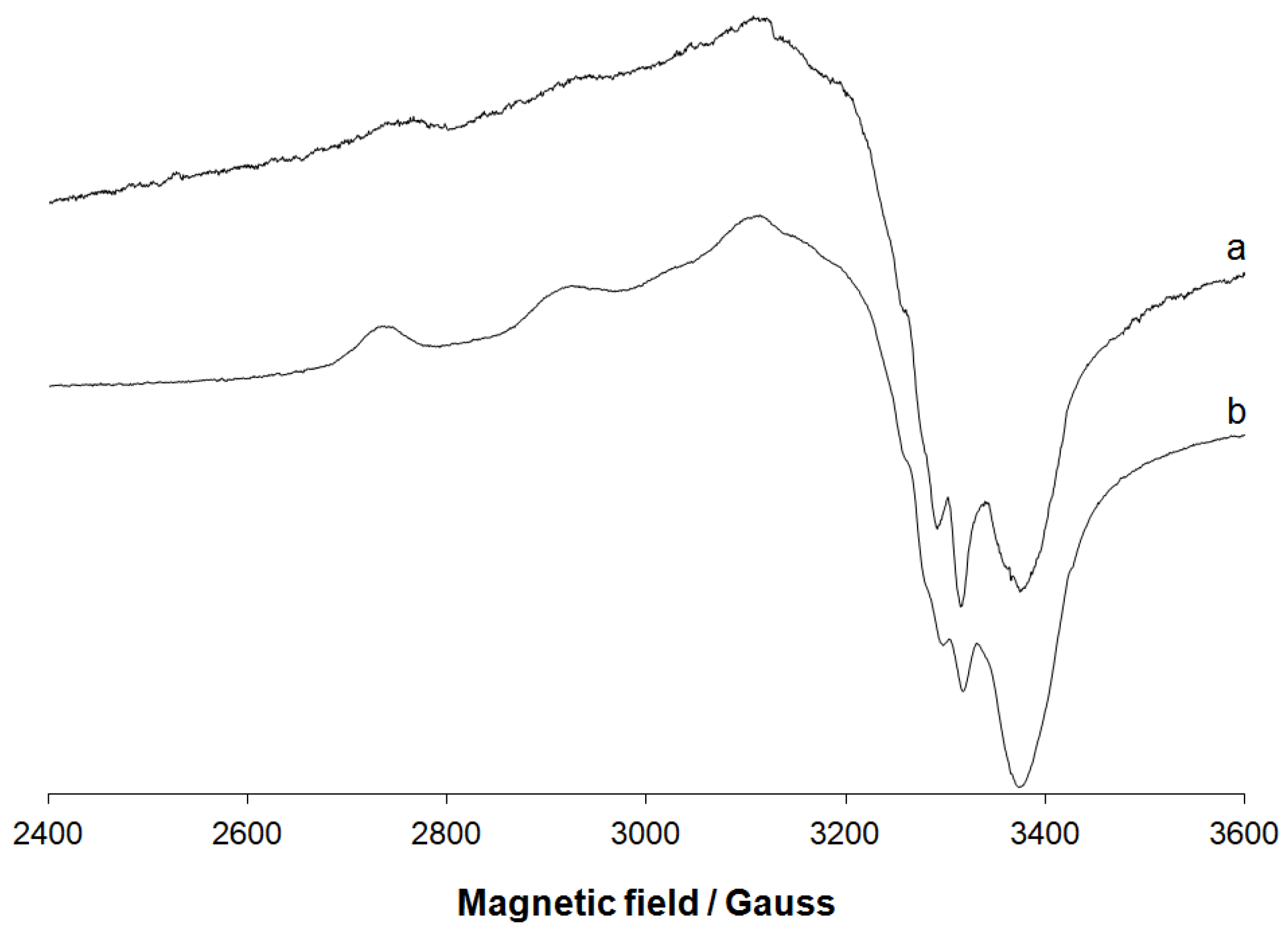
3.1.1. IR Spectral Studies
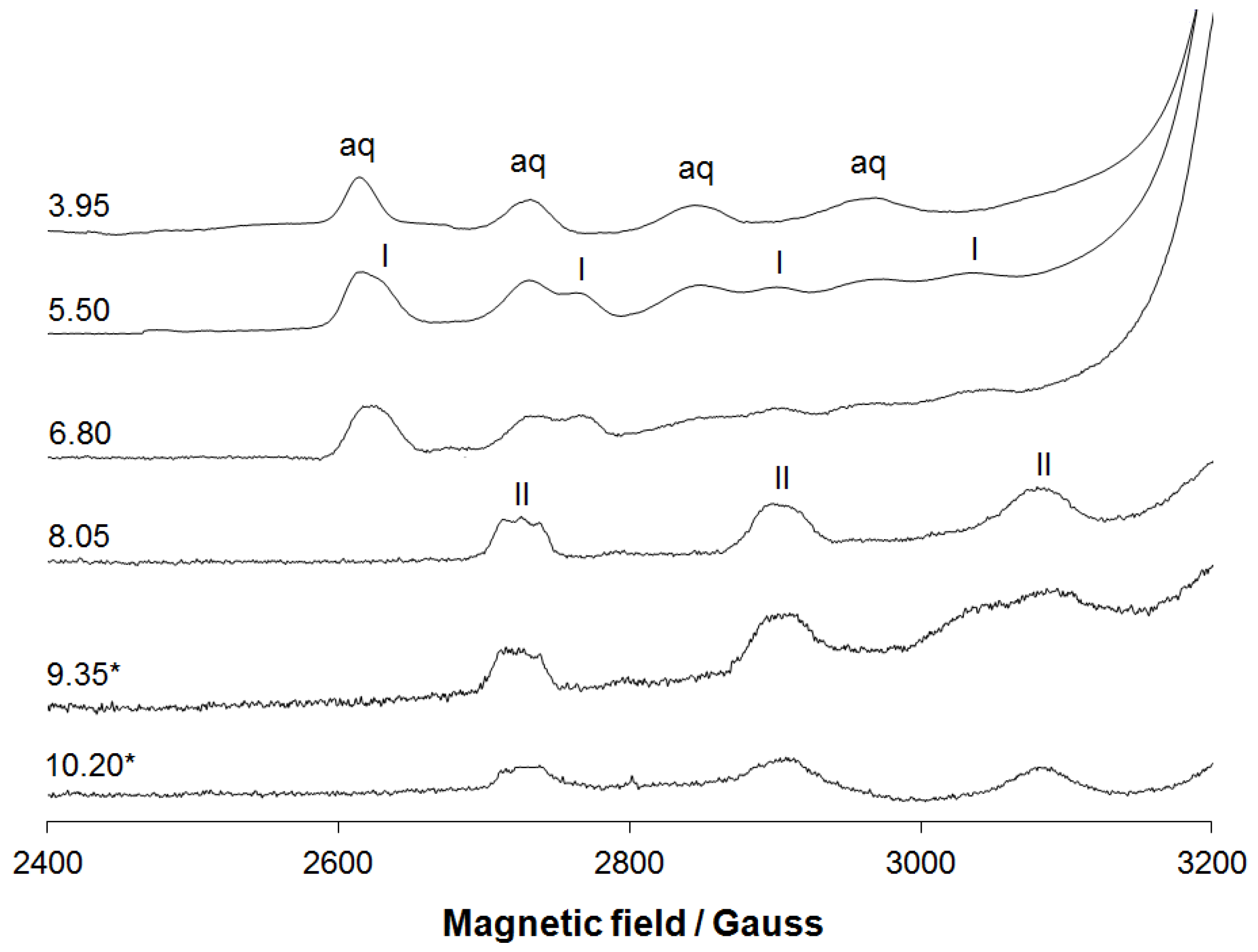
3.1.2. EPR Spectral Studies in Solid State and Solution
3.2. Interaction of the Compounds with CT DNA
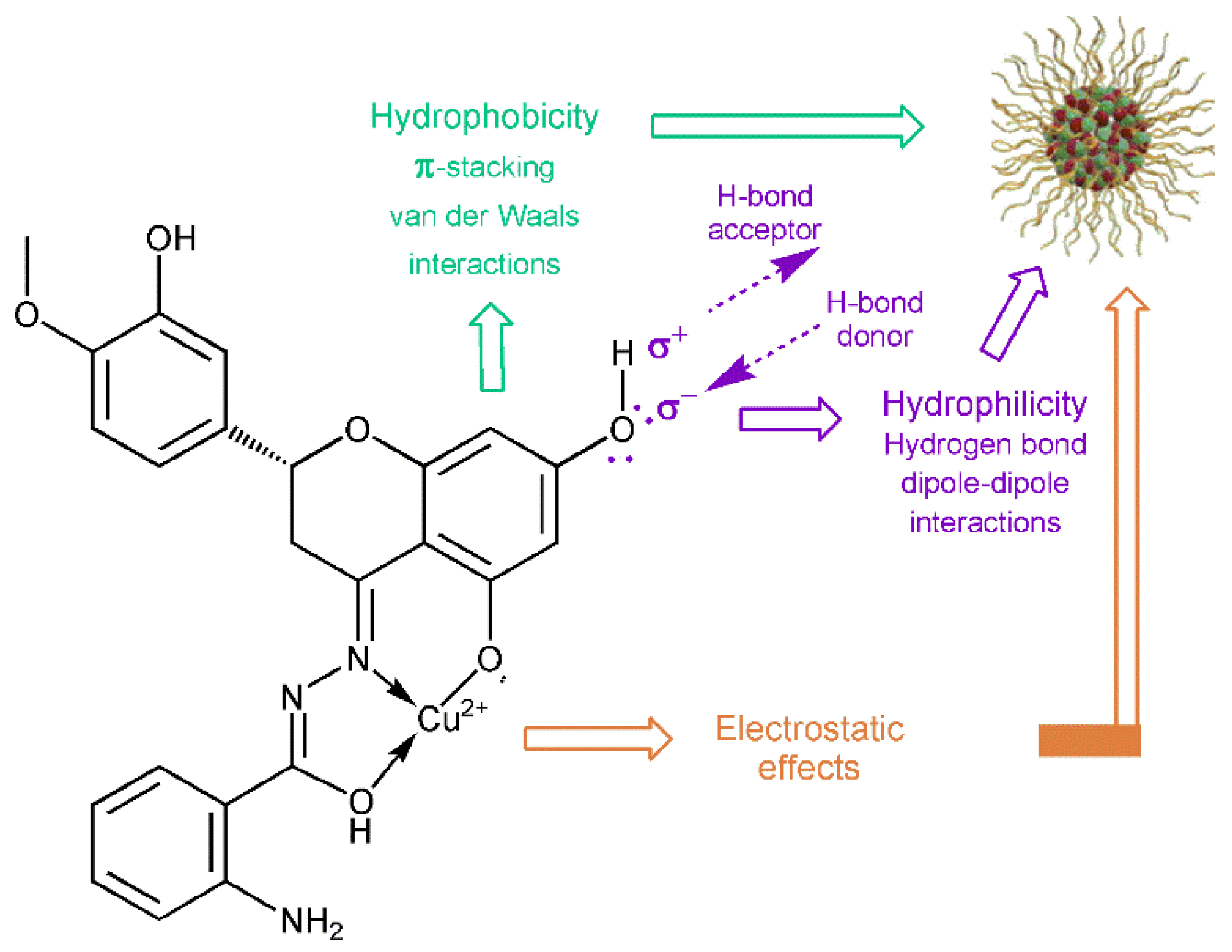
3.3. Antibacterial Activity of the HABH and CuHABH Complexes
3.4. Antibiofilm Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Galiè, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the Food Industry: Health Aspects and Control Methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, M.; Benoliel, C.; Drider, D.; Dhulster, P.; Chihib, N.-E. Biofilm formation and persistence on abiotic surfaces in the context of food and medical environments. Arch. Microbiol. 2014, 196, 453–472. [Google Scholar] [CrossRef] [PubMed]
- Colagiorgi, A.; Bruini, I.; Di Ciccio, P.A.; Zanardi, E.; Ghidini, S.; Ianieri, A. Listeria monocytogenes Biofilms in the Wonderland of Food Industry. Pathogens 2017, 6, 41. [Google Scholar] [CrossRef] [Green Version]
- Cepas, V.; López, Y.; Muñoz, E.; Rolo, D.; Ardanuy, C.; Martí, S.; Xercavins, M.; Horcajada, J.P.; Bosch, J.; Soto, S.M. Relationship Between Biofilm Formation and Antimicrobial Resistance in Gram-Negative Bacteria. Microb. Drug Resist. 2019, 25, 72–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Pozo, J.L. Biofilm-related disease. Expert Rev. Anti-Infect. Ther. 2017, 16, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.-F. Biofilm-specific antibiotic resistance. Futur. Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Kong, Y.; Zhao, H.; Yan, X. Dependence of the Bacillus subtilis biofilm expansion rate on phenotypes and the morphology under different growing conditions. Dev. Growth Differ. 2019, 61, 431–443. [Google Scholar] [CrossRef]
- Ullah, A.; Munir, S.; Badshah, S.L.; Khan, N.; Ghani, L.; Poulson, B.G.; Emwas, A.-H.; Jaremko, M. Important Flavonoids and Their Role as a Therapeutic Agent. Molecules 2020, 25, 5243. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef]
- Rakesh, K.; Kumara, H.; Ullas, B.; Shivakumara, J.; Gowda, D.C. Amino acids conjugated quinazolinone-Schiff’s bases as potential antimicrobial agents: Synthesis, SAR and molecular docking studies. Bioorganic Chem. 2019, 90, 103093. [Google Scholar] [CrossRef]
- Oliveira, A.A.; de Oliveira, A.P.A.; Franco, L.L.; Ferencs, M.O.; Ferreira, J.; Bachi, S.M.P.S.; Speziali, N.L.; Farias, L.M.; Magalhães, P.P.; Beraldo, H. 5-Nitroimidazole-derived Schiff bases and their copper(II) complexes exhibit potent antimicrobial activity against pathogenic anaerobic bacteria. BioMetals 2018, 31, 571–584. [Google Scholar] [CrossRef]
- More, P.G.; Karale, N.N.; Lawand, A.S.; Narang, N.; Patil, R.H. Synthesis and anti-biofilm activity of thiazole Schiff bases. Med. Chem. Res. 2013, 23, 790–799. [Google Scholar] [CrossRef]
- Pervaiz, M.; Ahmad, I.; Yousaf, M.; Kirn, S.; Munawar, A.; Saeed, Z.; Adnan, A.; Gulzar, T.; Kamal, T.; Ahmad, A.; et al. Synthesis, spectral and antimicrobial studies of amino acid derivative Schiff base metal (Co, Mn, Cu, and Cd) complexes. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 206, 642–649. [Google Scholar] [CrossRef]
- Mumtaz, A.; Mahmud, T. Structural investigation of some novel synthesized Schiff base Transition metal complexes derived from drug together with Antimicrobial study. Pak. J. Pharm. Sci. 2019, 32, 963–967. [Google Scholar] [PubMed]
- Al-Amiery, A.A.; Kadhum, A.A.H.; Mohamad, A.B. Antifungal and Antioxidant Activities of Pyrrolidone Thiosemicarbazone Complexes. Bioinorg. Chem. Appl. 2012, 2012, 1–6. [Google Scholar] [CrossRef]
- Łodyga-Chruścińska, E.; Symonowicz, M.; Sykuła, A.; Bujacz, A.; Garribba, E.; Rowińska-Żyrek, M.; Ołdziej, S.; Klewicka, E.; Janicka, M.; Krolewska, K.; et al. Chelating ability and biological activity of hesperetin Schiff base. J. Inorg. Biochem. 2015, 143, 34–47. [Google Scholar] [CrossRef]
- Brodowska, K.; Correia, I.; Garribba, E.; Marques, F.; Klewicka, E.; Lodyga-Chruscinska, E.; Pessoa, J.C.; Dzeikala, A.; Chrusciński, L. Coordination ability and biological activity of a naringenin thiosemicarbazone. J. Inorg. Biochem. 2016, 165, 36–48. [Google Scholar] [CrossRef]
- Ramirez, T.; Shrestha, A.; Kishen, A. Inflammatory potential of monospecies biofilm matrix components. Int. Endod. J. 2019, 52, 1020–1027. [Google Scholar] [CrossRef]
- Fong, J.N.C.; Yildiz, F.H. Biofilm Matrix Proteins. Microbiol. Spectr. 2015, 3, 1–27. [Google Scholar] [CrossRef] [Green Version]
- Gannesen, A.V.; Zdorovenko, E.L.; Botchkova, E.A.; Hardouin, J.; Massier, S.; Kopitsyn, D.S.; Gorbachevskii, M.V.; Kadykova, A.A.; Shashkov, A.S.; Zhurina, M.V.; et al. Composition of the Biofilm Matrix of Cutibacterium acnes Acneic Strain RT5. Front. Microbiol. 2019, 10, 1284. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Z.-Y. DNA binding affinity and antioxidative activity of copper(II) and zinc(II) complexes with a novel hesperetin Schiff base ligand. Inorg. Chim. Acta 2009, 362, 4823–4831. [Google Scholar] [CrossRef]
- Sykuła, A. Kowalska-Baron, K. Gałęcki, P. Błazińska, E. Łodyga-Chruścińska, Structural and Spectral Investigation of a Series of Flavanone Derivatives. Molecules 2021, 26, 1298. [Google Scholar] [CrossRef]
- Gans, P.; Sabatini, A.; Vacca, A. SUPERQUAD: An improved general program for computation of formation constants from potentiometric data. J. Chem. Soc. Dalton Trans. 1985, 1195–1200. [Google Scholar] [CrossRef]
- WinEPR SimFonia, Version 1.25. Bruker Analytische Messtechnik GmbH: Karlshrue. 1996. Available online: https://www.bruker.com/en/products-and-solutions/mr/epr-instruments/epr-software/winepr.html (accessed on 26 December 2021).
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Fox Gaussian 09; revision D.01. Gaussian, Inc.: Wallingford, CT, USA, 2012. Available online: https://gaussian.com (accessed on 26 December 2021).
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef] [Green Version]
- Bühl, M.; Kabrede, H. Geometries of Transition-Metal Complexes from Density-Functional Theory. J. Chem. Theory Comput. 2006, 2, 1282–1290. [Google Scholar] [CrossRef]
- Buehl, M.; Reimann, C.; Pantazis, D.A.; Bredow, T.; Neese, F. Geometries of Third-Row Transition-Metal Complexes from Density-Functional Theory. J. Chem. Theory Comput. 2008, 4, 1449–1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Micera, G.; Garribba, E. The effect of the functional, basis set, and solvent in the simulation of the geometry and spectroscopic properties of V. Int. J. Quantum Chem. 2012, 112, 2486–2498. [Google Scholar] [CrossRef]
- Neese, F. ORCA – An Ab Initio, DFT and Semiempirical Program Package, Version 4.0, Max-Planck-Institute for Chemical Energy Conversion, Mülheim a. d. Ruhr. 2017. Available online: https://orcaforum.kofo.mpg.de/app.php/portal (accessed on 26 December 2021).
- Neese, F. Software update: The ORCA program system, version 4.0. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2018, 8, e1327. [Google Scholar] [CrossRef]
- Neese, F.; Wennmohs, F.; Becker, U.; Riplinger, C. The ORCA quantum chemistry program package. J. Chem. Phys. 2020, 152, 224108. [Google Scholar] [CrossRef]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996, 77, 3865–3868. [Google Scholar] [CrossRef] [Green Version]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized Gradient Approximation Made Simple [Phys. Rev. Lett. 77, 3865 (1996)]. Phys. Rev. Lett. 1997, 78, 1396. [Google Scholar] [CrossRef] [Green Version]
- Sciortino, G.; Lubinu, G.; Maréchal, J.-D.; Garribba, E. DFT Protocol for EPR Prediction of Paramagnetic Cu(II) Complexes and Application to Protein Binding Sites. Magnetochemistry 2018, 4, 55. [Google Scholar] [CrossRef] [Green Version]
- Wolfe, G.H.; Shimer, T. Mechan, Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry 1987, 26, 6392–6396. [Google Scholar] [CrossRef]
- Boger, D.L.; Tse, W.C. Thiazole orange as the fluorescent intercalator in a high resolution fid assay for determining DNA binding affinity and sequence selectivity of small molecules. Bioorg. Med. Chem. 2001, 9, 2511–2518. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006; p. 11. [Google Scholar]
- Eftink, M.R. Fluorescence Quenching: Theory and Applications. In Topics in Fluorescence Spectroscopy; Lakowicz, J.R., Ed.; Springer: Boston, MA, USA, 2002; Volume 2, pp. 53–126. [Google Scholar] [CrossRef]
- Algar, W.R.; Massey, M.; Krull, U.J. Fluorescence Resonance Energy Transfer and Complex Formation between Thiazole Orange and Various Dye-DNA Conjugates: Implications in Signaling Nucleic Acid Hybridization. J. Fluoresc. 2006, 16, 555–567. [Google Scholar] [CrossRef]
- EUCAST. EUCAST Disk Diffusion Test Methodology. Available online: https://www.eucast.org/ast_of_bacteria/disk_diffusion_methodology/ (accessed on 26 November 2021).
- Piccirillo, C.; Demiray, S.; Ferreira, A.S.; Pintado, M.; Castro, P. Chemical composition and antibacterial properties of stem and leaf extracts from Ginja cherry plant. Ind. Crop. Prod. 2012, 43, 562–569. [Google Scholar] [CrossRef]
- Kregiel, D.; Rygala, A.; Kolesinska, B.; Nowacka, M.; Herc, A.S.; Kowalewska, A. Antimicrobial and Antibiofilm N-acetyl-L-cysteine Grafted Siloxane Polymers with Potential for Use in Water Systems. Int. J. Mol. Sci. 2019, 20, 2011. [Google Scholar] [CrossRef] [Green Version]
- Myszka, K.; Czaczyk, K. Effect of starvation stress on morphological changes and production of adhesive exopolysaccharide (EPS) by Proteus vulgaris. Acta Sci. Pol. Technol. Aliment. 2011, 10, 305–312. [Google Scholar]
- Nagesh, G.Y.; Mahadev, U.D.; Mruthyunjayaswamy, B.H.M. Mononuclear Metal (II) Schiff Base complexes derived from thiazole and o-vanillin moieties: Synthesis, characterization, Thermal Behaviour and Biological Evaluation. Int. J. Pharm. Sci. Rev. Res. 2015, 31, 190–197. Available online: https://globalresearchonline.net/journalcontents/v31-1/38.pdf (accessed on 22 November 2021).
- Chandra, S.; Gupta, L.K. Electronic, EPR, magnetic and mass spectral studies of mono and homo-binuclear Co(II) and Cu(II) complexes with a novel macrocyclic ligand. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2005, 62, 1102–1106. [Google Scholar] [CrossRef] [PubMed]
- Peisach, J.; Blumberg, W. Structural implications derived from the analysis of electron paramagnetic resonance spectra of natural and artificial copper proteins. Arch. Biochem. Biophys. 1974, 165, 691–708. [Google Scholar] [CrossRef]
- Garribba, E.; Micera, G. The Determination of the Geometry of Cu(II) Complexes: An EPR Spectroscopy Experiment. J. Chem. Educ. 2006, 83, 1229–1232. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, D. Studies of EPR and ENDOR spectra of14N and15N label bis(2-hydroxyacetophenyl ketoxime)-63Cu(II) and-65Cu(II) complexes in disordered system. Appl. Magn. Reson. 1996, 10, 103–116. [Google Scholar] [CrossRef]
- Cunningham, K.L.; McNett, K.M.; Pierce, R.A.; Davis, K.A.; Harris, H.H.; Falck, A.D.M.; McMillin, D.R. EPR Spectra, Luminescence Data, and Radiationless Decay Processes of Copper(II) Porphyrins. Inorg. Chem. 1997, 36, 608–613. [Google Scholar] [CrossRef]
- Klement, R.; Stock, F.; Elias, H.; Paulus, H.; Pelikán, P.; Valko, M.; Mazúr, M. Copper(II) complexes with derivatives of salen and tetrahydrosalen: A spectroscopic, electrochemical and structural study. Polyhedron 1999, 18, 3617–3628. [Google Scholar] [CrossRef]
- Thakurta, S.; Chakraborty, J.; Rosair, G.; Tercero, J.; El Fallah, M.S.; Garribba, E.; Mitra, S. Synthesis of Two New Linear Trinuclear CuIIComplexes: Mechanism of Magnetic Coupling through Hybrid B3LYP Functional and CShM Studies. Inorg. Chem. 2008, 47, 6227–6235. [Google Scholar] [CrossRef]
- Neese, F. Metal and ligand hyperfine couplings in transition metal complexes: The effect of spin–orbit coupling as studied by coupled perturbed Kohn–Sham theory. J. Chem. Phys. 2003, 118, 3939–3948. [Google Scholar] [CrossRef]
- Sanna, D.; Ágoston, C.G.; Micera, G.; Sóvágó, I. The effect of the ring size of fused chelates on the thermodynamic and spectroscopic properties of peptide complexes of copper(II). Polyhedron 2001, 20, 3079–3090. [Google Scholar] [CrossRef]
- Zeglis, B.M.; Pierre, V.C.; Barton, J.K. Metallo-intercalators and metallo-insertors. Chem. Commun. 2007, 44, 4565–4579. [Google Scholar] [CrossRef] [Green Version]
- Pyle, A.M.; Rehmann, J.P.; Meshoyrer, R.; Kumar, C.V.; Turro, N.J.; Barton, J.K. Mixed-ligand complexes of ruthenium(II): Factors governing binding to DNA. J. Am. Chem. Soc. 1989, 111, 3051–3058. [Google Scholar] [CrossRef]
- Gupta, R.K.; Pandey, R.; Sharma, G.; Prasad, R.; Koch, B.; Srikrishna, S.; Li, P.-Z.; Xu, Q.; Pandey, D.S. DNA Binding and Anti-Cancer Activity of Redox-Active Heteroleptic Piano-Stool Ru(II), Rh(III), and Ir(III) Complexes Containing 4-(2-Methoxypyridyl)phenyldipyrromethene. Inorg. Chem. 2013, 52, 3687–3698. [Google Scholar] [CrossRef]
- Mancin, F.; Scrimin, P.; Tecilla, P.; Tonellato, U. Artificial metallonucleases. Chem. Commun. 2005, 36, 2540–2548. [Google Scholar] [CrossRef]
- Lang, K.; Kubát, P.; Lhoták, P.; Mosinger, J.; Wagnerová, D.M. Photophysical Properties and Photoinduced Electron Transfer within Host-Guest Complexes of 5,10,15,20-Tetrakis(4-N-methylpyridyl)porphyrin with Water-soluble Calixarenes and Cyclodextrins. Photochem. Photobiol. 2007, 74, 558–565. [Google Scholar] [CrossRef]
- Chikira, M.; Tomizawa, Y.; Fukita, D.; Sugizaki, T.; Sugawara, N.; Yamazaki, T.; Sasano, A.; Shindo, H.; Palaniandavar, M.; Antholine, W.E. DNA-fiber EPR study of the orientation of Cu(II) complexes of 1,10-phenanthroline and its derivatives bound to DNA: Mono(phenanthroline)-copper(II) and its ternary complexes with amino acids. J. Inorg. Biochem. 2002, 89, 163–173. [Google Scholar] [CrossRef]
- Silva, G.L.; Ediz, V.; Yaron, D.; Armitage, B.A. Experimental and Computational Investigation of Unsymmetrical Cyanine Dyes: Understanding Torsionally Responsive Fluorogenic Dyes. J. Am. Chem. Soc. 2007, 129, 5710–5718. [Google Scholar] [CrossRef] [Green Version]
- Bunkenborg, J.; Gadjev, N.I.; Deligeorgiev, T.; Jacobsen, J.P. Concerted Intercalation and Minor Groove Recognition of DNA by a Homodimeric Thiazole Orange Dye. Bioconjugate Chem. 2000, 11, 861–867. [Google Scholar] [CrossRef]
- Nygren, J.; Svanvik, N.; Kubista, M. The interactions between the fluorescent dye thiazole orange and DNA. Biopolymers 1998, 46, 39–51. [Google Scholar] [CrossRef]
- Privat, E.; Melvin, T.; Mérola, F.; Schweizer, G.; Prodhomme, S.; Asseline, U.; Vigny, P. Fluorescent properties of oligonucleo-tide-conjugated thiazole orange probes. Photochem. Photobiol. 2002, 75, 201–210. [Google Scholar] [CrossRef]
- Muskalska, B.; Szymczak, B. Progress in research on the genus Listeria. Post. Mikrobiol. 2015, 54, 123–132. [Google Scholar]
- Brown, M.R.W.; Williams, P. The influence of environment on envelope properties affecting survival of bacteria in infections. Ann. Rev. Microbiol. 1985, 39, 527–556. [Google Scholar] [CrossRef] [PubMed]
- Manning, A.J.; Kuehn, M.J. Functional Advantages Conferred by Extracellular Prokaryotic Membrane Vesicles. J. Mol. Microbiol. Biotechnol. 2013, 23, 131–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaar, V.; Nordström, T.; Mörgelin, M.; Riesbeck, K. Moraxella catarrhalisOuter Membrane Vesicles Carry β-Lactamase and Promote Survival ofStreptococcus pneumoniaeandHaemophilus influenzaeby Inactivating Amoxicillin. Antimicrob. Agents Chemother. 2011, 55, 3845–3853. [Google Scholar] [CrossRef] [Green Version]
- Wasążnik, A.; Grinholc, M.; Bielawski, K.P. Active efflux as the multidrug resistance mechanism. Postepy Hig Med Dosw. 2009, 63, 123–133. [Google Scholar]
- Tintino, S.R.; Morais-Tintino, C.D.; Campina, F.F.; Costa, M.S.; Menezes, I.R.A.; de Matos, Y.M.L.S.; Calixto-Júnior, J.T.; Pereira, P.S.; Siqueira-Junior, J.P.; Leal-Balbino, T.C.; et al. Tannic acid affects the phenotype of Staphylococcus aureus resistant totetracycline and erythromycin by inhibition of efflux pumps. Bioorganic Chemistry. 2017, 74, 197–200. [Google Scholar] [CrossRef]
- Dos Santos, J.F.; Tintino, S.R.; de Freitas, T.S.; Campina, F.F.; Menezes, I.R.D.A.; Siqueira-Júnior, J.P.; Coutinho, H.D.; Cunha, F.A. In vitro e in silico evaluation of the inhibition of Staphylococcus aureus efflux pumps by caffeic and gallic acid. Comp. Immunol. Microbiol. Infect. Dis. 2018, 57, 22–28. [Google Scholar] [CrossRef]
- Figueredo, F.G.; Ramos, I.T.D.L.; Paz, J.A.; Silva, T.M.; Câmara, C.A.; Oliveira-Tintino, C.D.D.M.; Tintino, S.R.; Farias, P.; de Menezes, I.R.A.; Coutinho, H.D.M.; et al. Effect of hydroxyamines derived from lapachol and norlachol against Staphylococcus aureus strains carrying the NorA efflux pump. Infect. Genet. Evol. 2020, 84, 104370. [Google Scholar] [CrossRef]
- Menezes, I.R.A.; Coutinho, H.D.M.; Pinheiro, P.G.; Santiago, G.M.P.; da Silva, F.E.F.; de Araújo, A.C.J.; de Oliveira, C.R.T.; Freitas, P.R.; Rocha, J.E.; Neto, J.B.D.A.; et al. Antibacterial activity and inhibition against Staphylococcus aureus NorA efflux pump by ferulic acid and its esterified derivatives. Asian Pac. J. Trop. Biomed. 2021, 11, 405. [Google Scholar] [CrossRef]
- Dos Santos, J.F.S.; Tintino, S.R.; da Silva, A.R.P.; Barbosa, C.R.d.S.; Scherf, J.R.; Silveira, Z.d.S.; de Freitas, T.S.; Neto, L.J.d.L.; Barros, L.M.; Menezes, I.R.d.A.; et al. Enhancement of the antibiotic activity by quercetin against Staphylococcus aureus efflux pumps. J. Bioenerg. Biomembr. 2021, 53, 157–167. [Google Scholar] [CrossRef]
- Absolom, D.R.; Lamberti, F.V.; Policova, Z.; Zingg, W.; van Oss, C.J.; Neumann, A.W. Surface thermodynamics of bacterial adhesion. Appl. Environ. Microbiol. 1983, 46, 90–97. [Google Scholar] [CrossRef] [Green Version]
- Ruan, B.; Wu, P.; Liu, J.; Jiang, L.; Wang, H.; Qiao, J.; Zhu, N.; Dang, Z.; Luo, H.; Yi, X. Adhesion of Sphingomonas sp. GY2B onto montmorillonite: A combination study by thermodynamics and the extended DLVO theory. Colloids Surf. B Biointerfaces 2020, 192, 111085. [Google Scholar] [CrossRef]
- O’Gara, J.P. icaand beyond: Biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 2007, 270, 179–188. [Google Scholar] [CrossRef] [Green Version]
- Duvernoy, M.-C.; Mora, T.; Ardré, M.; Croquette, V.; Bensimon, D.; Quilliet, C.; Ghigo, J.-M.; Balland, M.; Beloin, C.; Lecuyer, S.; et al. Asymmetric adhesion of rod-shaped bacteria controls microcolony morphogenesis. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kregiel, D. Adhesion of Aeromonas hydrophila to glass surfaces modified with organosilanes. Food Technol. Biotechnol. 2013, 51, 345–351. [Google Scholar]
- Nowacka, M.; Rygała, A.; Kręgiel, D.; Kowalewska, A. New Antiadhesive Hydrophobic Polysiloxanes. Molecules 2021, 26, 814. [Google Scholar] [CrossRef]
- Sánchez-López, E.; Gomes, D.; Esteruelas, G.; Bonilla, L.; Lopez-Machado, A.L.; Galindo, R.; Cano, A.; Espina, M.; Ettcheto, M.; Camins, A.; et al. Metal-Based Nanoparticles as Antimicrobial Agents: An Overview. Nanomaterials 2020, 10, 292. [Google Scholar] [CrossRef] [Green Version]
- Abbaszadegan, A.; Ghahramani, Y.; Gholami, A.; Hemmateenejad, B.; Dorostkar, S.; Nabavizadeh, M.; Sharghi, H. The Effect of Charge at the Surface of Silver Nanoparticles on Antimicrobial Activity against Gram-Positive and Gram-Negative Bacteria: A Preliminary Study. J. Nanomater. 2015, 2015, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Correa, M.G.; Martínez, F.B.; Vidal, C.P.; Streitt, C.; Escrig, J.; De Dicastillo, C.L. Antimicrobial metal-based nanoparticles: A review on their synthesis, types and antimicrobial action. Beilstein J. Nanotechnol. 2020, 11, 1450–1469. [Google Scholar] [CrossRef]
- Simon, A.T.; Dutta, D.; Chattopadhyay, A.; Ghosh, S.S. Copper Nanocluster-Doped Luminescent Hydroxyapatite Nanoparticles for Antibacterial and Antibiofilm Applications. ACS Omega 2019, 4, 4697–4706. [Google Scholar] [CrossRef]
- Subramaniyan, S.B.; Vijayakumar, S.; Megarajan, S.; Kamlekar, R.K.; Anbazhagan, V. Remarkable Effect of Jacalin in Diminishing the Protein Corona Interference in the Antibacterial Activity of Pectin-Capped Copper Sulfide Nanoparticles. ACS Omega 2019, 4, 14049–14056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haeili, M.; Moore, C.; Davis, C.J.C.; Cochran, J.B.; Shah, S.; Shrestha, T.B.; Zhang, Y.; Bossmann, S.H.; Benjamin, W.H.; Kutsch, O.; et al. Copper Complexation Screen Reveals Compounds with Potent Antibiotic Properties against Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2014, 58, 3727–3736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brahma, U.; Kothari, R.; Sharma, P.; Bhandari, V. Antimicrobial and anti-biofilm activity of hexadentated macrocyclic complex of copper (II) derived from thiosemicarbazide against Staphylococcus aureus. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Duarte, S.; Gregoire, S.; Singh, A.P.; Vorsa, N.; Schaich, K.; Bowen, W.H.; Koo, H. Inhibitory effects of cranberry polyphenols on formation and acidogenicity ofStreptococcus mutansbiofilms. FEMS Microbiol. Lett. 2006, 257, 50–56. [Google Scholar] [CrossRef] [Green Version]
- Abuelsaad, A.S.; Allam, G.; Al-Solumani, A.A. Hesperidin inhibits inflammatory response induced by Aeromonas hydrophila infec-tion and alters CD4+/CD8+ T cell ratio. Med. Inflamm. 2014, 2014, 393217. [Google Scholar] [CrossRef] [Green Version]
- Alalwan, H.K.A.; Rajendran, R.; Lappin, D.F.; Combet, E.; Shahzad, M.; Robertson, D.; Nile, C.; Williams, C.; Ramage, G. The Anti-Adhesive Effect of Curcumin on Candida albicans Biofilms on Denture Materials. Front. Microbiol. 2017, 8, 659. [Google Scholar] [CrossRef]
- Lee, J.-H.; Park, J.-H.; Cho, M.H.; Lee, J. Flavone Reduces the Production of Virulence Factors, Staphyloxanthin and α-Hemolysin, in Staphylococcus aureus. Curr. Microbiol. 2012, 65, 726–732. [Google Scholar] [CrossRef]
- Lee, J.-H.; Regmi, S.C.; Kim, J.-A.; Cho, M.H.; Yun, H.; Lee, C.-S.; Lee, J. Apple Flavonoid Phloretin Inhibits Escherichia coli O157:H7 Biofilm Formation and Ameliorates Colon Inflammation in Rats. Infect. Immun. 2011, 79, 4819–4827. [Google Scholar] [CrossRef] [Green Version]
- Kaur, A.; Sharma, P.; Capalash, N. Curcumin alleviates persistence of Acinetobacter baumannii against colistin. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Muthusamy, B.; Shanmugam, G. Analysis of flavonoid content, antioxidant, antimicrobial and antibiofilm activity of in vitro hairy root extract of radish (Raphanus sativus L.). Plant Cell Tissue Organ. Cult. (PCTOC) 2020, 140, 619–633. [Google Scholar] [CrossRef]
- Lopes, L.A.A.; Rodrigues, J.B.D.S.; Magnani, M.; de Souza, E.L.; de Siqueira-Júnior, J.P. Inhibitory effects of flavonoids on biofilm formation by Staphylococcus aureus that overexpresses efflux protein genes. Microb. Pathog. 2017, 107, 193–197. [Google Scholar] [CrossRef]



| Species | gx | gy | gz | PD(gz) a | Ax(Cu) b | Ax(Cu) b | Ax(Cu) b | PD(Az) a |
|---|---|---|---|---|---|---|---|---|
| Powder | c | c | 2.232 | −1.0 | c | c | −193.4 | 3.9 |
| DMF | 2.040 | 2.060 | 2.240 | −1.4 | −11.0 | −14.0 | −192.5 | 4.4 |
| DMSO | 2.038 | 2.061 | 2.241 | −1.4 | −12.0 | −13.0 | −194.0 | 3.6 |
| Aqueous solution d | 2.039 | 2.061 | 2.245 | −1.6 | −11.5 | −13.0 | −188.8 | 6.4 |
| DFT calculated e | 2.061 | 2.062 | 2.209 | – | −7.7 | −18.5 | −200.9 | – |
| Compound | Kb [M−1] | λmax [nm] | (Afree − Abound)/Afree [%]max (+)Hyperchromism/ (−)Hypochromism |
|---|---|---|---|
| HESP | 1.20 (±0.15) × 104 | 323 | +5.57 |
| HABH | 4.00 (±0.22) × 105 | 330 | −41.78 |
| CuHABH | 5.00 (±0.18) × 105 | 371 | −56.52 |
| Compound | KSV [M−1] | C50 [μM] | Kapp [M−1] * | Kq [M−1] | n | Ka [M−1] ** |
|---|---|---|---|---|---|---|
| Hesperetin | 3.73 (±0.06) × 103 | nd *** | nd *** | 1.43 (±0.20) × 1012 | 0.55 ± 0.04 | 1.58 (±0.20) × 103 |
| HABH | 3.88 (±0.13) × 104 | 40.4 | 1.86 × 106 | 1.49 (±0.15) × 1013 | 1.01 (±0.03) | 2.78 (±0.15) × 104 |
| CuHABH | 1.31 (±0.06) × 105 | 38.4 | 1.95 × 106 | 5.04 (±0.14) × 1013 | 1.63 (±0.03) | 6.74 (±0.14) × 104 |
| Compound 0.1 μM | Zone of inhibition [mm] | ||||||
|---|---|---|---|---|---|---|---|
| Salmonella Choleraesuis ATCC 7001 | Salmonella Typhimurium ATCC 14028 | Escherichia coli ATCC 10536 | Staphylococcus aureus ATCC 25923 | Staphylococcus aureus ATCC 27734 | Listeria monocytogenes ATCC 19111 | Listeria monocytogenes ATCC 19115 | |
| CuHABH | 0.0 ± 0.0 aA | 0.0 ± 0.0 aA | 0.0 ± 0.0 aA | 10.5 ± 0.50 aA | 10.5 ± 0.50 aA | 0.0 ± 0.0 aA | 12.5 ± 0.50 aA |
| HABH | 0.0 ± 0.0 aA | 0.0 ± 0.0 aA | 0.0 ± 0.0 aA | 11.5 ± 0.50 aA | 12.0 ± 0.0 bA | 0.0 ± 0.0 aA | 13.5 ± 0.50 bA |
| HESP | 0.0 ± 0.0 aA | 0.0 ± 0.0 aA | 0.0 ± 0.0 aA | 9.0 ± 0.0 bA | 9.0 ± 0.50 cA | 0.0 ± 0.0 aA | 14.5 ± 0.50 cA |
| DMSO | 0.0 ± 0.0 aA | 0.0 ± 0.0 aA | 0.0 ± 0.0 aA | 0.0 ± 0.0 cA | 0.0 ± 0.0 dA | 0.0 ± 0.0 aA | 0.0 ± 0.0 dA |
| Vancomycin | 10.5 ± 0.00B | Nd | 14.5 ± 0.50B | 16.00 ± 0.00 B | 17.5 ± 0.00 B | 20.0 ± 0.80 B | 30.5 ± 0.50 B |
| Ampicillin | Nd | 12.60 ± 0.90 B | Nd | Nd | Nd | 21.0 ± 0.80 B | 16.5 ± 0.50 C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sykuła, A.; Łodyga-Chruścińska, E.; Garribba, E.; Kręgiel, D.; Dzeikala, A.; Klewicka, E.; Piekarska-Radzik, L. From the Physicochemical Characteristic of Novel Hesperetin Hydrazone to Its In Vitro Antimicrobial Aspects. Molecules 2022, 27, 845. https://doi.org/10.3390/molecules27030845
Sykuła A, Łodyga-Chruścińska E, Garribba E, Kręgiel D, Dzeikala A, Klewicka E, Piekarska-Radzik L. From the Physicochemical Characteristic of Novel Hesperetin Hydrazone to Its In Vitro Antimicrobial Aspects. Molecules. 2022; 27(3):845. https://doi.org/10.3390/molecules27030845
Chicago/Turabian StyleSykuła, Anna, Elżbieta Łodyga-Chruścińska, Eugenio Garribba, Dorota Kręgiel, Aliaksandr Dzeikala, Elżbieta Klewicka, and Lidia Piekarska-Radzik. 2022. "From the Physicochemical Characteristic of Novel Hesperetin Hydrazone to Its In Vitro Antimicrobial Aspects" Molecules 27, no. 3: 845. https://doi.org/10.3390/molecules27030845








