Design of Turmeric Rhizome Extract Nano-Formula for Delivery to Cancer Cells
Abstract
1. Introduction
2. Results and Discussion
2.1. Curcuminoids Content of Turmeric Rhizome Fractions
2.2. Determination of the Optimal Turmeric Rhizome Extract Nanoparticles Formulation
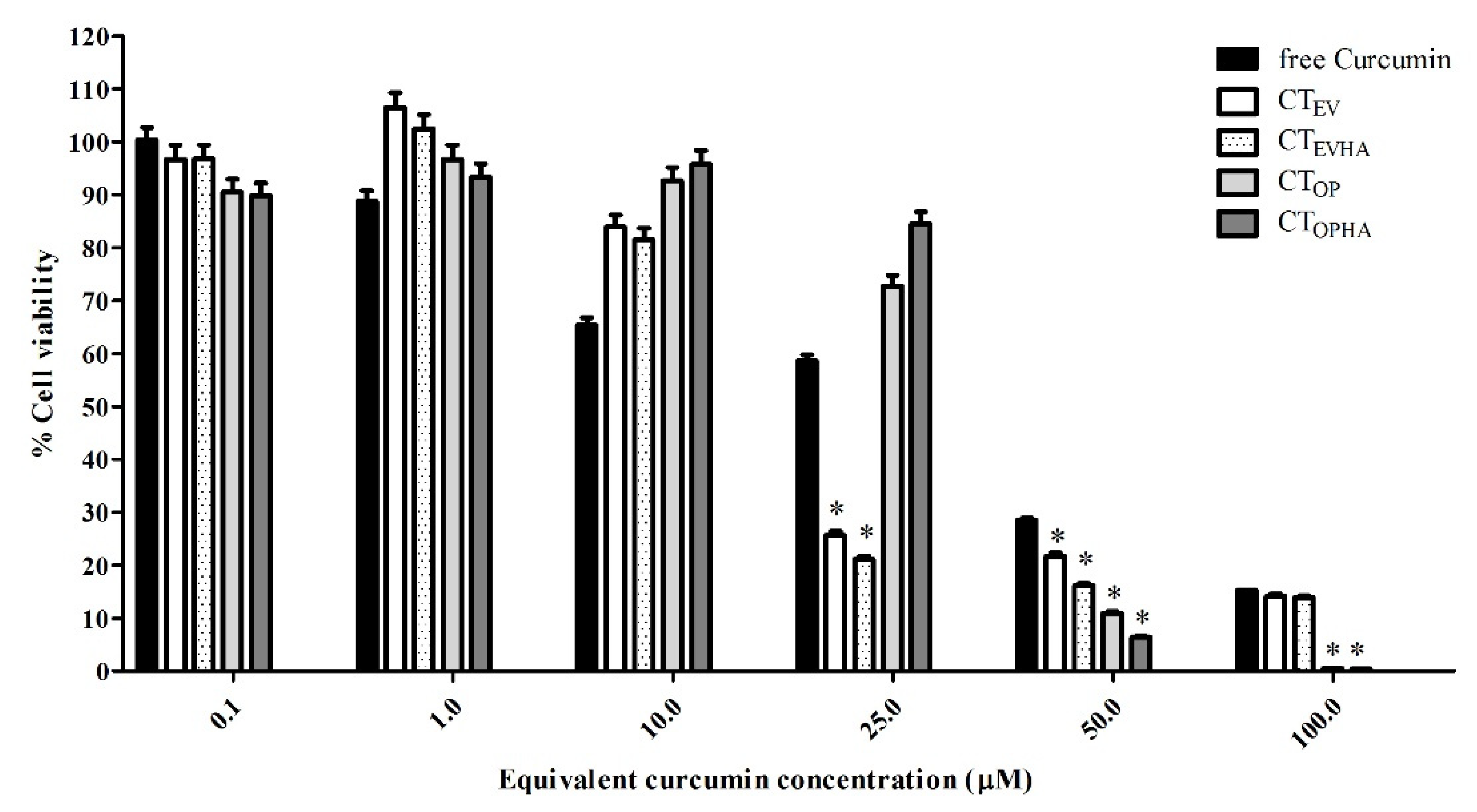
2.3. Cytotoxicity of Turmeric Rhizome Extract Nanoparticles in HepG2 Cells
3. Materials and Methods
3.1. Materials
3.2. Preparation of Turmeric Rhizome Fraction
3.3. Characterization and Curcuminoids Content Analysis of Turmeric Rhizome Extracts
3.4. Preparation of Turmeric Rhizome Extract Nanoparticles
3.5. Characterization and Curcuminoids Content Analysis of Turmeric Rhizome Extract Nanoparticles
3.6. Determination of the Optimal Turmeric Rhizome Extract Nanoparticles Formulation
3.7. Preparation, Characterization, and Cytotoxicity Test of the Optimal Turmeric Rhizome Extract Nanoparticles
3.7.1. Cell Culture
3.7.2. MTT Assay (Cytotoxicity Assay)
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Farrell, K.T. Spices, Condiments, and Seasoning, 2nd ed.; Van Nostrand Reinhold: New York, NY, USA, 1990; pp. 203–205. [Google Scholar]
- Nair, K.P.P. The Agronomy and Economy of Turmeric and Ginger: The Invaluable Medicinal Spice Crops; Elsevier: London, UK, 2013; pp. 1–224. [Google Scholar]
- Nabavi, S.F.; Daglia, M.; Moghaddam, A.H.; Habtemariam, S.; Nabavi, S.M. Curcumin and Liver Disease: From Chemistry to Medicine. Compr. Rev. Food Sci. Food Saf. 2014, 13, 62–77. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, B.B.; Surh, Y.J.; Shishodia, S. The Molecular Targets and Therapeutic Uses of Curcumin in Health and Disease; Springer Science+Business Media: New York, NY, USA, 2007. [Google Scholar]
- Basnet, P.; Skalko-Basnet, N. Curcumin: A Challenge in Cancer Treatment. J. Nepal Pharm. Assoc. 2012, 26, 19–47. [Google Scholar] [CrossRef]
- Atsumi, T.; Tonosaki, K.; Fujisawa, S. Comparative cytotoxicity and ROS generation by curcumin and tetrahydrocurcumin following visible-light irradiation or treatment with horseradish peroxidase. Anticancer. Res. 2007, 27, 363–372. [Google Scholar] [PubMed]
- Sandur, S.K.; Pandey, M.K.; Sung, B. Curcumin, demethoxycurcumin, bisdemethoxycurcumin, tetrahydrocurcumin and turmerones differentially regulate anti-inflammatory and anti-proliferative responses through a ROS-independent mecha-nism. Carcinogenesis 2007, 28, 1765–1773. [Google Scholar] [CrossRef]
- Ravindran, J.; Subbaraju, G.V.; Ramani, M.V. Bisdemethylcurcumin and structurally related hispolon analogues of curcumin ex-hibit enhanced prooxidant, anti-proliferative and anti-inflammatory activities in vitro. Biochem. Pharmacol. 2010, 79, 1658–1666. [Google Scholar] [CrossRef]
- Yue, G.G.; Chan, B.C.; Hon, P.M. Evaluation of in vitro anti-proliferative and immunomodulatory activities of compounds isolated from Curcuma longa. Food Chem. Toxicol. 2010, 48, 2011–2020. [Google Scholar] [CrossRef]
- Sharma, R.; Gescher, A.; Steward, W. Curcumin: The story so far. Eur. J. Cancer 2005, 41, 1955–1968. [Google Scholar] [CrossRef]
- Cheng, A.L.; Hsu, C.-H.; Lin, J.K.; Hsu, M.M.; Ho, Y.-F.; Shen, T.S.; Ko, J.Y.; Lin, J.T.; Lin, B.-R.; Ming-Shiang, W.; et al. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high-risk or pre-malignant lesions. Anticancer Res. 2001, 21, 2895–2900. [Google Scholar]
- Nair, H.B.; Sung, B.; Yadav, V.R.; Kannappan, R.; Chaturvedi, M.M.; Aggarwal, B.B. Delivery of antiinflammatory nutraceuticals by nanoparticles for the prevention and treatment of cancer. Biochem. Pharmacol. 2010, 80, 1833–1843. [Google Scholar] [CrossRef]
- Naksuriya, O.; Okonogi, S.; Schiffelers, R.; Hennink, W.E. Curcumin nanoformulations: A review of pharmaceutical properties and preclinical studies and clinical data related to cancer treatment. Biomaterials 2014, 35, 3365–3383. [Google Scholar] [CrossRef]
- Yallapu, M.M.; Jaggi, M.; Chauhan, S.C. Curcumin nanoformulations: A future nanomedicine for cancer. Drug Discov. Today 2012, 17, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Xie, J.; Chen, H.; Gu, S.; Zhao, R.; Shao, J.; Jia, L. Nanotechnology-based intelligent drug design for cancer metastasis treatment. Biotechnol. Adv. 2014, 32, 761–777. [Google Scholar] [CrossRef] [PubMed]
- Hull, L.; Farrell, D.; Grodzinski, P. Highlights of recent developments and trends in cancer nanotechnology research—View from NCI Alliance for Nanotechnology in Cancer. Biotechnol. Adv. 2014, 32, 666–678. [Google Scholar] [CrossRef] [PubMed]
- Kateb, B.; Chiu, K.; Black, K.L. Nanoplatforms for constructing new approaches to cancer treatment, imaging, and drug delivery: What should be the policy? NeuroImage 2011, 54 (Suppl. S1), S106–S124. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Pattni, B.S.; Abouzeid, A.H.; Torchilin, V.P. Nanopreparations to overcome multidrug resistance in cancer. Adv. Drug Deliv. Rev. 2013, 65, 1748–1762. [Google Scholar] [CrossRef]
- Patra, H.K.; Turner, A.P. The potential legacy of cancer nanotechnology: Cellular selection. Trends Biotechnol. 2014, 32, 21–31. [Google Scholar] [CrossRef]
- da Rocha, E.L.; Porto, L.; Rambo, C. Nanotechnology meets 3D in vitro models: Tissue engineered tumors and cancer therapies. Mater. Sci. Eng. C 2014, 34, 270–279. [Google Scholar] [CrossRef]
- Anand, P.; Nair, H.B.; Sung, B. Design of curcumin-loaded PLGA nanoparticles formulation with enhanced cellular up-take, and increased bioactivity in vitro and superior bioavailability in vivo. Biochem. Pharmacol. 2010, 79, 330–338. [Google Scholar] [CrossRef]
- Anitha, A.; Maya, S.; Deepa, N. Efficient water soluble O-carboxymethyl chitosan nanocarrier for the delivery of curcumin to cancer cells. Carbohydr. Polym. 2011, 83, 452–461. [Google Scholar] [CrossRef]
- Cui, J.; Yu, B.; Zhao, Y.; Zhu, W.; Li, H.; Lou, H.; Zhai, G. Enhancement of oral absorption of curcumin by self-microemulsifying drug delivery systems. Int. J. Pharm. 2009, 371, 148–155. [Google Scholar] [CrossRef]
- Khalil, N.M.; do Nascimento, T.C.; Casa, D.M. Pharmacokinetics of curcumin-loaded PLGA and PLGA-PEG blend nanoparticles after oral administration in rats. Colloids Surf. B 2013, 101, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Konwarh, R.; Saikia, J.P.; Karak, N.; Konwar, B.K. ‘Poly(ethylene glycol)-magnetic nanoparticles-curcumin’ trio: Directed morphogenesis and synergistic free-radical scavenging. Colloids Surf. B Biointerfaces 2010, 81, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-H.; Chang, F.-Y.; Hung, D.-K. Terpene microemulsions for transdermal curcumin delivery: Effects of terpenes and cosurfactants. Colloids Surf. B Biointerfaces 2011, 82, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, C.; Sahoo, S.K. The in vitro stability and in vivo pharmacokinetics of curcumin prepared as an aqueous nanoparticulate formulation. Biomaterials 2010, 31, 6597–6611. [Google Scholar] [CrossRef] [PubMed]
- Nair, K.L.; Thulasidasan, A.K.; Deepa, G. Purely aqueous PLGA nanoparticulate formulations of curcumin exhibit en-hanced anticancer activity with dependence on the combination of the carrier. Int. J. Pharm. 2012, 425, 44–52. [Google Scholar] [CrossRef]
- Shaikh, J.; Ankola, D.; Beniwal, V.; Singh, D.; Kumar, M.R. Nanoparticle encapsulation improves oral bioavailability of curcumin by at least 9-fold when compared to curcumin administered with piperine as absorption enhancer. Eur. J. Pharm. Sci. 2009, 37, 223–230. [Google Scholar] [CrossRef]
- Song, L.; Shen, Y.; Hou, J. Polymeric micelles for parenteral delivery of curcumin: Preparation, characterization and in vitro evaluation. Colloids Surf. A 2011, 390, 25–32. [Google Scholar] [CrossRef]
- Tsai, Y.-M.; Chien, C.-F.; Lin, L.-C.; Tsai, T.-H. Curcumin and its nano-formulation: The kinetics of tissue distribution and blood–brain barrier penetration. Int. J. Pharm. 2011, 416, 331–338. [Google Scholar] [CrossRef]
- Tsai, Y.M.; Jan, W.C.; Chien, C.F. Optimised nano-formulation on the bioavailability of hydrophobic polyphenol, curcumin, in freely-moving rats. Food Chem. 2011, 127, 918–925. [Google Scholar] [CrossRef]
- Wan, Z.; Ke, D.; Hong, J.; Ran, Q.; Wang, X.; Chen, Z.; An, X.; Shen, W. Comparative study on the interactions of cationic gemini and single-chain surfactant micelles with curcumin. Colloids Surf. A Physicochem. Eng. Asp. 2012, 414, 267–273. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, Y.; Wang, Y.-W.; Huang, M.-T.; Ho, C.-T.; Huang, Q. Enhancing anti-inflammation activity of curcumin through O/W nanoemulsions. Food Chem. 2008, 108, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Yallapu, M.M.; Gupta, B.K.; Jaggi, M.; Chauhan, S.C. Fabrication of curcumin encapsulated PLGA nanoparticles for improved therapeutic effects in metastatic cancer cells. J. Colloid Interface Sci. 2010, 351, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Zanotto-Filho, A.; Coradini, K.; Braganhol, E.; Schröder, R.; de Oliveira, C.M.; Simões-Pires, A.; Battastini, A.M.O.; Pohlmann, A.; Guterres, S.; Forcelini, C.M.; et al. Curcumin-loaded lipid-core nanocapsules as a strategy to improve pharmacological efficacy of curcumin in glioma treatment. Eur. J. Pharm. Biopharm. 2013, 83, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Du, J.; Duan, Y. Curcumin loaded mixed micelles composed of Pluronic P123 and F68: Preparation, optimiza-tion and in vitro characterization. Colloids Surf. B 2012, 97, 101–108. [Google Scholar] [CrossRef]
- Jin, Y.-J.; Ubonvan, T.; Kim, D.-D. Hyaluronic Acid in Drug Delivery Systems. J. Pharm. Investig. 2010, 40, 33–43. [Google Scholar] [CrossRef]
- Mizrahy, S.; Raz, S.R.; Hasgaard, M. Hyaluronan-coated nanoparticles: The influence of the molecular weight on CD44-hyaluronan interactions and on the immune response. Control. Release 2011, 156, 231–238. [Google Scholar] [CrossRef]
- Oyarzun-Ampuero, F.A.; Rivera-Rodriguez, G.; Alonso, M.J.; Torres, D. Hyaluronan nanocapsules as a new vehicle for intracellular drug delivery. Eur. J. Pharm. Sci. 2013, 49, 483–490. [Google Scholar] [CrossRef]
- Necas, J.; Bartosikova, L.; Brauner, P.; Kolar, J. Hyaluronic acid (hyaluronan): A review. Vet. Med. 2008, 53, 397–411. [Google Scholar] [CrossRef]
- Platt, V.M.; Szoka, F.C. Anticancer therapeutics: Targeting macromolecules and nanocarriers to hyaluronan or CD44, a hyaluronan receptor. Mol. Pharm. 2008, 5, 474–486. [Google Scholar] [CrossRef]
- Toole, B.P. Hyaluronan-CD44 Interactions in Cancer: Paradoxes and Possibilities. Clin. Cancer Res. 2009, 15, 7462–7468. [Google Scholar] [CrossRef]
- Anton, N.; Benoit, J.-P.; Saulnier, P. Design and production of nanoparticles formulated from nano-emulsion templates—A review. J. Control. Release 2008, 128, 185–199. [Google Scholar] [CrossRef]
- Solans, C.; Izquierdo, P.; Nolla, J. Nano-emulsions. Curr. Opin. Colloid Interface Sci. 2005, 10, 102–110. [Google Scholar] [CrossRef]
- Grillo, I. Small-angle neutron scattering study of a world-wide known emulsion: Le Pastis. Colloids Surf. A Physicochem. Eng. Asp. 2003, 225, 153–160. [Google Scholar] [CrossRef]
- Vitale, S.A.; Katz, J.L. Liquid droplet dispersions formed by homogeneous liquid-liquid nucleation: “the Ouzo effect”. Langmuir 2003, 19, 4105–4110. [Google Scholar] [CrossRef]
- Carteau, D.; Bassani, D.; Pianet, I. The “Ouzo effect”: Following the spontaneous emulsification of trans-anethole in water by NMR. C. R. Chim. 2008, 11, 493–498. [Google Scholar] [CrossRef]
- Scholten, E.; van der Linden, E.; This, H. The Life of an Anise-Flavored Alcoholic Beverage: Does Its Stability Cloud or Confirm Theory? Langmuir 2008, 24, 1701–1706. [Google Scholar] [CrossRef]
- Aubry, J.; Ganachaud, F.; Addad, J.-P.C.; Cabane, B. Nanoprecipitation of Polymethylmethacrylate by Solvent Shifting:1. Boundaries. Langmuir 2009, 25, 1970–1979. [Google Scholar] [CrossRef] [PubMed]
- Beck-Broichsitter, M.; Rytting, E.; Lebhardt, T.; Wang, X.; Kissel, T. Preparation of nanoparticles by solvent displacement for drug delivery: A shift in the “ouzo region” upon drug loading. Eur. J. Pharm. Sci. 2010, 41, 244–253. [Google Scholar] [CrossRef]
- Mora-Huertas, C.E.; Fessi, H.; Elaissari, A. Influence of process and formulation parameters on the formation of submicron par-ticles by solvent displacement and emulsification-diffusion methods critical comparison. Adv. Colloid Interface Sci. 2011, 163, 90–122. [Google Scholar] [CrossRef] [PubMed]
- Botet, R. The “ouzo effect”, recent developments and application to therapeutic drug carrying. J. Phys. Conf. Ser. 2012, 352, 12047. [Google Scholar] [CrossRef]
- Solans, C.; Solé, I. Nano-emulsions: Formation by low-energy methods. Curr. Opin. Colloid Interface Sci. 2012, 17, 246–254. [Google Scholar] [CrossRef]
- Lepeltier, E.; Bourgaux, C.; Couvreur, P. Nanoprecipitation and the “Ouzo effect”: Application to drug delivery devices. Adv. Drug Deliv. Rev. 2014, 71, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Jirawongse, V.A. Khamin chan. In Thai Herbal Pharmacopoeia; Jirawongse, V.A., Ed.; Prachachon: Bangkok, Thailand, 2020; Volume I, pp. 142–149. [Google Scholar]
- Wichitnithad, W.; Jongaroonngamsang, N.; Pummangura, S. A simple isocratic HPLC method for the simultaneous determina-tion of curcuminoids in commercial turmeric extracts. Phytochem. Anal. 2009, 20, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yuan, W.; Deng, G. Chemical composition and product quality control of turmeric (Curcuma longa L.). Pharm. Crops. 2011, 2, 28–54. [Google Scholar] [CrossRef]
- Grynkiewicz, G.; Ślifirski, P. Curcumin and curcuminoids in quest for medicinal status. Acta Biochim. Pol. 2012, 59. [Google Scholar] [CrossRef]
- Pothirat, W.; Gritsanapan, W. Quantitative analysis of curcumin, demethocycurcumin and bisdemethoxycurcumin in the crude curcuminoid extract from Curcuma longa in Thailand by TLC-densitometry. MUJPS 2005, 32, 23–30. [Google Scholar]
- Chopra, R.N.; Chopra, I.C.; Hemda, K.L. Chopra’s Indigenous Drugs of India, 2nd ed.; Academic: Calcutta, India, 1958. [Google Scholar]
- Natural and Non-Prescription Health Products Directorate; Health Canada: Ottawa, ON, Canada, 2015.
- Vu, M.N.; Rajasekhar, P.; Poole, D.P.; Khor, S.Y.; Truong, N.P.; Nowell, C.J.; Quinn, J.F.; Whittaker, M.R.; Veldhuis, N.A.; Davis, T.P. Rapid Assessment of Nanoparticle Extravasation in a Microfluidic Tumor Model. ACS Appl. Nano Mater. 2019, 2, 1844–1856. [Google Scholar] [CrossRef]
- Shi, J.; Kantoff, P.W.; Wooster, R.; Farokhzad, O.C. Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 2017, 17, 20–37. [Google Scholar] [CrossRef]
- Shukla, T.; Upmanyu, N.; Pandey, S.P.; Sudheesh, M.S. Site-specific drug delivery, targeting, and gene therapy. In Nanoarchitectonics in Biomedicine; Grumezescu, A.M., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2019; pp. 473–505. [Google Scholar]
- Greish, K. Enhanced permeability and retention (EPR) effect for anticancer nanomedicine drug targeting. In Cancer Nanotechnology—Methods and Protocols; Grobmyer, S.R., Moudgil, B.M., Eds.; Humana Press, c/o Springer Science+Business Media, LLC: New York, NY, USA, 2010; pp. 25–37. [Google Scholar]
- Perez-Herrero, E.; Fernandez-Medarde, A. Advanced targeted therapies in cancer: Drug nanocarriers, the future of chemotherapy. Eur. J. Pharm. Biopharm. 2015, 93, 52–79. [Google Scholar] [CrossRef]
- Demir, E.A.; Demir, S.; Aliyazicioglu, Y. In vitro cytotoxic effect of ethanol and dimethyl sulfoxide on various human cell lines. KSU J. Agric. Nat. 2020, 23, 1119–1124. [Google Scholar]
- Nguyen, S.T.; Nguyen, H.T.-L.; Truong, K.D. Comparative cytotoxic effects of methanol, ethanol and DMSO on human cancer cell lines. Biomed. Res. Ther. 2020, 7, 3855–3859. [Google Scholar] [CrossRef]
- Castaneda, F.; Rosin-Steiner, S. Low concentration of ethanol induce apoptosis in HepG2 cells: Role of various signal transduc-tion pathways. Int. J. Med. Sci. 2006, 3, 160–167. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pastorino, J.G.; Hoek, J.B. Ethanol potentiates tumor necrosis factor-α cytotoxicity in hepatoma cells and primary rat hepatocytes by promoting induction of the mitochondrial permeability transition. Hepatology 2000, 31, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Cederbaum, A.I. Ethanol Cytotoxicity to a Transfected HepG2 Cell Line Expressing Human Cytochrome P4502E1. J. Biol. Chem. 1996, 271, 23914–23919. [Google Scholar] [CrossRef]
- Smith, W.F. Experimental Design for Formulation; American Statistical Association: Alexandria, VA, USA, 2005; pp. 1–347. [Google Scholar]
- Piriyaprasarth, S.; Sriamornsak, P.; Juttulapa, M.; Puttipipatkhachorn, S. Modeling of Drug Release from Matrix Tablets with Process Variables of Microwave-Assisted Modification of Arrowroot Starch Using Artificial Neural Network. Adv. Mater. Res. 2010, 152–153, 1700–1703. [Google Scholar] [CrossRef]
- Chittasupho, C.; Jaturanpinyo, M.; Mangmool, S. Pectin nanoparticle enhances cytotoxicity of methotrexate against hepG2 cells. Drug Deliv. 2012, 20, 1–9. [Google Scholar] [CrossRef]
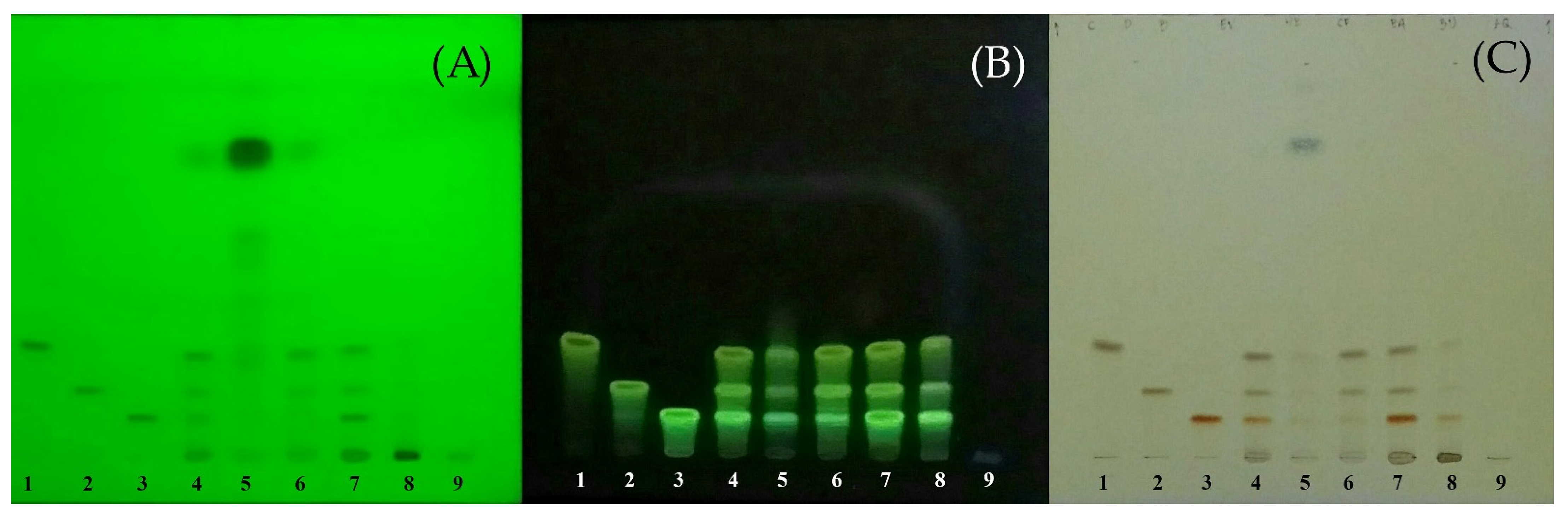
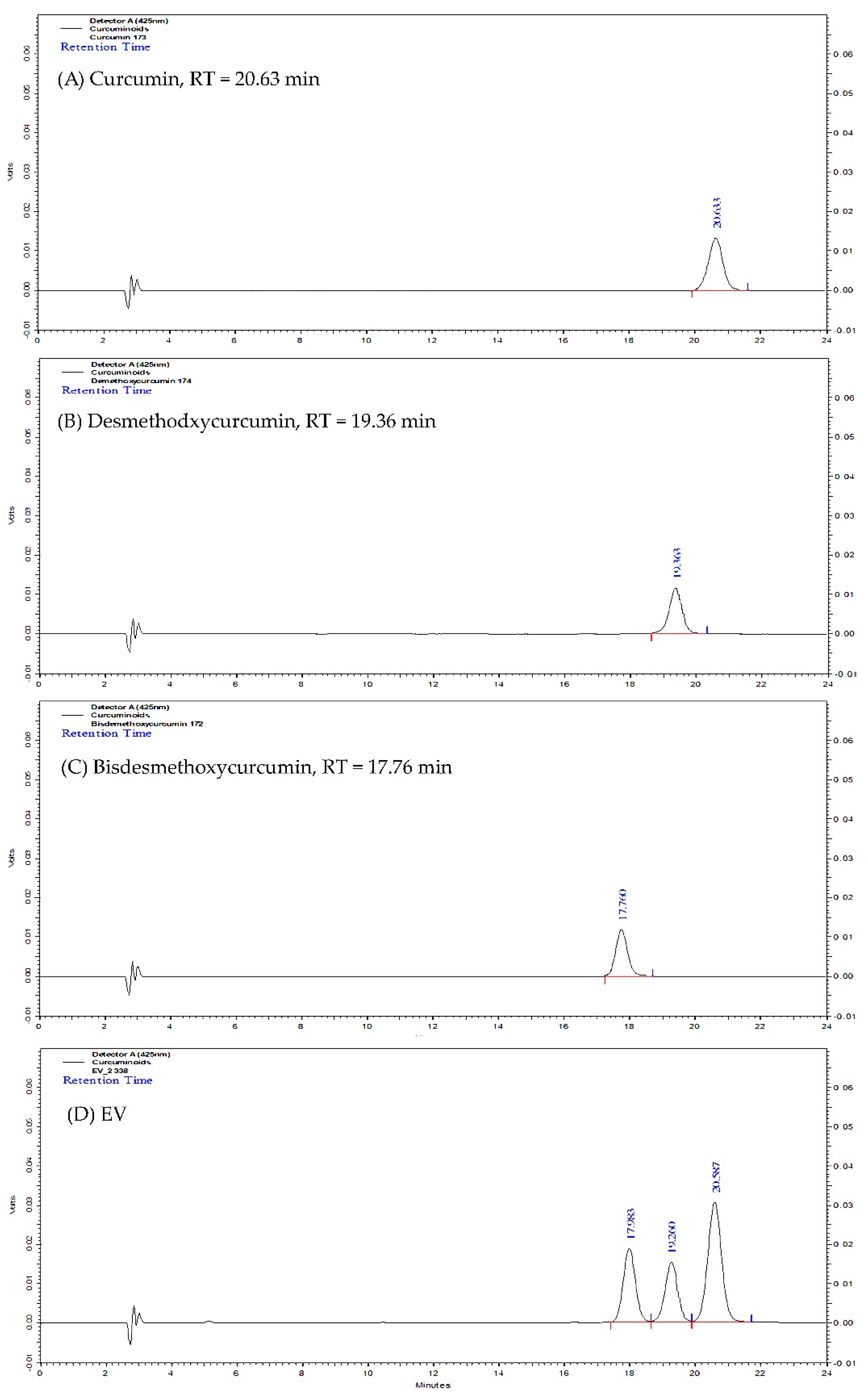
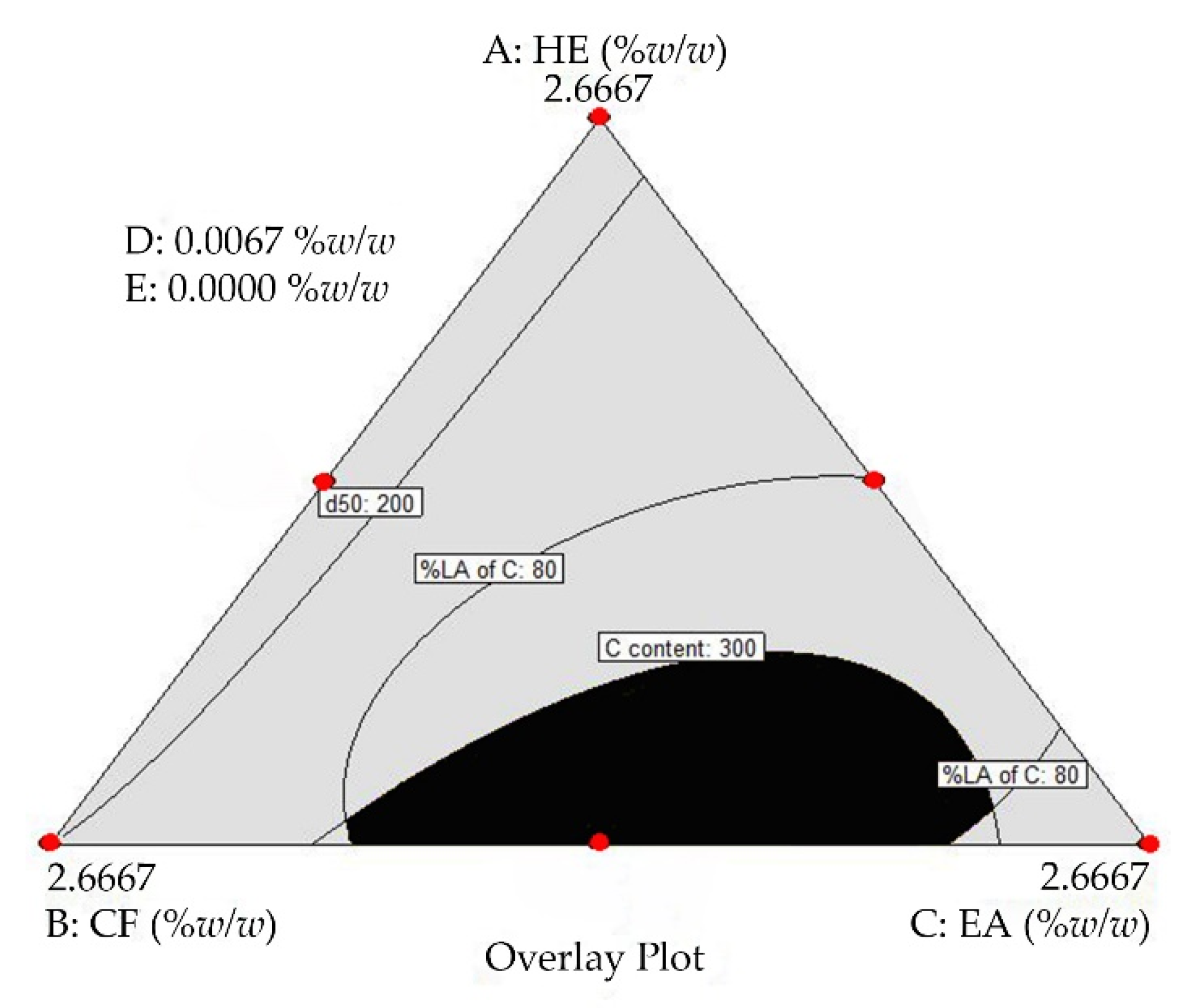
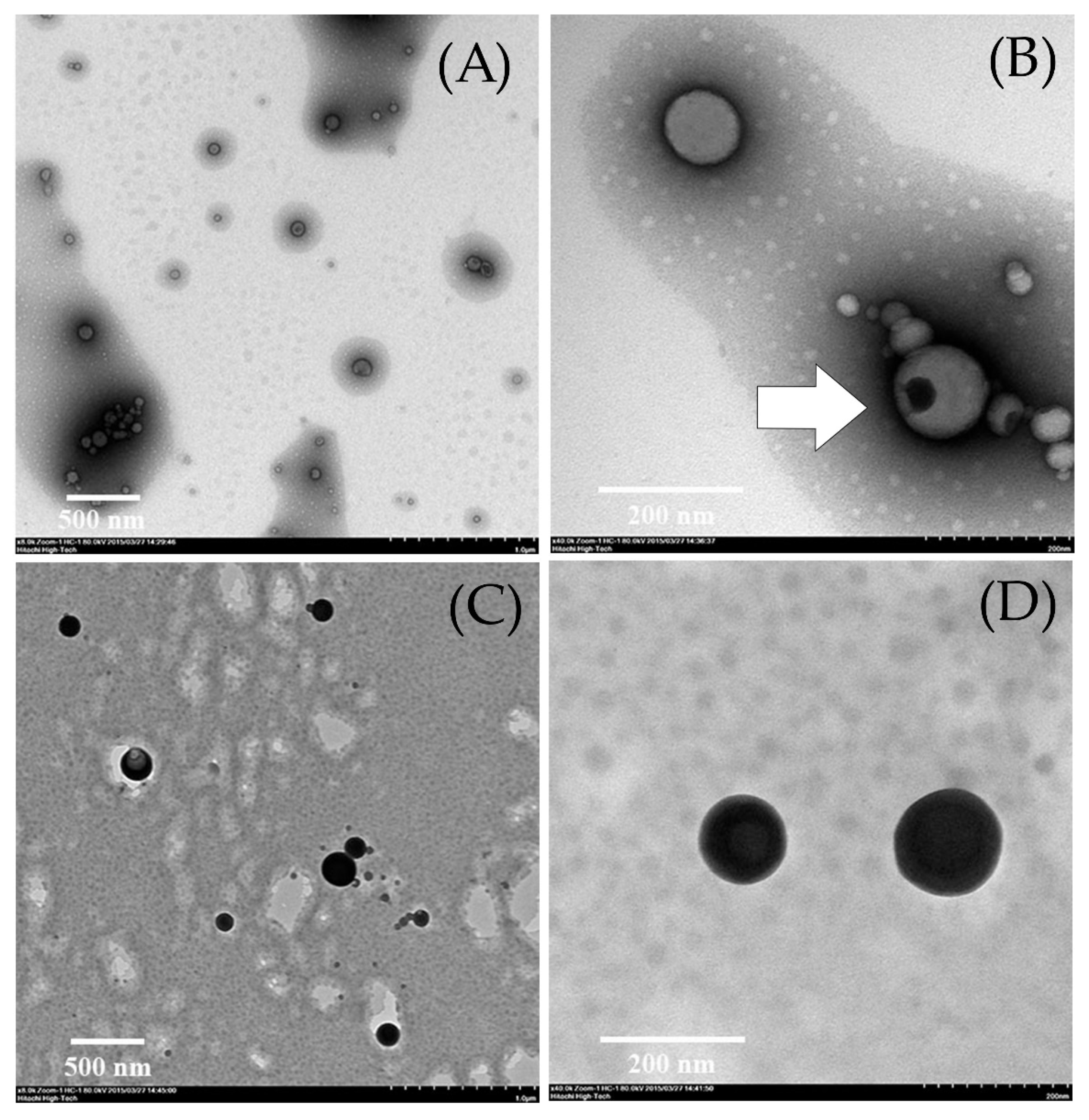
| Fraction | Curcuminoids Content (mg/g of Dried Extract) | |||
|---|---|---|---|---|
| CM | DCM | BDCM | Total Curcuminoids | |
| EV | 147.97 ± 1.24 | 68.64 ± 0.57 | 69.39 ± 0.55 | 285.99 ± 2.35 |
| HE | 2.21 ± 0.02 | 1.12 ± 0.01 | 1.54 ± 0.01 | 4.88 ± 0.04 |
| CF | 114.05 ± 1.59 | 52.26 ± 0.75 | 13.72 ± 0.13 | 180.04 ± 2.47 |
| EA | 132.09 ± 1.82 | 88.42 ± 1.31 | 200.89 ± 3.77 | 421.41 ± 6.84 |
| BU | 4.38 ± 1.82 | 4.18 ± 1.74 | 3.58 ± 1.50 | 12.14 ± 5.06 |
| AQ | ND | ND | ND | ND |
| Regression Model | Min | Max | r2 | Predicted r2 | RMSE | Predicted RMSE |
|---|---|---|---|---|---|---|
| CM content (Ym1) | 0.00 ± 0.00 | 348.67 ± 6.08 | 0.9603 | 0.8673 | 1.14 | 49.11 |
| %LA of CM (Ym2) | 0.00 ± 0.00 | 112.02 ± 9.71 | 0.9480 | 0.7140 | 0.75 | 17.38 |
| Z-average (Ym5) | 144.5 ± 1.3 | 281.3 ± 4.4 | 0.9635 | 0.9120 | 8.00 | 12.00 |
| d50 (Ym6) | 152.3 ± 0.6 | 477.3 ± 25.2 | 0.9003 | 0.9891 | 0.97 | 26.30 |
| d90 (Ym7) | 238.7 ± 7.1 | 989.7 ± 151.5 | 0.8961 | 0.9992 | 1.80 | 88.24 |
| Dependent Variables | Acceptance Limit | Initial | 3 Months |
|---|---|---|---|
| CM content (µM) | ≥300 µM | 357.48 ± 8.39 | 358.84 ± 4.65 |
| %LA of CM (%LA) | 80–120 %LA | 92.74 ± 2.18 | 93.09 ± 1.21 |
| Z-average (nm) | ≤200 nm | 159.6 ± 1.7 | 166.6 ± 0.6 |
| d50 (nm) | ≤200 nm | 169.7 ± 2.1 | 177.0 ± 1.0 |
| d90 (nm) | ≤400 nm | 272.3 ± 9.1 | 276.7 ± 2.5 |
| Curcumin Concentration (µM) | % Cell Viability | ||||
|---|---|---|---|---|---|
| Free Curcumin | CTEV | CTEVHA | CTOP | CTOPHA | |
| 0.1 | 100.34 ± 2.31 | 96.70 ± 2.62 | 96.76 ± 2.62 | 90.49 ± 2.45 | 89.82 ± 2.43 |
| 1.0 | 88.80 ± 1.99 | 106.38 ± 2.88 | 102.40 ± 2.78 | 96.74 ± 2.62 | 93.34 ± 2.53 |
| 10.0 | 65.39 ± 1.36 | 83.92 ± 2.27 | 81.47 ± 2.21 | 92.63 ± 2.51 | 95.76 ± 2.60 |
| 25.0 | 58.61 ± 1.17 | 25.69 ± 0.70 | 21.19 ± 0.57 | 72.77 ± 1.97 | 84.49 ± 2.29 |
| 50.0 | 28.55 ± 0.36 | 21.78 ± 0.59 | 16.17 ± 0.44 | 10.95 ± 0.30 | 6.45 ± 0.17 |
| 100.0 | 15.28 ± 0.00 | 14.18 ± 0.38 | 13.92 ± 0.38 | 0.60 ± 0.02 | 0.43 ± 0.01 |
| Variables | Actual Variables | Coded Variables | |||
|---|---|---|---|---|---|
| Unit | Low | High | Low | High | |
| Mixture components | |||||
| 2.5 %w/w HE | %w/w | 0 | 2.6667 | 0 | 1 |
| 2.5 %w/w CF | %w/w | 0 | 2.6667 | 0 | 1 |
| 2.5 %w/w EA | %w/w | 0 | 2.6667 | 0 | 1 |
| Process variables | |||||
| External CM | %w/w | 0 | 0.0067 | −1 | 1 |
| 0.1 %w/w NaHA | %w/w | 0 | 3.3333 | −1 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Auychaipornlert, S.; Lawanprasert, P.P.; Piriyaprasarth, S.; Sithisarn, P.; Mangmool, S. Design of Turmeric Rhizome Extract Nano-Formula for Delivery to Cancer Cells. Molecules 2022, 27, 896. https://doi.org/10.3390/molecules27030896
Auychaipornlert S, Lawanprasert PP, Piriyaprasarth S, Sithisarn P, Mangmool S. Design of Turmeric Rhizome Extract Nano-Formula for Delivery to Cancer Cells. Molecules. 2022; 27(3):896. https://doi.org/10.3390/molecules27030896
Chicago/Turabian StyleAuychaipornlert, Sakchai, Pojawon Prayurnprohm Lawanprasert, Suchada Piriyaprasarth, Pongtip Sithisarn, and Supachoke Mangmool. 2022. "Design of Turmeric Rhizome Extract Nano-Formula for Delivery to Cancer Cells" Molecules 27, no. 3: 896. https://doi.org/10.3390/molecules27030896
APA StyleAuychaipornlert, S., Lawanprasert, P. P., Piriyaprasarth, S., Sithisarn, P., & Mangmool, S. (2022). Design of Turmeric Rhizome Extract Nano-Formula for Delivery to Cancer Cells. Molecules, 27(3), 896. https://doi.org/10.3390/molecules27030896






