Selenocystine-Derived Label-Free Fluorescent Schiff Base Nanocomplex for siRNA Delivery Synergistically Kills Cancer Cells
Abstract
:1. Introduction
2. Results
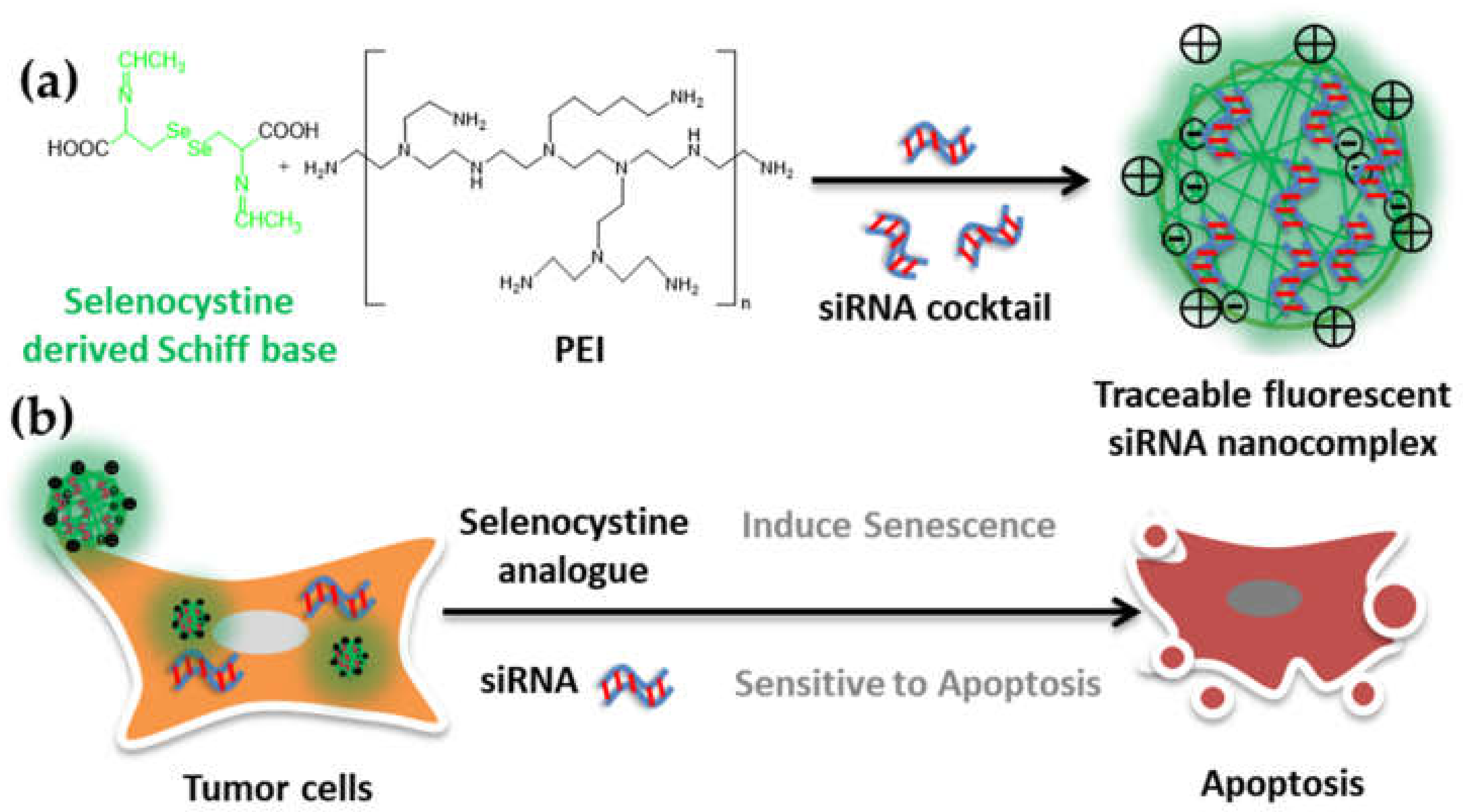
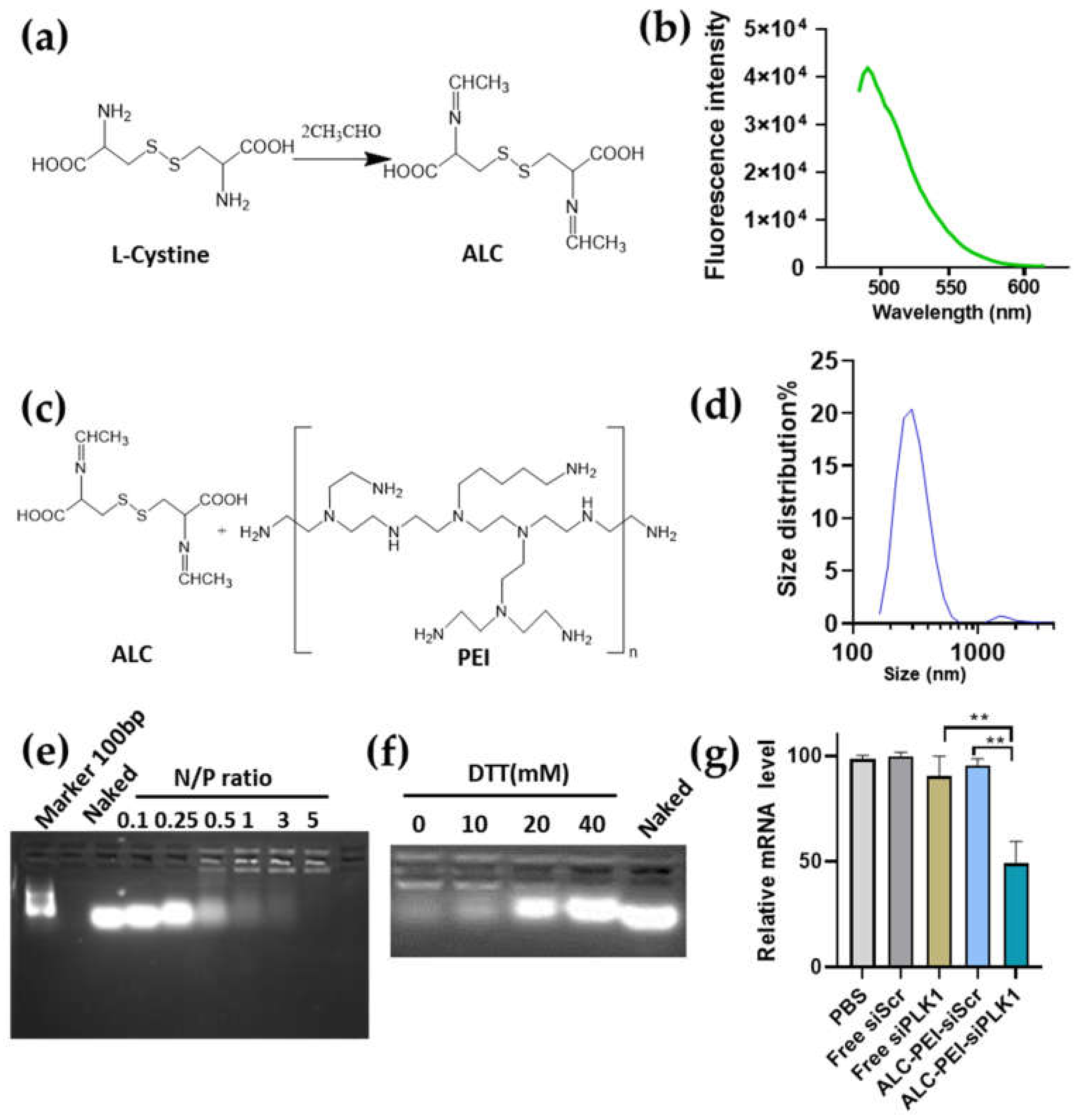
2.1. Developing Schiff Base Fluorescence Vector for siRNA
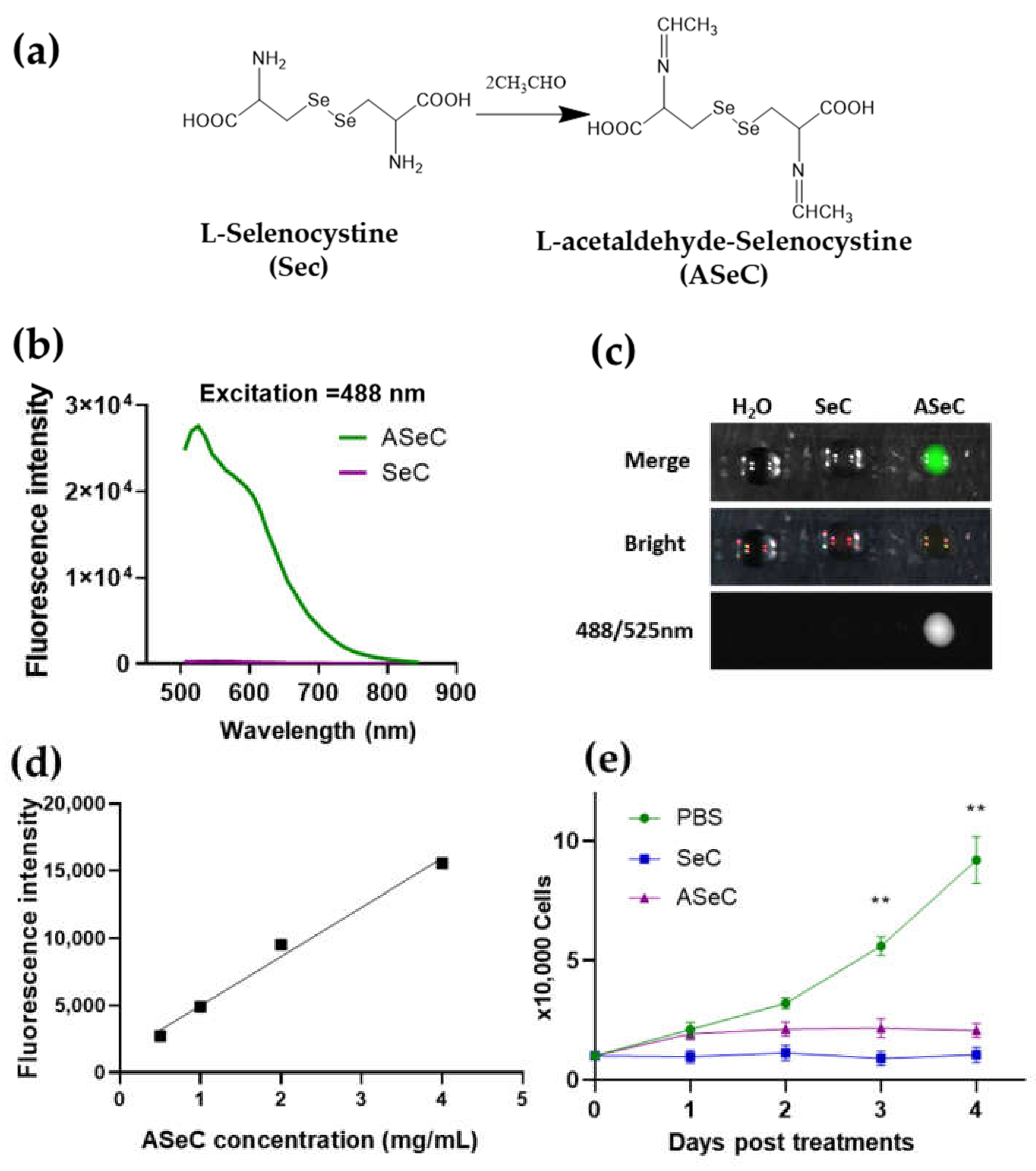
2.2. Selenocystine-Derived Schiff Base Exhibits a Fluorescent Activity
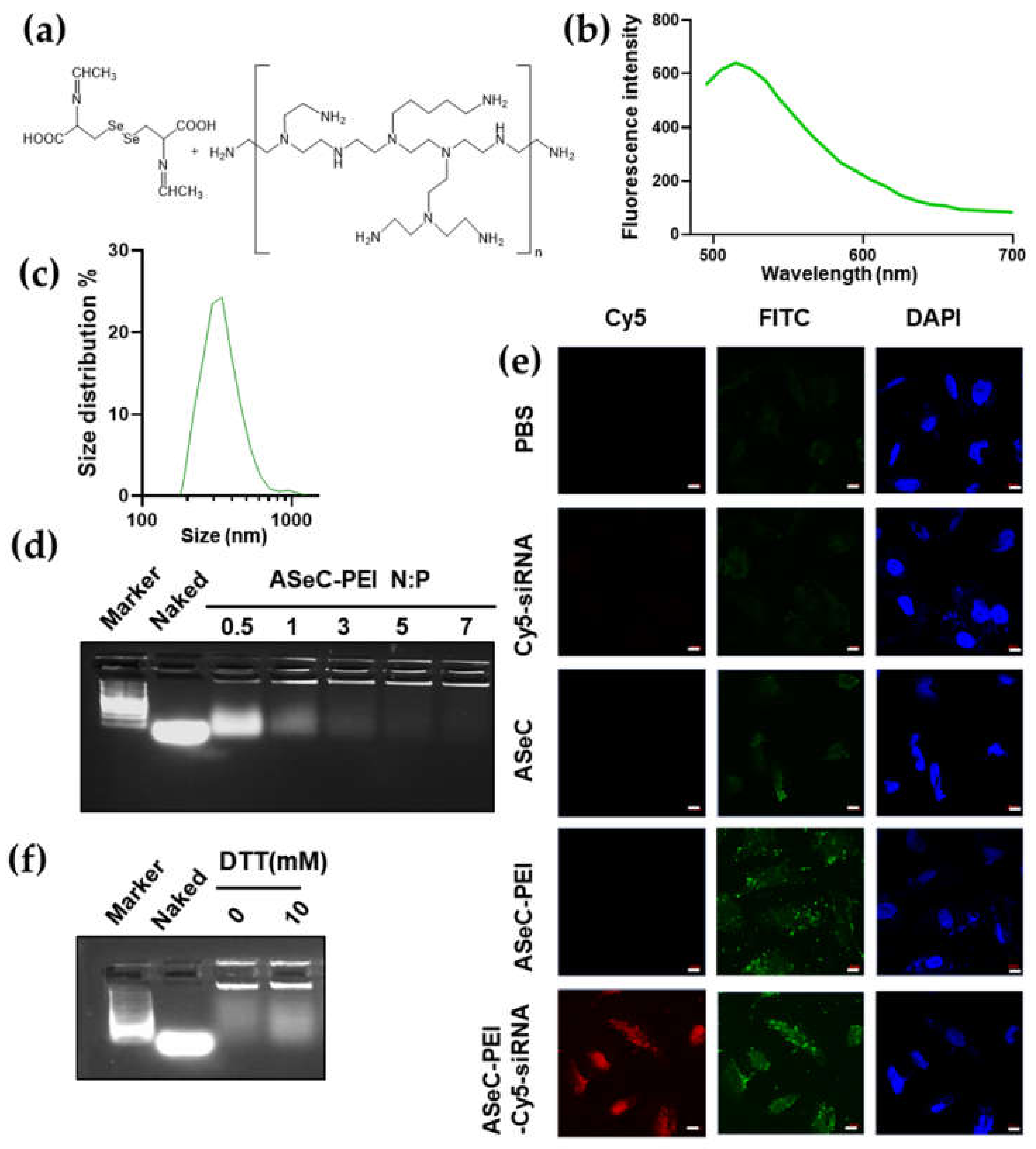
2.3. siRNA Delivery Vector Development by Using Selenocystine-Derived Schiff Base
2.4. siRNA-Chemo Synergic Killing of Tumor Cells
3. Materials and Methods
3.1. Chemicals
3.2. Synthesis Procedures
3.2.1. Synthesis of Acetaldehyde L-Cystine (ALC) and Acetaldehyde L-Selenocystine (ASeC)
3.2.2. Synthesis of ALC-PEI and ASeC-PEI
3.3. Determination of Maximal Emission and Excitation for Compounds, and the Fluorescence Dot Assay
3.4. siRNA Upload by ALC/ASeC-PEI
3.5. Gel Retardation Assay
3.6. Cell Survival Assay
3.7. Confocal Imaging
3.8. qPCR Assay
3.9. Western Blot Assay
3.10. Statics Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Wu, H.; Zhu, H.; Li, X.; Liu, Z.; Zheng, W.; Chen, T.; Yu, B.; Wong, K.-H. Induction of Apoptosis and Cell Cycle Arrest in A549 Human Lung Adenocarcinoma Cells by Surface-Capping Selenium Nanoparticles: An Effect Enhanced by Polysaccharide–Protein Complexes from Polyporus rhinocerus. J. Agric. Food Chem. 2013, 61, 9859–9866. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Zheng, W.; Fu, X.; Li, X.; Wong, Y.-S.; Chen, T. Strategy to enhance the therapeutic effect of doxorubicin in human hepatocellular carcinoma by selenocystine, a synergistic agent that regulates the ROS-mediated signaling. Oncotarget 2014, 5, 2853–2863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Wong, Y.-S. Selenocystine Induces S-Phase Arrest and Apoptosis in Human Breast Adenocarcinoma MCF-7 Cells by Modulating ERK and Akt Phosphorylation. J. Agric. Food Chem. 2008, 56, 10574–10581. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Pérez, M.; Ali, W.; Marć, M.A.; Handzlik, J.; Domínguez-Álvarez, E. Selenides and Diselenides: A Review of Their Anticancer and Chemopreventive Activity. Molecules 2018, 23, 628. [Google Scholar] [CrossRef] [Green Version]
- Radomska, D.; Czarnomysy, R.; Radomski, D.; Bielawski, K. Selenium Compounds as Novel Potential Anticancer Agents. Int. J. Mol. Sci. 2021, 22, 1009. [Google Scholar] [CrossRef]
- Misra, S.; Boylan, M.; Selvam, A.; Spallholz, J.E.; Björnstedt, M. Redox-Active Selenium Compounds—From Toxicity and Cell Death to Cancer Treatment. Nutr. 2015, 7, 3536–3556. [Google Scholar] [CrossRef] [Green Version]
- Sanmartín, C.; Plano, D.; Sharma, A.K.; Palop, J.A. Selenium Compounds, Apoptosis and Other Types of Cell Death: An Overview for Cancer Therapy. Int. J. Mol. Sci. 2012, 13, 9649–9672. [Google Scholar] [CrossRef]
- Pons, D.G.; Moran, C.; Alorda-Clara, M.; Oliver, J.; Roca, P.; Sastre-Serra, J. Micronutrients Selenomethionine and Selenocysteine Modulate the Redox Status of MCF-7 Breast Cancer Cells. Nutr. 2020, 12, 865. [Google Scholar] [CrossRef] [Green Version]
- Kuršvietienė, L.; Mongirdienė, A.; Bernatonienė, J.; Šulinskienė, J.; Stanevičienė, I. Selenium anticancer properties and impact on cellular redox status. Antioxidants 2020, 9, 80. [Google Scholar] [CrossRef] [Green Version]
- Misra, S.; Björnstedt, M. Selenocystine and Cancer. In Selenium; Michalke, B., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 271–286. [Google Scholar]
- Liu, C.; Lai, H.; Chen, T. Boosting Natural Killer Cell-Based Cancer Immunotherapy with Selenocystine/Transforming Growth Factor-Beta Inhibitor-Encapsulated Nanoemulsion. ACS Nano 2020, 14, 11067–11082. [Google Scholar] [CrossRef]
- Hatem, E.; El Banna, N.; Huang, M.E. Multifaceted Roles of Glutathione and Glutathione-Based Systems in Carcinogenesis and Anticancer Drug Resistance. Antioxid. Redox Signal. 2017, 27, 1217–1234. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, C.; Wei, T.; Gayet, O.; Loncle, C.; Borge, L.; Dusetti, N.; Ma, X.; Marson, D.; Laurini, E.; et al. Dendrimeric nanosystem consistently circumvents heterogeneous drug response and resistance in pancreatic cancer. Explor. 2021, 1, 21–34. [Google Scholar] [CrossRef]
- Guo, S.; Li, K.; Hu, B.; Li, C.; Zhang, M.; Hussain, A.; Wang, X.; Cheng, Q.; Yang, F.; Ge, K.; et al. Membrane-destabilizing ionizable lipid empowered imaging-guided siRNA delivery and cancer treatment. Explor. 2021, 1, 35–49. [Google Scholar] [CrossRef]
- Huang, H.; Dong, C.; Chang, M.; Ding, L.; Chen, L.; Feng, W.; Chen, Y. Mitochondria-specific nanocatalysts for chemotherapy-augmented sequential chemoreactive tumor therapy. Explor. 2021, 1, 50–60. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Z.; Pan, Z.; Liu, Y. Advanced bioactive nanomaterials for biomedical applications. Explor. 2021, 1, 20210089. [Google Scholar] [CrossRef]
- Saw, P.E.; Song, E.W. siRNA therapeutics: A clinical reality. Sci. China. Life Sci. 2020, 63, 485–500. [Google Scholar] [CrossRef]
- Singh, A.; Trivedi, P.; Jain, N.K. Advances in siRNA delivery in cancer therapy. Artif. Cells Nanomed. Biotechnol. 2018, 46, 274–283. [Google Scholar] [CrossRef]
- An, H.-W.; Mamuti, M.; Wang, X.; Yao, H.; Wang, M.-D.; Zhao, L.; Li, L.-L. Rationally designed modular drug delivery platform based on intracellular peptide self-assembly. Exploration 2021, 1, 20210153. [Google Scholar] [CrossRef]
- Xie, A.; Hanif, S.; Ouyang, J.; Tang, Z.; Kong, N.; Kim, N.Y.; Qi, B.; Patel, D.; Shi, B.; Tao, W. Stimuli-responsive prodrug-based cancer nanomedicine. EBioMedicine 2020, 56, 102821. [Google Scholar] [CrossRef]
- Jiang, T.; Qiao, Y.; Ruan, W.; Zhang, D.; Yang, Q.; Wang, G.; Chen, Q.; Zhu, F.; Yin, J.; Zou, Y.; et al. Cation-Free siRNA Micelles as Effective Drug Delivery Platform and Potent RNAi Nanomedicines for Glioblastoma Therapy. Adv. Mater. 2021, 33, 2104779. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, H.; Wei, D.; Zhang, X.; Wang, J.; Wu, X.; Chang, J. Mitochondria-targeted nanoparticles in treatment of neurodegenerative diseases. Explor. 2021, 1, 20210115. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, F.; Liu, Y.; Zheng, M.; Wang, Y.; Zhang, D.; Anraku, Y.; Zou, Y.; Li, J.; Wu, H.; et al. Blood-brain barrier penetrating siRNA nanomedicine for Alzheimer’s disease therapy. Sci. Adv. 2020, 6, eabc7031. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Tao, W.; Zou, Y.; Farokhzad, O.C.; Shi, B. Nanotechnology-Based Strategies for siRNA Brain Delivery for Disease Therapy. Trends Biotechnol. 2018, 36, 562–575. [Google Scholar] [CrossRef] [PubMed]
- Wahane, A.; Waghmode, A.; Kapphahn, A.; Dhuri, K.; Gupta, A.; Bahal, R. Role of Lipid-Based and Polymer-Based Non-Viral Vectors in Nucleic Acid Delivery for Next-Generation Gene Therapy. Molecules 2020, 25, 2866. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, S.; Wang, B.; Cui, S.; Yan, J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release Off. J. Control. Release Soc. 2006, 114, 100–109. [Google Scholar] [CrossRef]
- Zheng, M.; Jiang, T.; Yang, W.; Zou, Y.; Wu, H.; Liu, X.; Zhu, F.; Qian, R.; Ling, D.; McDonald, K.; et al. The siRNAsome: A Cation-Free and Versatile Nanostructure for siRNA and Drug Co-delivery. Angew. Chem. (Int. Ed. Engl.) 2019, 58, 4938–4942. [Google Scholar] [CrossRef]
- Saw, P.E.; Yao, H.; Lin, C.; Tao, W.; Farokhzad, O.C.; Xu, X. Stimuli-Responsive Polymer-Prodrug Hybrid Nanoplatform for Multistage siRNA Delivery and Combination Cancer Therapy. Nano Lett. 2019, 19, 5967–5974. [Google Scholar] [CrossRef]
- Tu, L.; Liao, Z.; Luo, Z.; Wu, Y.-L.; Herrmann, A.; Huo, S. Ultrasound-controlled drug release and drug activation for cancer therapy. Exploration 2021, 1, 20210023. [Google Scholar] [CrossRef]
- Zheng, M.; Liu, Y.; Wang, Y.; Zhang, D.; Zou, Y.; Ruan, W.; Yin, J.; Tao, W.; Park, J.B.; Shi, B. ROS-Responsive Polymeric siRNA Nanomedicine Stabilized by Triple Interactions for the Robust Glioblastoma Combinational RNAi Therapy. Adv. Mater. 2019, 31, 1903277. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, D.; Zou, Y.; Wang, Z.; Hao, M.; Zheng, M.; Xue, X.; Pan, X.; Lu, Y.; Wang, J.; et al. Developing a pH-sensitive Al(OH)3 layer-mediated UCNP@Al(OH)3/Au nanohybrid for photothermal therapy and fluorescence imaging in vivo. J. Mater. Chem. B 2018, 6, 7862–7870. [Google Scholar] [CrossRef]
- Li, L.; Zou, J.; Dai, Y.; Fan, W.; Niu, G.; Yang, Z.; Chen, X. Burst release of encapsulated annexin A5 in tumors boosts cytotoxic T-cell responses by blocking the phagocytosis of apoptotic cells. Nat. Biomed. Eng. 2020, 4, 1102–1116. [Google Scholar] [CrossRef] [PubMed]
- Luby, B.M.; Charron, D.M.; MacLaughlin, C.M.; Zheng, G. Activatable fluorescence: From small molecule to nanoparticle. Adv. Drug Deliv. Rev. 2017, 113, 97–121. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, Y.; Hsu, S.-h. Hydrogels Based on Schiff Base Linkages for Biomedical Applications. Molecules 2019, 24, 3005. [Google Scholar] [CrossRef] [Green Version]
- Troschke, E.; Oschatz, M.; Ilic, I.K. Schiff-bases for sustainable battery and supercapacitor electrodes. Explor. 2021, 1, 20210128. [Google Scholar] [CrossRef]
- Shi, B.; Zhang, H.; Qiao, S.Z.; Bi, J.; Dai, S. Intracellular microenvironment-responsive label-free autofluorescent nanogels for traceable gene delivery. Adv. Healthc. Mater. 2014, 3, 1839–1848. [Google Scholar] [CrossRef]
- Shi, B.; Du, X.; Chen, J.; Fu, L.; Morsch, M.; Lee, A.; Liu, Y.; Cole, N.; Chung, R. Multifunctional Hybrid Nanoparticles for Traceable Drug Delivery and Intracellular Microenvironment-Controlled Multistage Drug-Release in Neurons. Small 2017, 13, 1603966. [Google Scholar] [CrossRef]
- Long, M.; Wu, J.; Hao, J.; Liu, W.; Tang, Y.; Li, X.; Su, H.; Qiu, W. Selenocystine-induced cell apoptosis and S-phase arrest inhibit human triple-negative breast cancer cell proliferation. Vitr. Cell. Dev. Biol.—Anim. 2015, 51, 1077–1084. [Google Scholar] [CrossRef]
- Su, J.; Lai, H.; Chen, J.; Li, L.; Wong, Y.-S.; Chen, T.; Li, X. Natural Borneol, a Monoterpenoid Compound, Potentiates Selenocystine-Induced Apoptosis in Human Hepatocellular Carcinoma Cells by Enhancement of Cellular Uptake and Activation of ROS-Mediated DNA Damage. PLoS ONE 2013, 8, e63502. [Google Scholar] [CrossRef] [PubMed]
- Hoefig, C.S.; Renko, K.; Köhrle, J.; Birringer, M.; Schomburg, L. Comparison of different selenocompounds with respect to nutritional value vs. toxicity using liver cells in culture. J. Nutr. Biochem. 2011, 22, 945–955. [Google Scholar] [CrossRef]
- Chen, T.; Wong, Y.-S. Selenocystine induces reactive oxygen species–mediated apoptosis in human cancer cells. Biomed. Pharmacother. 2009, 63, 105–113. [Google Scholar] [CrossRef]
- Yao, Y.; Zhang, H.; Wang, Z.; Ding, J.; Wang, S.; Huang, B.; Ke, S.; Gao, C. Reactive oxygen species (ROS)-responsive biomaterials mediate tissue microenvironments and tissue regeneration. J. Mater. Chem. B 2019, 7, 5019–5037. [Google Scholar] [CrossRef] [PubMed]
- Vasileiou, P.V.S.; Evangelou, K.; Vlasis, K.; Fildisis, G.; Panayiotidis, M.I.; Chronopoulos, E.; Passias, P.-G.; Kouloukoussa, M.; Gorgoulis, V.G.; Havaki, S. Mitochondrial Homeostasis and Cellular Senescence. Cells 2019, 8, 686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasanna, P.G.; Citrin, D.E.; Hildesheim, J.; Ahmed, M.M.; Venkatachalam, S.; Riscuta, G.; Xi, D.; Zheng, G.; Deursen, J.V.; Goronzy, J.; et al. Therapy-Induced Senescence: Opportunities to Improve Anticancer Therapy. JNCI: J. Natl. Cancer Inst. 2021, 113, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Mongiardi, M.P.; Pellegrini, M.; Pallini, R.; Levi, A.; Falchetti, M.L. Cancer Response to Therapy-Induced Senescence: A Matter of Dose and Timing. Cancers 2021, 13, 484. [Google Scholar] [CrossRef] [PubMed]
- Faget, D.V.; Ren, Q.; Stewart, S.A. Unmasking senescence: Context-dependent effects of SASP in cancer. Nat. Rev. Cancer 2019, 19, 439–453. [Google Scholar] [CrossRef]
- Brattinga, B.; van Leeuwen, B.L. Senescent Cells: A Potential Target for New Cancer Therapies in Older Oncologic Patients. Cancers 2021, 13, 278. [Google Scholar] [CrossRef]
- Triana-Martínez, F.; Loza, M.I.; Domínguez, E. Beyond Tumor Suppression: Senescence in Cancer Stemness and Tumor Dormancy. Cells 2020, 9, 346. [Google Scholar] [CrossRef] [Green Version]
- Saleh, T.; Bloukh, S.; Carpenter, V.J.; Alwohoush, E.; Bakeer, J.; Darwish, S.; Azab, B.; Gewirtz, D.A. Therapy-Induced Senescence: An “Old” Friend Becomes the Enemy. Cancers 2020, 12, 822. [Google Scholar] [CrossRef] [Green Version]
- Yosef, R.; Pilpel, N.; Tokarsky-Amiel, R.; Biran, A.; Ovadya, Y.; Cohen, S.; Vadai, E.; Dassa, L.; Shahar, E.; Condiotti, R.; et al. Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat. Commun. 2016, 7, 11190. [Google Scholar] [CrossRef]
- Mas-Bargues, C.; Borrás, C.; Viña, J. Bcl-xL as a Modulator of Senescence and Aging. Int. J. Mol. Sci. 2021, 22, 1527. [Google Scholar] [CrossRef]
- Luo, D.; Cheng, S.C.-S.; Xie, H.; Xie, Y. Effects of Bcl-2 and Bcl-XL protein levels on chemoresistance of hepatoblastoma HepG2 cell line. Biochem. Cell Biol. 2000, 78, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.Y.; Zhong, M.; Feng, L.F.; Zhu, B.Y.; Tang, S.S.; Liao, D.F. Bcl-XL small interfering RNA enhances sensitivity of Hepg2 hepatocellular carcinoma cells to 5-fluorouracil and hydroxycamptothecin. Acta Biochim. Et Biophys. Sin. 2006, 38, 704–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.J.-F.; Gong, H.-Y.; Tseng, H.-C.; Wang, W.-L.; Wu, J.-L. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem. Biophys. Res. Commun. 2008, 375, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yin, J.; Yu, J.; Xiang, Q.; Liu, Y.; Tang, S.; Liao, D.; Zhu, B.; Zu, X.; Tang, H.; et al. miRNA-195 sensitizes human hepatocellular carcinoma cells to 5-FU by targeting BCL-w. Oncol. Rep. 2012, 27, 250–257. [Google Scholar]
- Chen, J.; Jin, S.; Abraham, V.; Huang, X.; Liu, B.; Mitten, M.J.; Nimmer, P.; Lin, X.; Smith, M.; Shen, Y.; et al. The Bcl-2/Bcl-X/Bcl-w Inhibitor, Navitoclax, Enhances the Activity of Chemotherapeutic Agents in Vitro and in Vivo. Mol. Cancer Ther. 2011, 10, 2340. [Google Scholar] [CrossRef] [Green Version]
- Hampl, V.; Martin, C.; Aigner, A.; Hoebel, S.; Singer, S.; Frank, N.; Sarikas, A.; Ebert, O.; Prywes, R.; Gudermann, T.; et al. Depletion of the transcriptional coactivators megakaryoblastic leukaemia 1 and 2 abolishes hepatocellular carcinoma xenograft growth by inducing oncogene-induced senescence. EMBO Mol. Med. 2013, 5, 1367–1382. [Google Scholar] [CrossRef]
- Karpel-Massler, G.; Ishida, C.T.; Bianchetti, E.; Zhang, Y.; Shu, C.; Tsujiuchi, T.; Banu, M.A.; Garcia, F.; Roth, K.A.; Bruce, J.N.; et al. Induction of synthetic lethality in IDH1-mutated gliomas through inhibition of Bcl-xL. Nat. Commun. 2017, 8, 1067. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Ran, R.; Wu, Z.; Li, X.; Zeng, Q.; Xia, R.; Wang, Y. Long Non-coding RNA X-Inactive Specific Transcript Mediates Cell Proliferation and Intrusion by Modulating the miR-497/Bcl-w Axis in Extranodal Natural Killer/T-cell Lymphoma. Front Cell Dev. Biol 2020, 8, 1480. [Google Scholar] [CrossRef]
- Tognon, R.; Gasparotto, E.P.L.; Neves, R.P.; Nunes, N.S.; Ferreira, A.F.; Palma, P.V.B.; Kashima, S.; Covas, D.T.; Santana, M.; Souto, E.X.; et al. Deregulation of apoptosis-related genes is associated with PRV1 overexpression and JAK2 V617F allele burden in Essential Thrombocythemia and Myelofibrosis. J. Hematol. Oncol. 2012, 5, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.A.; Yang, J.W.; Choi, S.; Jang, H.W. Nanoscale electrodeposition: Dimension control and 3D conformality. Explor. 2021, 1, 20210012. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Yang, H.; Liu, Q.; Pan, M.; Wang, D.; Pan, S.; Zhang, W.; Wei, J.; Zhao, X.; Ji, J. Selenocystine-Derived Label-Free Fluorescent Schiff Base Nanocomplex for siRNA Delivery Synergistically Kills Cancer Cells. Molecules 2022, 27, 1302. https://doi.org/10.3390/molecules27041302
Liu Y, Yang H, Liu Q, Pan M, Wang D, Pan S, Zhang W, Wei J, Zhao X, Ji J. Selenocystine-Derived Label-Free Fluorescent Schiff Base Nanocomplex for siRNA Delivery Synergistically Kills Cancer Cells. Molecules. 2022; 27(4):1302. https://doi.org/10.3390/molecules27041302
Chicago/Turabian StyleLiu, Yang, Haoying Yang, Qian Liu, Mingming Pan, Danli Wang, Shiyuan Pan, Weiran Zhang, Jinfeng Wei, Xiaowei Zhao, and Junfeng Ji. 2022. "Selenocystine-Derived Label-Free Fluorescent Schiff Base Nanocomplex for siRNA Delivery Synergistically Kills Cancer Cells" Molecules 27, no. 4: 1302. https://doi.org/10.3390/molecules27041302
APA StyleLiu, Y., Yang, H., Liu, Q., Pan, M., Wang, D., Pan, S., Zhang, W., Wei, J., Zhao, X., & Ji, J. (2022). Selenocystine-Derived Label-Free Fluorescent Schiff Base Nanocomplex for siRNA Delivery Synergistically Kills Cancer Cells. Molecules, 27(4), 1302. https://doi.org/10.3390/molecules27041302




