Synthesis, Characterization, Biological Evaluation and DNA Interaction Studies of 4-Aminophenol Derivatives: Theoretical and Experimental Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. General
2.2. Instrumentation and Measurements
2.3. General Synthetic Procedure of Schiff Bases
2.4. Antimicrobial Activity
2.5. Antidiabetic Studies
2.5.1. α-amylase Inhibition Assay
2.5.2. α-glucosidase Inhibition Assay
2.6. DNA Interactions Studies
2.7. Molecular Docking Studies
3. Results and Discussion
3.1. Analysis of Physical Data
3.2. Microanalysis
3.3. Spectroscopic Studies
3.3.1. FT-IR (Fourier Transform Infrared) Spectroscopy
3.3.2. Nuclear Magnetic Resonance (NMR) Spectroscopy
- Proton (1H) and Carbon (13C)-NMR
3.4. Antimicrobial Studies
- The enzymatic degradation of the synthesized compound;
- An alteration of the bacterial protein targeted by the synthesized compound;
- A change in the membrane permeability to the tested compound, etc. [39].
3.5. Antidiabetic Studies
3.6. DNA Interaction Studies
3.7. Molecular Modeling Behavior
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Kabak, M.; Elmali, A.; Elerman, Y. Keto-enol tautomerism, conformations and structure of N-(2-hydroxy-5-methylphenyl), 2-hydroxybenzaldehydeimine. J. Mol. Struct. 1999, 477, 151–158. [Google Scholar] [CrossRef]
- Patel, P.; Thaker, B.; Zele, S. Preparation and Characterisation of Some Lanthanide Complexes Involving a Heterocyclic β-Diketone; NISCAIR-CSIR: New Delhi, India, 1999. [Google Scholar]
- Spichiger-Keller, U.E. Chemical Sensors and Biosensors for Medical and Biological Applications; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Lawrence, J.F.; Frei, R.W. Chemical Derivatization in Liquid Chromatography; Elsevier: Amsterdam, The Netherlands, 2000. [Google Scholar]
- Luo, X.-Y.; Zhao, J.-Z.; Lin, Y.-J.; Liu, Z.-Q. Antioxidative effect of Schiff bases with o-hydroxybenzylidene groupon free radical induced hemolysis of human red blood cell. Chem. Res. Chin. Univ. 2002, 18, 287–289. [Google Scholar]
- Solomon, K.; Lieberman, H.; Groundwater, P.; Hibbs, D.; Hursthouse, M. Molecular modeling and biological evaluation of a series of hydroxylated benzylideneanilines and benzylamines designed as tyrosine kinase inhibitors. Anti-Cancer Drug Des. 1997, 12, 635–647. [Google Scholar]
- Dholakiya, P.P.; Patel, M. Synthesis, spectroscopic studies, and antimicrobial activity of Mn (II), Co (II), Ni (II), Cu (II), and Cd (II) complexes with bidentate Schiff bases and 2, 2′-bipyridylamine. Synth. React. Inorg. Met.-Org. Chem. 2002, 32, 753–762. [Google Scholar] [CrossRef]
- Ashraf, M.A.; Mahmood, K.; Wajid, A.; Maah, M.J.; Yusoff, I. Synthesis, characterization and biological activity of Schiff bases. IPCBEE 2011, 10, 185. [Google Scholar]
- Li, S.; Chen, S.; Lei, S.; Ma, H.; Yu, R.; Liu, D. Investigation on some Schiff bases as HCl corrosioninhibitors for copper. Corros. Sci. 1999, 41, 1273–1287. [Google Scholar] [CrossRef]
- Ashassi-Sorkhabi, H.; Shabani, B.; Aligholipour, B.; Seifzadeh, D. The effect of some Schiff bases on the corrosion of aluminum in hydrochloric acid solution. Appl. Surf. Sci. 2006, 252, 4039–4047. [Google Scholar] [CrossRef]
- Quan, Z.; Chen, S.; Li, S. Protection of copper corrosion by modification of self-assembled films of Schiff bases with alkanethiol. Corros. Sci. 2001, 43, 1071–1080. [Google Scholar] [CrossRef]
- Abirami, M.; Nadaraj, V. Synthesis of schiff base under solvent-free condition: As a green approach. Int. J. Chem. Tech. Res. 2014, 6, 2534–2538. [Google Scholar]
- Sari, N.; Arslan, S.; Loğoğlu, E.; Şakiyan, İ. Antibacterial activites of some new amino acid-schi bases. Gazi Univ. J. Sci. 2010, 16, 283–288. [Google Scholar]
- Verma, M.; Pandeya, S.N.; Singh, K.N.; Stables, J.P. Anticonvulsant activity of Schiff bases of isatin derivatives. Acta Pharm. 2004, 54, 49–56. [Google Scholar] [PubMed]
- El-Ferjani, R.M.; Ahmad, M.; Harun, F.W.; Bulgasem, B.Y. Synthesis, Characterization and Antibacterial Activityof Schiff Bases Derived from 4-Dimethylaminobenzaldehyde with Some Amino Acids and 4-Aminoantipyrine toward Cu (II), Ni (II), Co (II), Cd (II) and Mn (II) Ions. IOSR J. Appl. Chem. 2017, 10, 6–13. [Google Scholar] [CrossRef]
- Masadeh, M.M.; Alzoubi, K.H.; Masadeh, M.M.; Aburashed, Z.O. Metformin as a Potential Adjuvant Antimicrobial Agent Against Multidrug Resistant Bacteria. Clin. Pharmacol. Adv. Appl. 2021, 13, 83. [Google Scholar] [CrossRef] [PubMed]
- Joseyphus, R.S.; Shiju, C.; Joseph, J.; Dhanaraj, C.J.; Arish, D. Synthesis and characterization of metal complexes of Schiff base ligand derived from imidazole-2-carboxaldehyde and 4-aminoantipyrine. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 133, 149–155. [Google Scholar] [CrossRef]
- Hranjec, M.; Starčević, K.; Pavelić, S.K.; Lučin, P.; Pavelić, K.; Zamola, G.K. Synthesis, spectroscopic characterization and antiproliferative evaluation in vitro of novel Schiff bases related to benzimidazoles. Eur. J. Med. Chem. 2011, 46, 2274–2279. [Google Scholar] [CrossRef]
- Ahmadi, F.; Alizadeh, A.A.; Shahabadi, N.; Rahimi-Nasrabadi, M. Study binding of Al–curcumin complex to ds-DNA, monitoring by multispectroscopic and voltammetric techniques. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2011, 79, 1466–1474. [Google Scholar] [CrossRef]
- Kashanian, S.; Javanmardi, S.; Chitsazan, A.; Paknejad, M.; Omidfar, K. Fluorometric study of fluoxetine DNA binding. J. Photochem. Photobiol. B Biol. 2012, 113, 1–6. [Google Scholar] [CrossRef]
- Shahabadi, N.; Mahdavi, M. DNA Interaction Studies of a Cobalt (II) Mixed-Ligand Complex Containing Two Intercalating Ligands: 4, 7-Dimethyl-1, 10-Phenanthroline and Dipyrido [3, 2-a:-c] phenazine. Int. Sch. Res. Not. 2013, 2013, 604218. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-L.; Hu, Y.-J.; Wang, H.; Yu, B.-Q.; Yue, H.-L. Molecular spectroscopy evidence of berberine binding to DNA: Comparative binding and thermodynamic profile of intercalation. Biomacromolecules 2012, 13, 873–880. [Google Scholar] [CrossRef]
- Sasikala, W.D.; Mukherjee, A. Molecular mechanism of direct proflavine–DNA intercalation: Evidence for drug-induced minimum base-stacking penalty pathway. J. Phys. Chem. B 2012, 116, 12208–12212. [Google Scholar] [CrossRef]
- Husain, M.A.; Yaseen, Z.; Rehman, S.U.; Sarwar, T.; Tabish, M. Naproxen intercalates with DNA and causes photocleavage through ROS generation. FEBS J. 2013, 280, 6569–6580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rescifina, A.; Zagni, C.; Varrica, M.G.; Pistarà, V.; Corsaro, A. Recent advances in small organic molecules as DNA intercalating agents: Synthesis, activity, and modeling. Eur. J. Med. Chem. 2014, 74, 95–115. [Google Scholar] [CrossRef] [PubMed]
- McKnight, R.E. Insights into the Relative DNA Binding Affinity and Preferred Binding Mode of Homologous Compounds Using Isothermal Titration Calorimetry (ITC). In Applications of Calorimetry in a Wide Context-Differential Scanning Calorimetry, Isothermal Titration Calorimetry and Microcalorimetry; InTech Publication: Rijeka, Croatia, 2013; pp. 129–152. [Google Scholar]
- Arrieta, A.; Lecea, B.; Palomo, C. Reagents and synthetic methods. Part 58. Synthesis of β-lactams from acetic acids and imines promoted by Vilsmeier type reagents. J. Chem. Soc. Perkin Trans. 1987, 1, 845–850. [Google Scholar] [CrossRef]
- Baluja, S.; Parikh, J.; Chanda, S.; Vaishnani, K. Evaluation of antibacterial activity of some Schiff bases. J. Indian Chem. Soc. 2009, 86, 1338–1342. [Google Scholar]
- Hegazy, G.H.; Ali, H.I. Design, synthesis, biological evaluation, and comparative Cox1 and Cox2 docking of p-substituted benzylidenamino phenyl esters of ibuprofenic and mefenamic acids. Bioorg. Med. Chem. 2012, 20, 1259–1270. [Google Scholar] [CrossRef]
- Radi, S.; Tighadouini, S.; Feron, O.; Riant, O.; Mabkhot, Y.N.; Al-Showiman, S.S.; Hadda, T.B.; El-Youbi, M.; Benabbes, R.; Saalaoui, E. One pot synthesis, antitumor, antibacterial and antifungal activities of some Sciff base heterocycles. Int. J. Pharm. 2015, 5, 39–45. [Google Scholar]
- Saeed, A.; Channar, P.A.; Larik, F.A.; Jabeen, F.; Muqadar, U.; Saeed, S.; Flörke, U.; Ismail, H.; Dilshad, E.; Mirza, B. Design, synthesis, molecular docking studies of organotin-drug derivatives as multi-target agents against antibacterial, antifungal, α-amylase, α-glucosidase and butyrylcholinesterase. Inorg. Chim. Acta 2017, 464, 204–213. [Google Scholar] [CrossRef]
- Arslantas, A.; Devrim, A.K.; Necefoglu, H. The interaction of sheep genomic DNA with a cobalt (II) complex containing p-Nitrobenzoate and N, N-Diethylnicotinamide ligands. Int. J. Mol. Sci. 2007, 8, 1225–1233. [Google Scholar] [CrossRef]
- Arunadevi, A.; Porkodi, J.; Ramgeetha, L.; Raman, N. Biological evaluation, molecular docking and DNA interaction studies of coordination compounds gleaned from a pyrazolone incorporated ligand. Nucleosides Nucleotides Nucleic Acids 2019, 38, 656–679. [Google Scholar] [CrossRef]
- Karrouchi, K.; Fettach, S.; Jotani, M.M.; Sagaama, A.; Radi, S.; Ghabbour, H.A.; Mabkhot, Y.N.; Himmi, B.; Faouzi, M.E.; Issaoui, N. Synthesis, crystal structure, hirshfeld surface analysis, DFT calculations, anti-diabetic activity and molecular docking studies of (E)-N’-(5-bromo-2-hydroxybenzylidene) isonicotinohydrazide. J. Mol. Struct. 2020, 1221, 128800. [Google Scholar] [CrossRef]
- Chemical Computing Group ULC. Molecular Operating Environment (MOE), 2013.08; Chemical Computing Group ULC: Montreal, QC, Canada, 2018. [Google Scholar]
- Pavan, F.R.; de Carvalho, G.S.G.; da Silva, A.D.; Leite, C.Q. Synthesis and anti–Mycobacterium tuberculosis evaluation of Aza-Stilbene derivatives. Sci. World J. 2011, 11, 1113–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raja, A.; Prabakaran, P.; Gajalakshmi, P. Isolation and screening of antibiotic producing psychrophilic actinomycetes and its nature from rothang hill soil against viridans Streptococcus sp. Res. J. Microbiol. 2010, 5, 44–49. [Google Scholar] [CrossRef] [Green Version]
- Özkay, Y.; Tunali, Y.; Karaca, H.; Işıkdağ, İ. Antimicrobial activity and a SAR study of some novel benzimidazole derivatives bearing hydrazone moiety. Eur. J. Med. Chem. 2010, 45, 3293–3298. [Google Scholar] [CrossRef] [PubMed]
- Dever, L.A.; Dermody, T.S. Mechanisms of bacterial resistance to antibiotics. Arch. Intern. Med. 1991, 151, 886–895. [Google Scholar] [CrossRef]
- Ventola, A.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277. [Google Scholar]
- Lv, F.; Liang, H.; Yuan, Q.; Li, C. In vitro antimicrobial effects and mechanism of action of selected plant essential oil combinations against four food-related microorganisms. Food Res. Int. 2011, 44, 3057–3064. [Google Scholar] [CrossRef]
- Rota, M.C.; Herrera, A.; Martínez, R.M.; Sotomayor, J.A.; Jordán, M.J. Antimicrobial activity and chemical composition of Thymus vulgaris, Thymus zygis and Thymus hyemalis essential oils. Food Control 2008, 19, 681–687. [Google Scholar] [CrossRef]
- Maah, M.J. Spectral investigation of the activities of amino substituted bases. Int. J. Chem. Eng. Appl. 2011, 2, 260–263. [Google Scholar]
- Mohamed, M.; Abdelakder, H.; Abdellah, B. Microwave assisted synthesis of 4-aminophenol Schiff bases: DFT computations, QSAR/Drug-likeness proprieties and antibacterial screening. J. Mol. Struct. 2021, 1241, 130666. [Google Scholar] [CrossRef]
- Bang, K.-H.; Lee, D.-W.; Park, H.-M.; Rhee, Y.H. Inhibition of fungal cell wall synthesizing enzymes by trans-cinnamaldehyde. Biosci. Biotechnol. Biochem. 2000, 64, 1061–1063. [Google Scholar] [CrossRef]
- Wang, S.-Y.; Chen, P.-F.; Chang, S.-T. Antifungal activities of essential oils and their constituents from indigenous cinnamon (Cinnamomum osmophloeum) leaves against wood decay fungi. Bioresour. Technol. 2005, 96, 813–818. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Duan, X.; Jing, G.; OuYang, Q.; Tao, N. Cinnamaldehyde inhibits the mycelial growth of Geotrichum citri-aurantii and induces defense responses against sour rot in citrus fruit. Postharvest Biol. Technol. 2017, 129, 23–28. [Google Scholar] [CrossRef]
- Williams, A.K.; Dasilva, S.C.; Bhatta, A.; Rawal, B.; Liu, M.; Korobkova, E.A. Determination of the drug–DNA binding modes using fluorescence-based assays. Anal. Biochem. 2012, 422, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Akdi, K.; Vilaplana, R.A.; Kamah, S.; González-Vílchez, F. Effects of Tris and Hepes buffers on the interaction of palladium–diaminopropane complexes with DNA. J. Inorg. Biochem. 2005, 99, 1360–1368. [Google Scholar] [CrossRef]
- Doshi, R.; Day, P.J.; Carampin, P.; Blanch, E.; Stratford, I.J.; Tirelli, N. Spectrophotometric analysis of nucleic acids: Oxygenation-dependant hyperchromism of DNA. Anal. Bioanal. Chem. 2010, 396, 2331–2339. [Google Scholar] [CrossRef] [PubMed]
- Arjmand, F.; Parveen, S.; Afzal, M.; Toupet, L.; Hadda, T.B. Molecular drug design, synthesis and crystal structure determination of CuII–SnIV heterobimetallic core: DNA binding and cleavage studies. Eur. J. Med. Chem. 2012, 49, 141–150. [Google Scholar] [CrossRef]
- Song, Y.; Zhong, D.; Luo, J.; Tan, H.; Chen, S.; Li, P.; Wang, L.; Wang, T. Binding characteristics and interactive region of 2-phenylpyrazolo [1, 5-c] quinazoline with DNA. Luminescence 2014, 29, 1141–1147. [Google Scholar] [CrossRef]
- Strekowski, L.; Wilson, B. Noncovalent interactions with DNA: An overview. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2007, 623, 3–13. [Google Scholar] [CrossRef]
- Tewey, K.; Rowe, T.; Yang, L.; Halligan, B.; Liu, L.-F. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science 1984, 226, 466–468. [Google Scholar] [CrossRef] [PubMed]
- Ross, W.E.; Bradley, M.O. DNA double-strand breaks in mammalian cells after exposure to intercalating agents. Biochim. Biophys. Acta-Nucleic Acids Protein Synth. 1981, 654, 129–134. [Google Scholar] [CrossRef]
- Severini, A.; Morgan, A.R. An assay for proteinases and their inhibitors based on DNA/ethidium bromide fluorescence. Anal. Biochem. 1991, 193, 83–89. [Google Scholar] [CrossRef]
- De Almeida, S.M.; Lafayette, E.A.; Da Silva, L.P.; Amorim, C.A.; De Oliveira, T.B.; Ruiz, A.L.; De Carvalho, J.E.; De Moura, R.O.; Beltrão, E.I.; De Lima, M.D.; et al. Synthesis, DNA binding, and antiproliferative activity of novel acridine-thiosemicarbazone derivatives. Int. J. Mol. Sci. 2015, 16, 13023–13042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, A.; Zaheer, M.; Qureshi, R.; Akhter, Z.; Nazar, M.F. Voltammetric and spectroscopic investigations of 4-nitrophenylferrocene interacting with DNA. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2010, 75, 1082–1087. [Google Scholar] [CrossRef] [PubMed]
- Tahir, S.; Badshah, A.; Hussain, R.A.; Tahir, M.N.; Tabassum, S.; Patujo, J.A.; Rauf, M.K. DNA-binding studies and biological activities of new nitrosubstituted acyl thioureas. J. Mol. Struct. 2015, 1099, 215–225. [Google Scholar] [CrossRef]
- Pratviel, G.; Bernadou, J.; Meunier, B. DNA and RNA cleavage by metal complexes. Adv. Inorg. Chem. 1998, 45, 251–312. [Google Scholar]
- Shahabadi, N.; Kashanian, S.; Khosravi, M.; Mahdavi, M. Multispectroscopic DNA interaction studies of a water-soluble nickel (II) complex containing different dinitrogen aromatic ligands. Transit. Met. Chem. 2010, 35, 699–705. [Google Scholar] [CrossRef]
- Kumar, K.A.; Reddy, K.L.; Vidhisha, S.; Satyanarayana, S. Synthesis, characterization and DNA binding and photocleavage studies of [Ru(bpy)2BDPPZ]2+, [Ru(dmb)2BDPPZ]2+ and [Ru(phen)2BDPPZ]2+ complexes and their antimicrobial activity. Appl. Organomet. Chem. 2009, 23, 409–420. [Google Scholar] [CrossRef]
- González-Ruiz, V.; Olives, A.I.; Martín, M.A.; Ribelles, P.; Ramos, M.T.; Menéndez, J.C. An overview of analytical techniques employed to evidence drug-DNA interactions. Applications to the design of genosensors. Biomed. Eng. Trends Res. Technol. 2011, 32, 215–219. [Google Scholar]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug–DNA interactions and their study by UV–Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B Biol. 2013, 124, 1–19. [Google Scholar] [CrossRef]





| Schiff Base | Color | Melting Point (°C) | Solubility |
|---|---|---|---|
| 4-chloro-2-(((4-hydroxyphenyl)imino)methyl)phenol (S-1) | Orange Yellow | 243–245 °C | Water, Alcohols (Hot state), Acetone and DMSO |
| 4-((4-(dimethylamino)benzylidene)amino)phenol (S-2) | Light Yellow | 181–183 °C 182.7 °C [36] | Water, Alcohols (Hot state), Ethyl acetate and DMSO |
| 4-((3-nitrobenzylidene)amino)phenol (S-3) | Pale Yellow | 162–164 °C | Water, Alcohols, Ethyl acetate and DMSO |
| 4-((thiophen-2-ylmethylene)amino)phenol (S-4) | Dull Yellow | 203–205 °C 204–205 °C [30] | Acetone (Hot), Ethanol and DMSO |
| 4-(((E)-3-phenylallylidene)amino)phenol (S-5) | Yellow | 210–212 °C | Acetone, Ethanol (Hot) and DMSO |
| Schiff Base | C (%) | H (%) | N (%) | Mol. Formula | Mwt (g/mol) | Yield (%) |
|---|---|---|---|---|---|---|
| S-1 | 63.04 63.28 | 4.07 3.99 | 5.66 5.50 | C13H10ClNO2 | 247.68 | 78 |
| S-2 | 74.97 75.21 | 6.71 6.64 | 11.66 11.51 | C15H16N2O | 240.31 | 79 |
| S-3 | 64.46 64.71 | 4.16 4.08 | 11.56 11.41 | C13H10N2O3 | 242.23 | 73 |
| S-4 | 65.00 65.23 | 4.46 4.35 | 17.05 16.89 | C11H9NOS | 203.26 | 87 |
| S-5 | 80.69 80.93 | 5.87 5.76 | 6.27 6.12 | C15H13NO | 223.28 | 84 |
| Bold: Experimental values | ||||||
| No. | Reagents | IR Values ν (cm−1) | Schiff Base | IR Values ν (cm−1) |
|---|---|---|---|---|
| 1 | 5-chlorosalisaldehyde | 3195 (sp2 C–H Str.), 1663 (C=O), 2916 and 2854 (C–H aliphatic), 1615 and 1543 (C=C str.), 1477, 1354, 1270, 1208, 1112, 953, 906, 828, 776 (C–Cl Str.), 714, 644 and 565 | S-1 | 1654 (C=N Str.), 1618 and 1564 (C=C), 1510, 1486, 1455, 1374, 1319, 1292, 1268, 1231, 1163, 1146, 1109, 1075, 990, 952, 918, 871, 844, 827, 799, 776 (C–Cl Str.), 731, 704, 674, 633 and 589 |
| 2 | 4-N,N-dimethylaminobenzaldehyde | 1659 (C=O), 1628 (C=C), 1589, 1547, 1368, 1230 1160 (C–N), 1064, 810, 726 and 594. | S-2 | 1608 (C=N), 1581 (C=C), 1533, 1503, 1489, 1442, 1367, 1319, 1234, 1200, 1170 (C–N), 1105, 1064, 1007, 976, 939, 830, 803, 786, 728, 660, 643, 585 and 562. |
| 3 | 3-nitrobenzaldehyde | 3068, 2878, 1701, 1689, 1613, 1581, 1530 (N=O Asymm. Str.), 1470, 1455, 1396, 1348 (N=O symm. str.), 1275, 1199, 1102, 1085, 1076, 1007, 932, 917, 822, 810, 728, 676, 666 and 525. | S-3 | 3073, 1654 (C=N), 1618, 1581 (C=C aromatic), 1527, 1506, 1479, 1445, 1377, 1347, 1316, 1272, 1228, 1190, 1163, 1102, 1078, 976, 952, 929, 898, 874, 830, 813, 793, 735, 711 and 674 |
| 4 | 2-Thiophencarboxyaldehyde | 1654 (C=O), 1518 (C=C), 1416, 1234, 1210, 1045, 863 (C–S–C), 722 (C–S) and 660 | S-4 | 1608 (C=N), 1509 (C=C), 142, 1234, 1220, 1045, 864 (C–S–C), 721 (C–S) and 709 |
| 5 | Cinnamaldehyde | 1668 (C=O), 1624 (C=C), 1449, 1294, 1119, 970, 744 and 686 | S-5 | 1643 (C=N), 1628 (C=C), 1450, 1298, 1120, 1024, 968, 752 and 691 |
| 6 | 4-aminophenol | 3338 (O–H str.), 3282 (N–H Str.), 2572, 1613 (N–H Bend.), 1508, 1471, 1384 (O–H Bend.), 1235(C–N Str.), 1168, 1091 (C–O Str.), 966, 810 and 748. | - | - |
| Schiff Bases | 1H-NMR | 13C-NMR |
|---|---|---|
| 4-chloro-2-(((4-hydroxyphenyl)imino)methyl)phenol (S-1) | 1H-NMR (DMSO, TMS, 400 MHz and d ppm): 13.40 and 9.72 (1H and –OH), 8.89 (1H and s), 7.68 (1H and dd), 7.40–7.38 (2H and ddd), 6.98 (1H and dd), 6.86-6.85 (2H and ddd) and 6.83 (1H and dd) | 13C-NMR (DMSO, TMS, 100 MHz and d ppm): 162.26 (–C=N), 161.35 and 158.46 (–C–OH), 148.90 (C–N=), 130.90 (C–Cl), 129.47, 128.59, 122.61, 118,.98 and 115.02 (Aromatic Carbons) |
| 4-((4-(dimethylamino)benzylidene)amino)phenol (S-2) | 1H-NMR (DMSO, TMS, 400 MHz and d ppm): 9.31 (1H and –OH), 8.39 (1H), 7.71–7.68 (2H and ddd), 7.10–7.07 (2H and ddd), 6.78–6.74(2H and ddd) and 3.05 (6H). | 13C-NMR (DMSO, TMS, 125 MHz and d ppm): 157.46 (–C=N), 155.78 (C–OH), 152.49 (N-C= (aromatic)), 144.11, 130.22, 124.80, 122.44, 116.07, 111.98 (other Aromatic Carbons) and 40.16 (–CH3). |
| 4-((3-nitrobenzylidene)amino)phenol (S-3) | 1H-NMR (DMSO, TMS, 400 MHz and d ppm): δ 9.62 (1H and -OH), 8.70(1H and s), 8.66 (1H and ddd), 8.31 (1H and td), 7.61 (1H and ddd), 7.31–7.28 (2H and ddd) and 6.84 (2H and ddd) | 13C-NMR (DMSO, 100 MHz and d ppm): 157.46 (–C=N), 155.35 (C–OH), 148.72(C–N=)–, 142.11 (C–NO2), 138.59, 134.68, 130.90, 125.47, 123.43, 122.68 and 116.26 (Aromatic Carbons). |
| 4-((thiophen-2-ylmethylene)amino)phenol (S-4) | 1H-NMR (DMSO, TMS, 400 MHz and d ppm): δ 9.49 (1H and -OH), 8.75 (1H and s (CH=N)), 7.74 (1H and dd), 7.59 (1H and dd), 7.20 (1H and dd), 7.16 (2H and ddd) and 6.79–6.77 (2H and ddd) | 13C-NMR (DMSO, 100 MHz and d ppm):157.46 (C–OH), 152.24 (–C=N), 149.79 (C–N=), 143.04 (C–S), 130.22, 129.40, 127.83 (C–S), 122.44 and 116.07. |
| 4-(((E)-3-phenylallylidene)amino)phenol (S-5) | 1H-NMR (DMSO, TMS, 400 MHz and d ppm): δ 9.48 (1H and -OH), 8.40–8.38 (1H and d), 7.66–7.64 (2H and tdd), 7.44–7.40 (2H and ddd), 7.38–7.34 (1H and tt), 7.29–7.25 (1H and d), 7.15–7.10 (2H and dd) and 6.79–6.76 (1H and dd) | 13C-NMR (DMSO, 100 MHz and d ppm): 157.58 (C–OH), 155.40 (–C=N), 149.73 (C–N=), 138.50 134.61, 131.13, 129.40 (=C–C (aliphatic)), 127.83, 122.81 and 116.18 (Other Aromatic Carbons) |
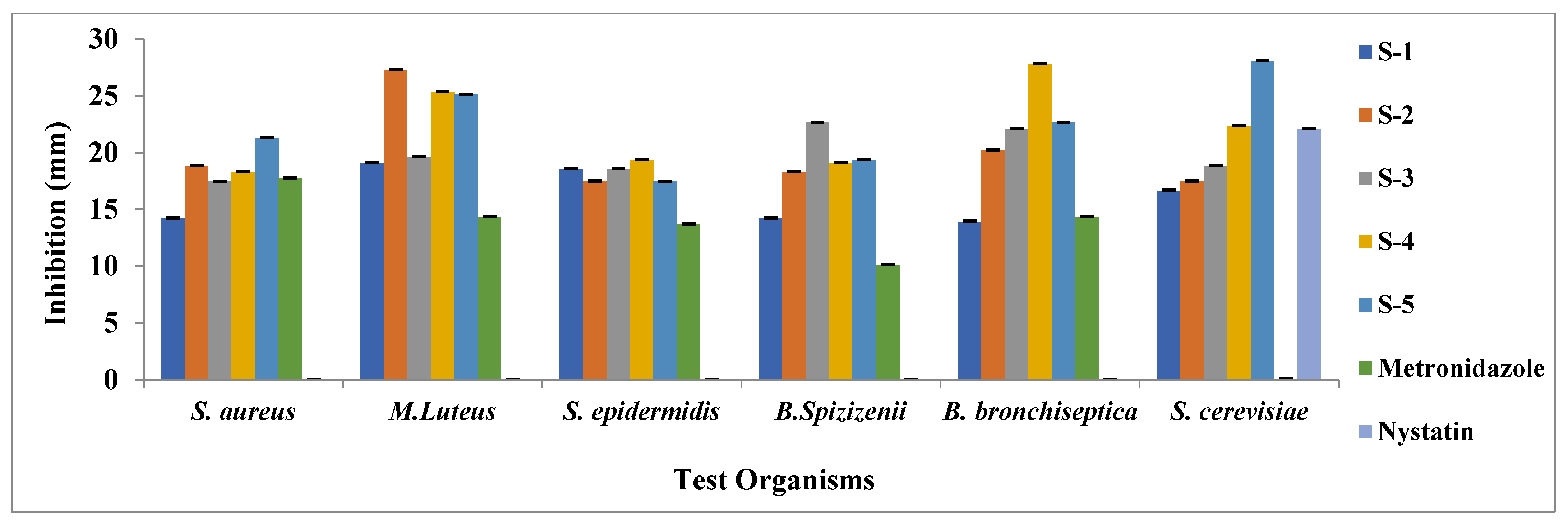
| Schiff Bases | S-1 | S-2 | S-3 | S-4 | S-5 | Metronidazole | Nystatin |
|---|---|---|---|---|---|---|---|
| S. aureus | 14.18 ± 0.08 | 18.82 ± 0.10 | 17.45 ± 0.08 | 18.27 ± 0.09 | 21.27 ± 0.05 | 17.73 ± 0.13 | − |
| M. Luteus | 19.09 ± 0.08 | 27.27 ± 0.08 | 19.64 ± 0.05 | 25.36 ± 0.11 | 25.09 ± 0.06 | 14.3 ± 0.07 | − |
| S. epidermidis | 18.55 ± 0.10 | 17.45 ± 0.08 | 18.55 ± 0.06 | 19.36 ± 0.07 | 17.45 ± 0.07 | 13.64 ± 0.03 | − |
| B. Spizizenii | 14.18 ± 0.12 | 18.27 ± 0.13 | 22.64 ± 0.09 | 19.09 ± 0.05 | 19.36 ± 0.04 | 10.09 ± 0.05 | − |
| B. bronchiseptica | 13.91 ± 0.07 | 20.18 ± 0.10 | 22.09 ± 0.08 | 27.82 ± 0.08 | 22.64 ± 0.08 | 14.33 ± 0.06 | − |
| S. cerevisiae | 16.64 ± 0.14 | 17.45 ± 0.06 | 18.82 ± 0.06 | 22.36 ± 0.06 | 28.09 ± 0.09 | − | 22.09 ± 0.07 |
| Average Percentage Inhibition | ||||||
|---|---|---|---|---|---|---|
| Conc. | S-1 | S-2 | S-3 | S-4 | S-5 | Acarbose |
| 500 ppm | 85.50 | 93.19 | 76.95 | 36.14 | 61.84 | 96.40 |
| 250 ppm | 61.00 | 69.00 | 56.95 | 27.82 | 51.33 | 94.64 |
| 125 ppm | 32.71 | 29.92 | 32.96 | 5.40 | 26.74 | 90.96 |
| IC50 ppm | 190.2 | 178.3 | 206.9 | 747.8 | 284.7 | 5.7 |
| Average Percentage Inhibition | ||||||
|---|---|---|---|---|---|---|
| Conc. | S-1 | S-2 | S-3 | S-4 | S-5 | Acarbose |
| 500 ppm | 76.67 | 58.87 | 60.98 | 46.44 | 73.70 | 91.94 |
| 250 ppm | 55.57 | 30.74 | 22.17 | 31.43 | 53.30 | 89.76 |
| 125 ppm | 35.72 | 20.83 | 4.24 | 15.85 | 26.85 | 87.54 |
| IC50 ppm | 202.5 | 403.4 | 417.1 | 558.2 | 238.8 | 0.4 |
| Schiff Base | Observations | Type of Interaction |
|---|---|---|
| S-1 | Hyperchromism | Intercalative |
| S-2 | Hyperchromism Bathochromic shift (202 nm–258 nm) | Intercalative |
| S-3 | Hyperchromism Bathochromic shift (201–213 nm) | Intercalative |
| S-4 | Hypochromism Bathochromic shift (341–346 nm) | Intercalative |
| S-5 | Hyperchromism | Intercalative |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rafique, B.; Kalsoom, S.; Sajini, A.A.; Ismail, H.; Iqbal, M. Synthesis, Characterization, Biological Evaluation and DNA Interaction Studies of 4-Aminophenol Derivatives: Theoretical and Experimental Approach. Molecules 2022, 27, 1352. https://doi.org/10.3390/molecules27041352
Rafique B, Kalsoom S, Sajini AA, Ismail H, Iqbal M. Synthesis, Characterization, Biological Evaluation and DNA Interaction Studies of 4-Aminophenol Derivatives: Theoretical and Experimental Approach. Molecules. 2022; 27(4):1352. https://doi.org/10.3390/molecules27041352
Chicago/Turabian StyleRafique, Bushra, Saima Kalsoom, Abdulrahim A. Sajini, Hammad Ismail, and Mudassir Iqbal. 2022. "Synthesis, Characterization, Biological Evaluation and DNA Interaction Studies of 4-Aminophenol Derivatives: Theoretical and Experimental Approach" Molecules 27, no. 4: 1352. https://doi.org/10.3390/molecules27041352
APA StyleRafique, B., Kalsoom, S., Sajini, A. A., Ismail, H., & Iqbal, M. (2022). Synthesis, Characterization, Biological Evaluation and DNA Interaction Studies of 4-Aminophenol Derivatives: Theoretical and Experimental Approach. Molecules, 27(4), 1352. https://doi.org/10.3390/molecules27041352






