Synthetic Diphenylacetylene-Based Retinoids Induce DNA Damage in Chinese Hamster Ovary Cells without Altering Viability
Abstract
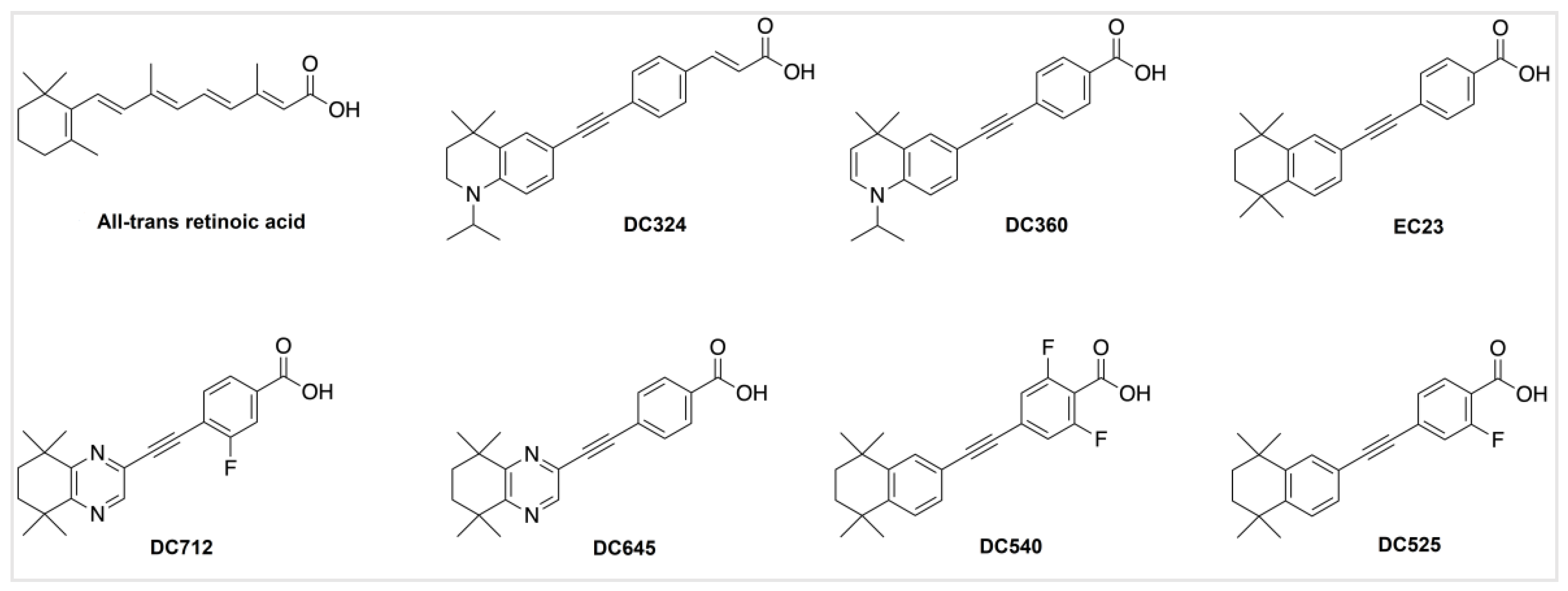
1. Introduction
2. Results
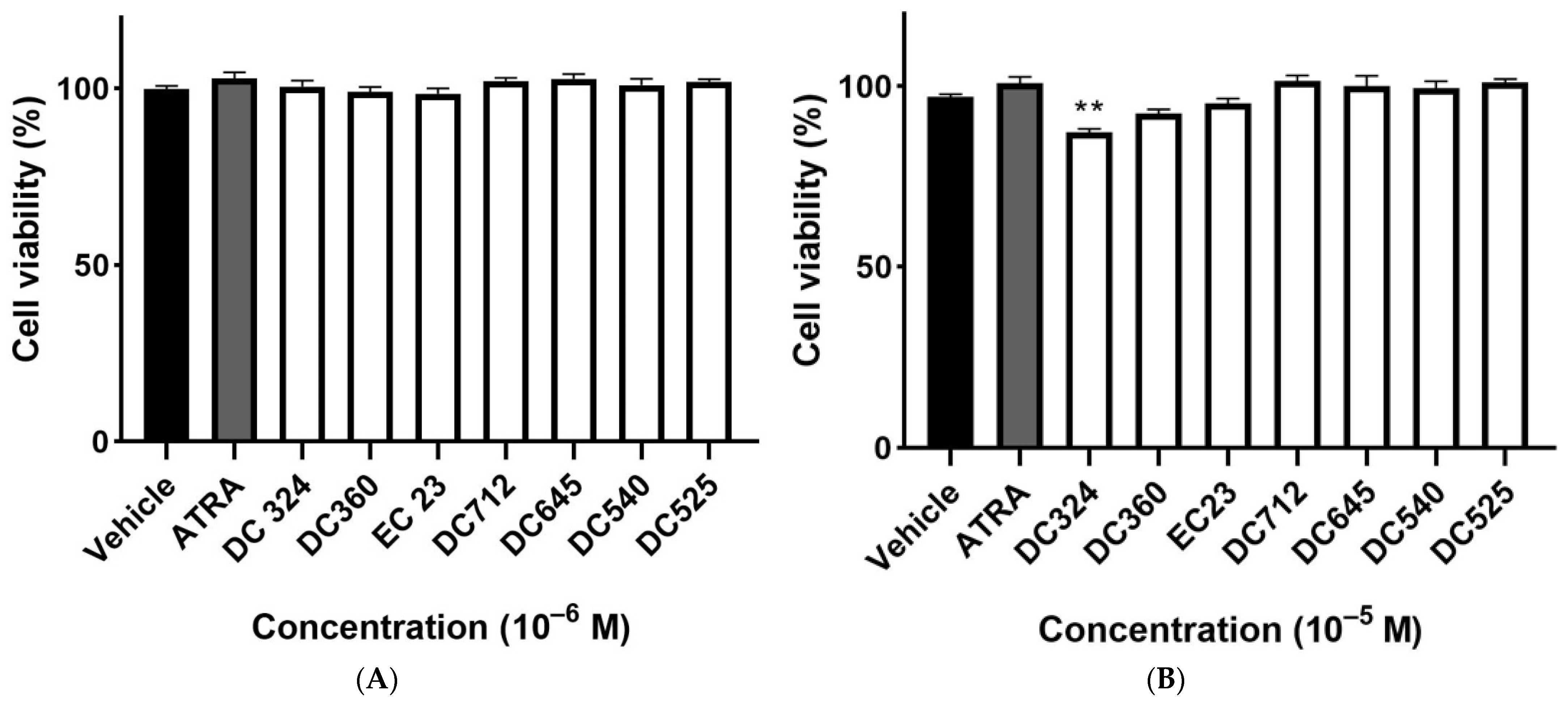
2.1. ATRA and Its Synthetic Derivatives Do Not Exert Cytotoxic Effects on CHO Cells
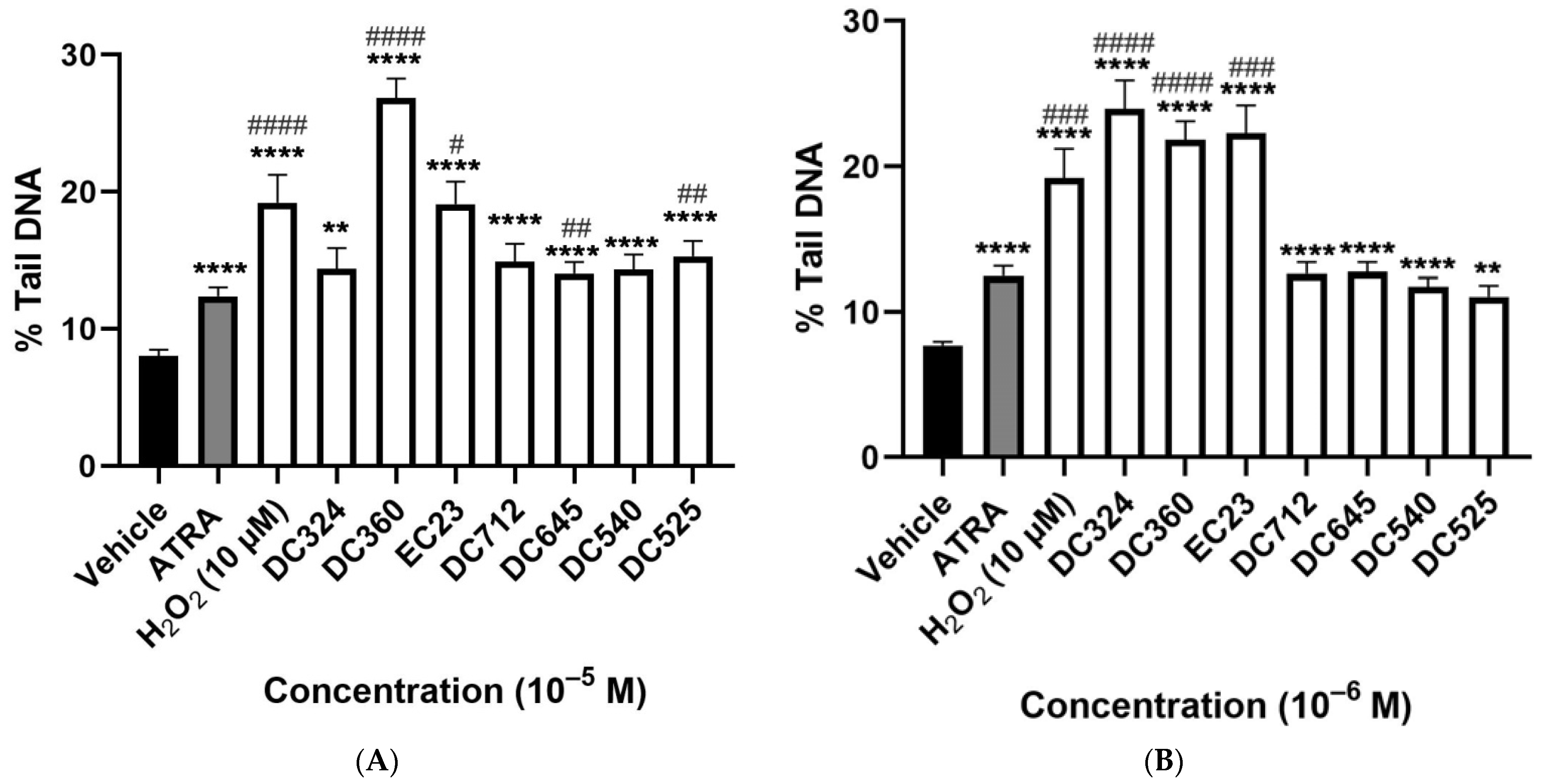
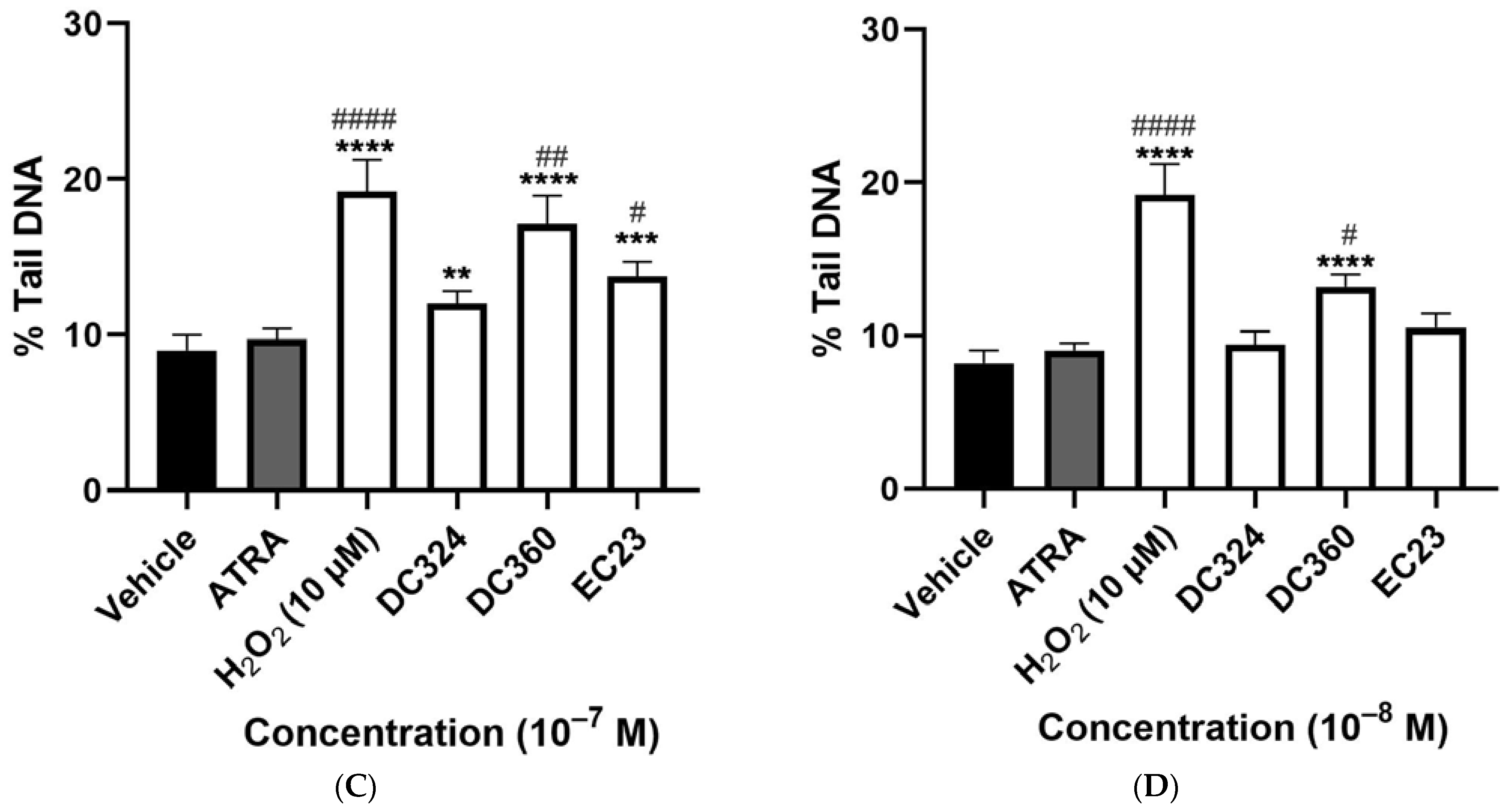
2.2. Synthetic Derivatives Induce Similar or Greater DNA Damage as Compared to ATRA
3. Discussion
4. Materials and Methods
4.1. Test Compounds
4.2. Cell Culture
4.3. Cell Viability ATP Assay
4.4. DNA Damage Comet Assay (Single Cell Gel Electrophoresis)
4.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Zasada, M.; Budzisz, E. Retinoids: Active molecules influencing skin structure formation in cosmetic and dermatological treatments. Adv. Dermatol. Allergol. 2019, 36, 392–397. [Google Scholar] [CrossRef] [PubMed]
- di Masi, A.; Leboffe, L.; De Marinis, E.; Pagano, F.; Cicconi, L.; Rochette-Egly, C.; Lo-Coco, F.; Ascenzi, P.; Nervi, C. Retinoic acid receptors: From molecular mechanisms to cancer therapy. Mol. Asp. Med. 2015, 41, 1–115. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, D.R.; Tomlinson, C.W.E.; Zhou, G.-L.; Holden, C.; Affleck, V.; Lamb, R.; Newling, K.; Ashton, P.; Valentine, R.; Redfern, C.; et al. Fluorescent Retinoic Acid Analogues as Probes for Biochemical and Intracellular Characterization of Retinoid Signaling Pathways. ACS Chem. Biol. 2019, 14, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Balaban, F.; Alagoz, Z.A.; Buyukbingol, E.; Iscan, M. Genotoxicity studies on benzimidazole retinoids. Die Pharm. 2005, 60, 861–868. [Google Scholar]
- Clemens, G.; Flower, K.R.; Gardner, P.; Henderson, A.P.; Knowles, J.P.; Marder, T.B.; Whiting, A.; Przyborski, S. Design and biological evaluation of synthetic retinoids: Probing length vs. stability vs. activity. Mol. BioSyst. 2013, 9, 3124–3134. [Google Scholar] [CrossRef] [PubMed]
- Christie, V.B.; Barnard, J.H.; Batsanov, A.S.; Bridgens, C.E.; Cartmell, E.B.; Collings, J.C.; Maltman, D.J.; Redfern, C.P.F.; Marder, T.B.; Przyborski, S.; et al. Synthesis and evaluation of synthetic retinoid derivatives as inducers of stem cell differentiation. Org. Biomol. Chem. 2008, 6, 3497–3507. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, C.W.E.; Chisholm, D.R.; Valentine, R.; Whiting, A.; Pohl, E. Novel Fluorescence Competition Assay for Retinoic Acid Binding Proteins. ACS Med. Chem. Lett. 2018, 9, 1297–1300. [Google Scholar] [CrossRef]
- Watanabe, T.; Pratt, R.M. Influence of retinoids on sister chromatid exchanges and chromosomes in cultured human embryonic palatal mesenchymal cells. Teratog. Carcinog. Mutagen. 1991, 11, 297–304. [Google Scholar] [CrossRef]
- Juhl, H.; Schürer, C.; Bartram, C.; Kohl, F.-V.; Melderis, H.; Wichert, P.; Rüdiger, H. Retinoids induce sister-chromatid exchanges in human diploid fibroblasts. Mutat. Res. Toxicol. 1978, 58, 317–320. [Google Scholar] [CrossRef]
- Tetzner, C.; Juhl, H.; Rüdiger, H. Sister-chromatid exchange induction by metabolically activated retinoids in human diploid fibroblast cultures. Mutat. Res. Toxicol. 1980, 79, 163–167. [Google Scholar] [CrossRef]
- Badr, F.M.; El-Habit, O.; Hamdy, M.; Hassan, G. The mutagenic versus protective role of vitamin A on the induction of chromosomal aberration in human lymphocyte cultures. Mutat. Res. Mol. Mech. Mutagen. 1998, 414, 157–163. [Google Scholar] [CrossRef]
- Dal-Pizzol, F.; Klamt, F.; Frota, M.L.; Moraes, L.F.; Moreira, J.C.F.; Benfato, M.S. Retinol supplementation induces DNA damage and modulates iron turnover in rat Sertoli cells. Free Radic. Res. 2000, 33, 677–687. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute (NCI). Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC Study) Population. Clinical Trial Registration NCT00342992. 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT00342992 (accessed on 21 March 2021).
- Lee, B.M.; Park, K.-K. Beneficial and adverse effects of chemopreventive agents. Mutat. Res. Mol. Mech. Mutagen. 2003, 523–524, 265–278. [Google Scholar] [CrossRef]
- Alakhras, R.S.; Stephanou, G.; Demopoulos, N.A.; Nikolaropoulos, S.S. Genotoxicity of all-trans retinoic acid (ATRA) and its steroidal analogue EA-4 in human lymphocytes and mouse cells in vitro. Cancer Lett. 2011, 306, 15–26. [Google Scholar] [CrossRef]
- Haffez, H.; Chisholm, D.R.; Valentine, R.; Pohl, E.; Redfern, C.; Whiting, A. The molecular basis of the interactions between synthetic retinoic acid analogues and the retinoic acid receptors. MedChemComm 2017, 8, 578–592. [Google Scholar] [CrossRef]
- Khatib, T.; Chisholm, D.R.; Whiting, A.; Platt, B.; McCaffery, P. Decay in Retinoic Acid Signaling in Varied Models of Alzheimer’s Disease and In-Vitro Test of Novel Retinoic Acid Receptor Ligands (RAR-Ms) to Regulate Protective Genes. J. Alzheimers Dis. 2020, 73, 935–954. [Google Scholar] [CrossRef]
- Haffez, H.; Khatib, T.; McCaffery, P.; Przyborski, S.; Redfern, C.; Whiting, A. Neurogenesis in Response to Synthetic Retinoids at Different Temporal Scales. Mol. Neurobiol. 2017, 55, 1942–1950. [Google Scholar] [CrossRef]
- Khatib, T.; Marini, P.; Nunna, S.; Chisholm, D.R.; Whiting, A.; Redfern, C.; Greig, I.R.; McCaffery, P. Genomic and non-genomic pathways are both crucial for peak induction of neurite outgrowth by retinoids. Cell Commun. Signal. 2019, 17, 40. [Google Scholar] [CrossRef]
- Clark, J.N.; Whiting, A.; McCaffery, P. Retinoic acid receptor-targeted drugs in neurodegenerative disease. Expert Opin. Drug Metab. Toxicol. 2020, 16, 1097–1108. [Google Scholar] [CrossRef]
- Ribeiro, D.A.; Scolastici, C.; de Lima, P.L.A.; Marques, M.E.A.; Salvadori, D.M.F. Genotoxicity of antimicrobial endodontic compounds by single cell gel (comet) assay in Chinese hamster ovary (CHO) cells. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2005, 99, 637–640. [Google Scholar] [CrossRef]
- Bajpayee, M.; Kumar, A.; Dhawan, A. The Comet Assay: Assessment of In Vitro and In Vivo DNA Damage. In Genotoxicity Assessment: Methods and Protocols; Dhawan, A., Bajpayee, M., Eds.; Springer: New York, NY, USA, 2019; pp. 237–257. [Google Scholar] [CrossRef]
- Atha, D.H.; Coskun, E.; Erdem, O.; Tona, A.; Reipa, V.; Nelson, B.C. Genotoxic Effects of Etoposide, Bleomycin, and Ethyl Methanesulfonate on Cultured CHO Cells: Analysis by GC-MS/MS and Comet Assay. J. Nucleic Acids 2020, 2020, e8810105. [Google Scholar] [CrossRef] [PubMed]
- Niles, R.M. Biomarker and animal models for assessment of retinoid efficacy in cancer chemoprevention. Acta Pharmacol. Sin. 2007, 28, 1383–1391. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hunsu, V.; Facey, C.; Fields, J.; Boman, B. Retinoids as Chemo-Preventive and Molecular-Targeted Anti-Cancer Therapies. Int. J. Mol. Sci. 2021, 22, 7731. [Google Scholar] [CrossRef] [PubMed]
- Penniston, K.L.; Tanumihardjo, S.A. The acute and chronic toxic effects of vitamin A. Am. J. Clin. Nutr. 2006, 83, 191–201. [Google Scholar] [CrossRef]
- Shih, M.Y.S.; Kane, M.A.; Zhou, P.; Yen, C.L.E.; Streeper, R.S.; Napoli, J.L.; Farese, R.V.; Shih, M.Y.S.; Kane, M.A.; Zhou, P.; et al. Retinol Esterification by DGAT1 Is Essential for Retinoid Homeostasis in Murine Skin. J. Biol. Chem. 2009, 284, 4292–4299. [Google Scholar] [CrossRef]
- Wood, C. In-Process Control Testing. In Separation Science and Technology; Ahuja, S., Scypinski, S., Eds.; Academic Press: Cambridge, MA, USA, 2011; Volume 10, pp. 397–427. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef]
- Basu, A.K. DNA Damage, Mutagenesis and Cancer. Int. J. Mol. Sci. 2018, 19, 970. [Google Scholar] [CrossRef]
- Huang, R.; Zhou, P.-K. DNA damage repair: Historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 254. [Google Scholar] [CrossRef]
- Chakarov, S.; Petkova, R.; Russev, G.C.; Zhelev, N. DNA damage and mutation. Types of DNA damage. BioDiscovery 2014, 11, e8957. [Google Scholar] [CrossRef]
- Surova, O.; Zhivotovsky, B. Various modes of cell death induced by DNA damage. Oncogene 2012, 32, 3789–3797. [Google Scholar] [CrossRef]
- Ohsaki, E.; Ueda, K. Interplay Between KSHV and the Host DNA Damage Response. Front. Cell. Infect. Microbiol. 2020, 10, 604351. [Google Scholar] [CrossRef] [PubMed]
- De Zio, D.; Cianfanelli, V.; Cecconi, F. New Insights into the Link Between DNA Damage and Apoptosis. Antioxidants Redox Signal. 2013, 19, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Tice, R.R.; Agurell, E.; Anderson, D.; Burlinson, B.; Hartmann, A.; Kobayashi, H.; Miyamae, Y.; Rojas, E.; Ryu, J.C.; Sasaki, Y.F. Single cell gel/comet assay: Guidelines for in vitro and in vivo genetic toxicology testing. Environ. Mol. Mutagen. 2020, 35, 206–221. [Google Scholar] [CrossRef]
- Burlinson, B. The In Vitro and In Vivo Comet Assays. In Genetic Toxicology; Parry, J.M., Parry, E.M., Eds.; Springer: New York, NY, USA, 2012; Volume 817, pp. 143–163. [Google Scholar] [CrossRef]
- Azqueta, A.; Collins, A. The essential comet assay: A comprehensive guide to measuring DNA damage and repair. Arch. Toxicol. 2013, 87, 949–968. [Google Scholar] [CrossRef] [PubMed]
- Arce, F.; Gätjens-Boniche, O.; Vargas, E.; Valverde, B.; Díaz, C. Apoptotic events induced by naturally occurring retinoids ATRA and 13-cis retinoic acid on human hepatoma cell lines Hep3B and HepG2. Cancer Lett. 2005, 229, 271–281. [Google Scholar] [CrossRef]
- Souček, K.; Pacherník, J.; Kubala, L.; Vondráček, J.; Hofmanová, J.; Kozubík, A. Transforming growth factor-β1 inhibits all-trans retinoic acid-induced apoptosis. Leuk. Res. 2006, 30, 607–623. [Google Scholar] [CrossRef]
- Alakhras, R.S.; Stephanou, G.; Demopoulos, N.A.; Grintzalis, K.; Georgiou, C.D.; Nikolaropoulos, S.S. DNA fragmentation induced by all-transretinoic acid and its steroidal analogue EA-4 in C2C12mouse and HL-60 human leukemic cellsin vitro. J. Appl. Toxicol. 2013, 34, 885–892. [Google Scholar] [CrossRef]
- Tokarz, P.; Piastowska-Ciesielska, A.W.; Kaarniranta, K.; Blasiak, J. All-Trans Retinoic Acid Modulates DNA Damage Response and the Expression of the VEGF-A and MKI67 Genes in ARPE-19 Cells Subjected to Oxidative Stress. Int. J. Mol. Sci. 2016, 17, 898. [Google Scholar] [CrossRef]
- Polyakov, N.E.; Leshina, T.V.; Konovalova, T.A.; Kispert, L.D. Carotenoids as scavengers of free radicals in a fenton reaction: Antioxidants or pro-oxidants? Free Radic. Biol. Med. 2001, 31, 398–404. [Google Scholar] [CrossRef]
- Oloris, S.C.S.; Dagli, M.L.Z.; Guerra, J.L. Effect of β-carotene on the development of the solid Ehrlich tumor in mice. Life Sci. 2002, 71, 717–724. [Google Scholar] [CrossRef]
- Murata, M.; Kawanishi, S. Oxidative DNA Damage by Vitamin A and Its Derivative via Superoxide Generation. J. Biol. Chem. 2000, 275, 2003–2008. [Google Scholar] [CrossRef] [PubMed]
- Mayne, S.T.; Graham, S.; Zheng, T. Dietary retinol: Prevention or promotion of carcinogenesis in humans? Cancer Causes Control 1991, 2, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Dal-Pizzol, F.; Klamt, F.; Benfato, M.S.; Bernard, E.A.; Moreira, J.C.F. Retinol supplementation induces oxidative stress and modulates antioxidant enzyme activities in rat sertoli cells. Free Radic. Res. 2001, 34, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Gutteridge, J.M.C. Oxidative stress and redox regulation: Adaptation, damage, repair, senescence, and death. In Free Radicals in Biology and Medicine, 5th ed.; Oxford University Press: Oxford, UK, 2015; Chapter 5; pp. 199–283. [Google Scholar]
- Moreira, J.; Dal-Pizzol, F.; Rocha, A.; Klamt, F.; Ribeiro, N.; Ferreira, C.; Bernard, E. Retinol-induced changes in the phosphorylation levels of histones and high mobility group proteins from Sertoli cells. Braz. J. Med. Biol. Res. 2000, 33, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Klamt, F.; Pizzol, F.D.; Roehrs, R.; de Oliveira, R.B.; Dalmolin, R.; Henriques, J.A.; de Andrades, H.H.R.; Ramos, A.L.L.D.P.; Saffi, J.; Moreira, J.C.F. Genotoxicity, recombinogenicity and cellular preneoplasic transformation induced by Vitamin a supplementation. Mutat. Res. Toxicol. Environ. Mutagen. 2003, 539, 117–125. [Google Scholar] [CrossRef]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Duellman, S.; Benink, H.A.; Worzella, T.J.; Minor, L. Cell Viability Assays. In Assay Guidance Manual; Markossian, S., Grossman, A., Brimacombe, K., Arkin, M., Auld, D., Austin, C.P., Baell, J., Chung, T.D.Y., Coussens, N.P., Dahlin, J.L., et al., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004. Available online: http://www.ncbi.nlm.nih.gov/books/NBK144065/ (accessed on 6 January 2022).
- Oancea, M.; Mazumder, S.; Crosby, M.E.; Almasan, A.; Qing, W. Apoptosis assays. Methods Mol. Med. 2006, 129, 279–290. [Google Scholar] [CrossRef]
- Lin, H.; Mei, N.; Manjanatha, M.G. In vitro comet assay for testing genotoxicity of chemicals. In Optimization in Drug Discovery, Methods in Pharmacology and Toxicology; Caldwell, G., Yan, Z., Eds.; Humana Press: Totowa, NJ, USA, 2014; Chapter 31; pp. 517–536. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, Y.; Yang, C. Evaluating In Vitro DNA Damage Using Comet Assay. J. Vis. Exp. 2017, 128, 56450. [Google Scholar] [CrossRef]
- Chisholm, D.R.; Zhou, G.-L.; Pohl, E.; Valentine, R.; Whiting, A. Practical synthetic strategies towards lipophilic 6-iodotetrahydroquinolines and -dihydroquinolines. Beilstein J. Org. Chem. 2016, 12, 1851–1862. [Google Scholar] [CrossRef]
- Whiting, A.; Chisholm, D.; Greig, I.; Khatib, T.; Mccaffery, P. Synthetic Retinoids for Use in Rar Activation. WO2020183173A2 17 September 2020. Available online: https://patents.google.com/patent/WO2020183173A2/en?oq=WO2020183173A2 (accessed on 1 September 2021).
- Collins, A.R. The Comet Assay for DNA Damage and Repair: Principles, Applications, and Limitations. Mol. Biotechnol. 2004, 26, 249. [Google Scholar] [CrossRef]
- Shaposhnikov, S.A.; Salenko, V.B.; Brunborg, G.; Nygren, J.; Collins, A.R. Single-cell gel electrophoresis (the comet assay): Loops or fragments? Electrophoresis 2008, 29, 3005–3012. [Google Scholar] [CrossRef]
- Collins, A.R.; Oscoz, A.A.; Brunborg, G.; Gaivao, I.; Giovannelli, L.; Kruszewski, M.; Smith, C.C.; Štětina, R. The comet assay: Topical issues. Mutagenesis 2008, 23, 143–151. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hudhud, L.; Chisholm, D.R.; Whiting, A.; Steib, A.; Pohóczky, K.; Kecskés, A.; Szőke, É.; Helyes, Z. Synthetic Diphenylacetylene-Based Retinoids Induce DNA Damage in Chinese Hamster Ovary Cells without Altering Viability. Molecules 2022, 27, 977. https://doi.org/10.3390/molecules27030977
Hudhud L, Chisholm DR, Whiting A, Steib A, Pohóczky K, Kecskés A, Szőke É, Helyes Z. Synthetic Diphenylacetylene-Based Retinoids Induce DNA Damage in Chinese Hamster Ovary Cells without Altering Viability. Molecules. 2022; 27(3):977. https://doi.org/10.3390/molecules27030977
Chicago/Turabian StyleHudhud, Lina, David R. Chisholm, Andrew Whiting, Anita Steib, Krisztina Pohóczky, Angéla Kecskés, Éva Szőke, and Zsuzsanna Helyes. 2022. "Synthetic Diphenylacetylene-Based Retinoids Induce DNA Damage in Chinese Hamster Ovary Cells without Altering Viability" Molecules 27, no. 3: 977. https://doi.org/10.3390/molecules27030977
APA StyleHudhud, L., Chisholm, D. R., Whiting, A., Steib, A., Pohóczky, K., Kecskés, A., Szőke, É., & Helyes, Z. (2022). Synthetic Diphenylacetylene-Based Retinoids Induce DNA Damage in Chinese Hamster Ovary Cells without Altering Viability. Molecules, 27(3), 977. https://doi.org/10.3390/molecules27030977






