Untargeted Metabolomics Analysis Revealed the Major Metabolites in the Seeds of four Polygonatum Species
Abstract
:1. Introduction
2. Results
2.1. Metabolite Profiles
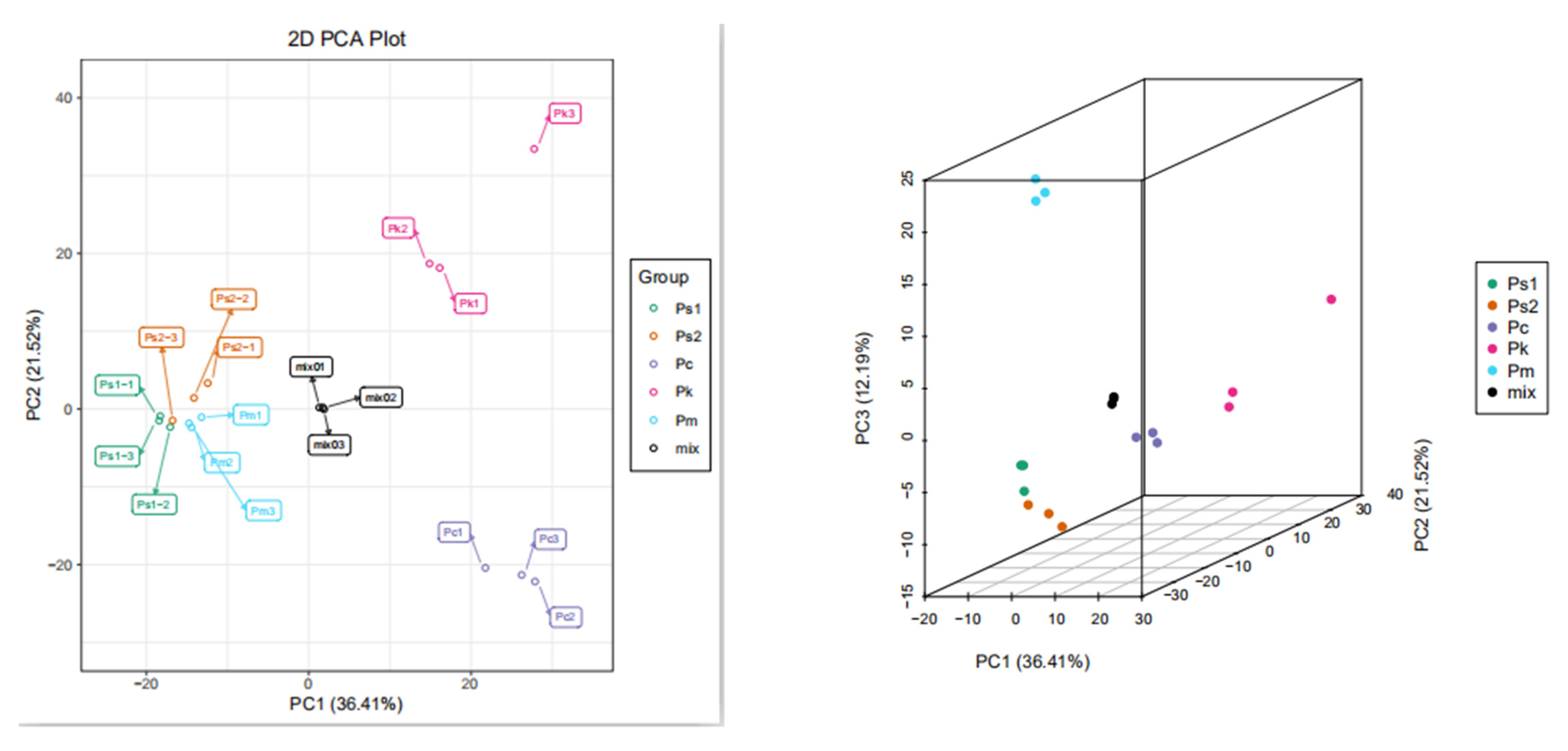
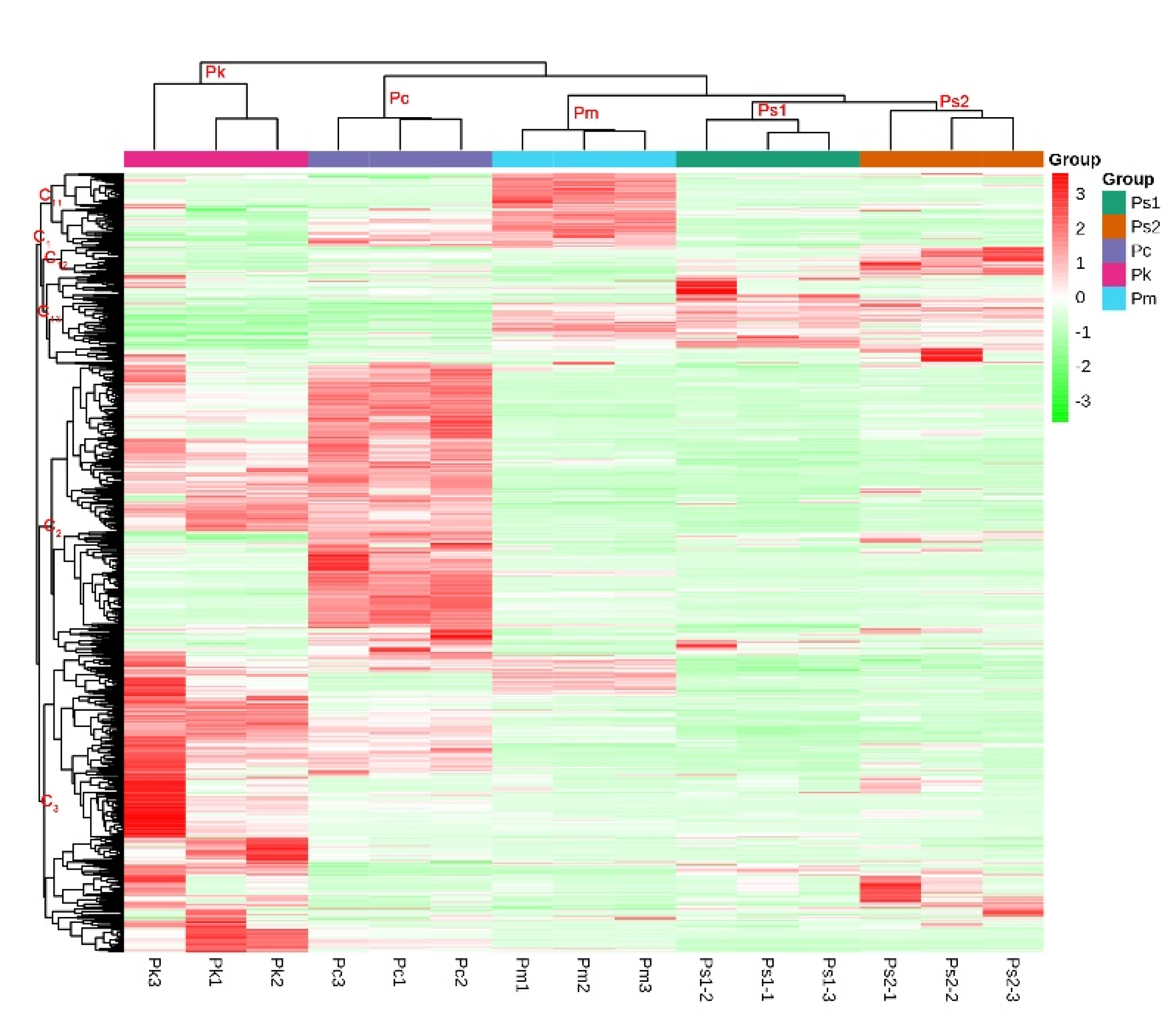
2.2. Metabolite Analyses via PCA as well as Hierarchical Cluster and Correlation Analyses
2.3. OPLS-DA Results
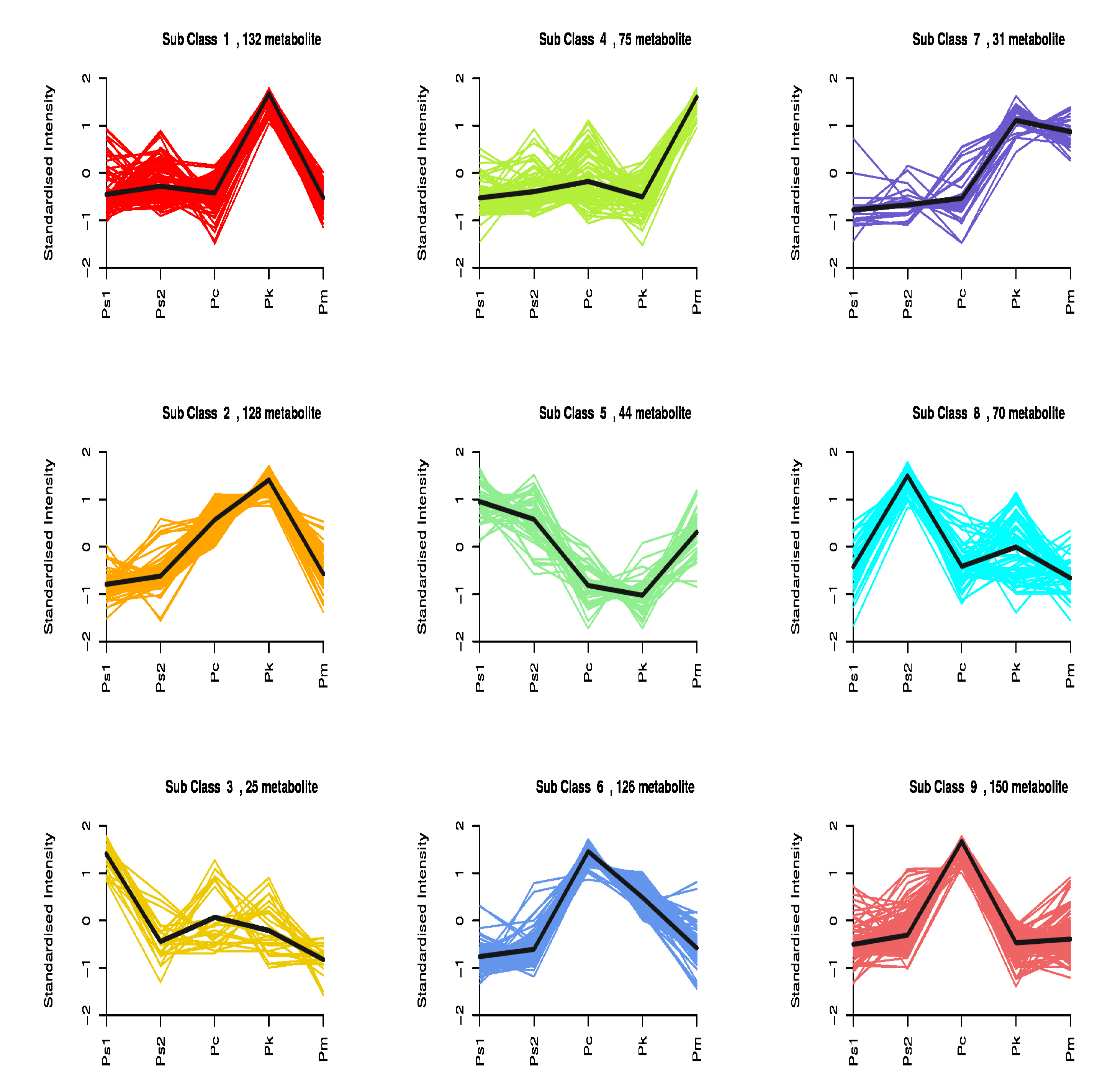
2.4. Analysis of Differentially Abundant Metabolites among the Seed Samples
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Sample Preparation
4.2. UPLC-ESI-Q TRAP-MS/MS Analysis
4.3. Qualitative and Quantitative Analyses of Metabolites
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ma, C.D.; Chang, H.; Yang, Y.C.; Sun, Y.; Wang, E.H. Textual research on genuineness of Shannxi Polygonati rhizoma. Mod China Med. 2018, 20, 887–898. [Google Scholar]
- Cui, X.; Wang, S.; Cao, H.; Guo, H.; Li, Y.J.; Xu, F.X.; Zheng, M.M.; Xi, X.Z.; Han, C.C. A Review: The Bioactivities and Pharmacological Applications of Polygonatum sibiricum polysaccharides. Molecules 2018, 23, 1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, W.; Wang, Y.F.; Zeng, L.X.; Dong, J.C.; Bi, Q.; Yang, X.X.; Che, Y.Y.; He, S.; Yu, J. Polysaccharides from Polygonatum kingianum improve glucose and lipid metabolism in rats fed a high fat diet. Biomed. Pharmacother. 2020, 25, 109910. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.Y.; Zhao, L.L.; Chen, S.B.; Hou, B.C.; Huang, J.; Hong, X.; Qing, L.; Fang, Y.; Tao, Z. Polygonatum sibiricum polysaccharides prevent LPS-induced acute lung injury by inhibiting inflammation via the TLR4/Myd88/NF-kappaB pathway. Exp. Ther. Med. 2020, 20, 3733–3739. [Google Scholar] [PubMed]
- Wang, G.; Fu, Y.; Li, J.; Li, Y.; Zhao, Q.; Hu, A.; Xu, C.; Shao, D.; Chen, W. Aqueous extract of Polygonatum sibiricum ameliorates ethanol-induced mice liver injury via regulation of the Nrf2/ARE pathway. J. Food Biochem. 2021, 45, e13537. [Google Scholar] [CrossRef]
- Zhao, P.; Zhao, C.C.; Gao, Q.Z.; Huang, L.Q.; Xiao, P.G.; Gao, W.Y. The genus Polygonatum: A review of ethnopharmacology, phytochemistry and pharmacology. J. Ethnopharmacol. 2018, 214, 274–291. [Google Scholar] [CrossRef]
- Zheng, S. Protective effect of Polygonatum sibiricum Polysaccharide on d-galactose-induced aging rats model. Sci. Rep. 2020, 10, 2246. [Google Scholar] [CrossRef] [Green Version]
- Khan, H.; Saeed, M.; Naveed, M.; Perviz, S. Phytochemical analysis, antibacterial, and antifungal assessment of aerial parts of Polygonatum verticillatum. Toxicol. Ind. Health 2016, 32, 841–847. [Google Scholar] [CrossRef]
- Huang, Z.Z.; Du, X.; Ma, C.D.; Zhang, R.R.; Gong, W.L.; Liu, F. Identifification of Antitumor Active Constituents in Polygonatum sibiricum Flower by UPLC-Q-TOF-MSE and Network Pharmacology. ACS Omega 2020, 5, 29755–29764. [Google Scholar] [CrossRef]
- Zhang, Z.R.; Huang, S.; Liu, J.J.; Zhang, X.F.; Si, J.P. Main nutrients and functional ingredients in flowers of Polygonatum cyrtonema and P. filipes. China J. Chin. Mater. Med. 2020, 45, 1329–1333. [Google Scholar]
- Huang, S.; Liu, J.J.; Zhang, X.F.; Duan, C.L.; Si, J.P. Main nutrients and functional ingredients in shoots of Polygonatum cyrtonema. China J. Chin. Mater. Med. 2020, 45, 1053–1058. [Google Scholar]
- Si, J.P.; Zhu, Y.X. Polygonati rhizome-a new high-quality crop with great potential and not occupying farmland (in Chinese). Sci. Sin. Vitae 2021, 11, 3–10. [Google Scholar]
- Wang, F.; Chen, L.; Chen, H.P.; Chen, S.W.; Liu, Y.P. Analysis of Flavonoid Metabolites in Citrus Peels (Citrus reticulata “Dahongpao”) Using UPLC-ESI-MS/MS. Molecules 2019, 24, 2680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nocedo-Mena, D.; Ríos, M.Y.; Ramírez-Cisneros, M.Á.; González-Maya, L.; Sánchez-Carranza, J.N.; Camacho-Corona, M.D.R. Metabolomic Profile and Cytotoxic Activity of Cissus incisa Leaves Extracts. Plants 2021, 10, 1389. [Google Scholar] [CrossRef]
- Omnia, H.A.; Eman, M.O.; John, R.F.; Samar, Y.D.; Sheila, M.P.; Justin, R.N.; Tanja, S.; Ahmed, T.; Usama, R.A. Metabolomics analysis and biological investigation of three Malvaceae plants. Phytochem Anal. 2020, 31, 204–214. [Google Scholar]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Hu, C.Y.; Xu, D.P.; Wu, Y.M.; Qu, S.Y. Triterpenoid saponins from the rhizome of Polygonatum sibiricum. J. Asian Nat. Prod. Res. 2010, 12, 801–808. [Google Scholar] [CrossRef]
- Zhou, X.L.; Zhang, Y.P.; Zhao, H.D.; Liang, J.S.; Zhang, Y.; Shi, S.Y. Antioxidant homoisoflflavonoids from Polygonatum odoratum. Food Chem. 2015, 186, 63–68. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, C.R.; Zhang, Y.J. Steroidal saponins from the rhizomes of Polygonatum prattii. J. Asian Nat. Prod. Res. 2016, 18, 268–273. [Google Scholar] [CrossRef]
- Tang, C.; Yu, Y.M.; Qi, Q.L.; Wu, X.D.; Wang, J.; Tang, S.G. Steroidal saponins from the rhizome of Polygonatum sibiricum. J. Asian Nat. Prod. Res. 2019, 21, 197–206. [Google Scholar] [CrossRef]
- Liao, D.Q.; An, R.P.; Wei, J.H.; Wang, D.M.; Li, X.E.; Qi, J.J. Transcriptome profiles revealed molecular mechanisms of alternating temperatures in breaking the epicotyl morphophysiological dormancy of Polygonatum sibiricum seeds. BMC Plant Biol. 2021, 21, 370. [Google Scholar] [CrossRef] [PubMed]
- Ying, H.; Shi, J.; Zhang, S.S.; Gesang, P.C.; Wang, S.; Zhao, F.; Cui, Y.; Zeng, X.L. Transcriptomic and metabolomic profiling provide novel insights into fruit development and flesh coloration in Prunus mira Koehne, a special wild peach species. BMC Plant Biol. 2019, 19, 463. [Google Scholar] [CrossRef] [PubMed]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Floden, A.; Schilling, E.E. Using phylogenomics to reconstruct phylogenetic relationships within tribe Polygonateae (Asparagaceae), with a special focus on Polygonatum. Mol. Phylogenetics Evol. 2018, 129, 202–213. [Google Scholar] [CrossRef]
- Meng, Y.; Nie, Z.L.; Deng, T.; Wen, J. Phylogenetics and evolution of phyllotaxy in the Solomon’s seal genus Polygonatum (Asparagaceae: Polygonateae). Bot. J. Linn. Soc. 2014, 176, 435–451. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Li, R.S.; Ren, L.; Gao, X.L.; Zhang, Y.G.; Ma, Z.M.; Ma, D.F.; Luo, Y.H. A comparative metabolomics study of flavonoids in sweet potato with different flesh colors (Ipomoea batatas (L.) Lam). Food Chem. 2018, 260, 124–134. [Google Scholar] [CrossRef]
- Wang, S.H.; Tu, H.; Wan, J.; Chen, W.; Liu, X.Q.; Luo, J.; Xu, J.; Zhang, H.Y. Spatio-temporal distribution and natural variation of metabolites in citrus fruits. Food Chem. 2016, 199, 8–17. [Google Scholar] [CrossRef]
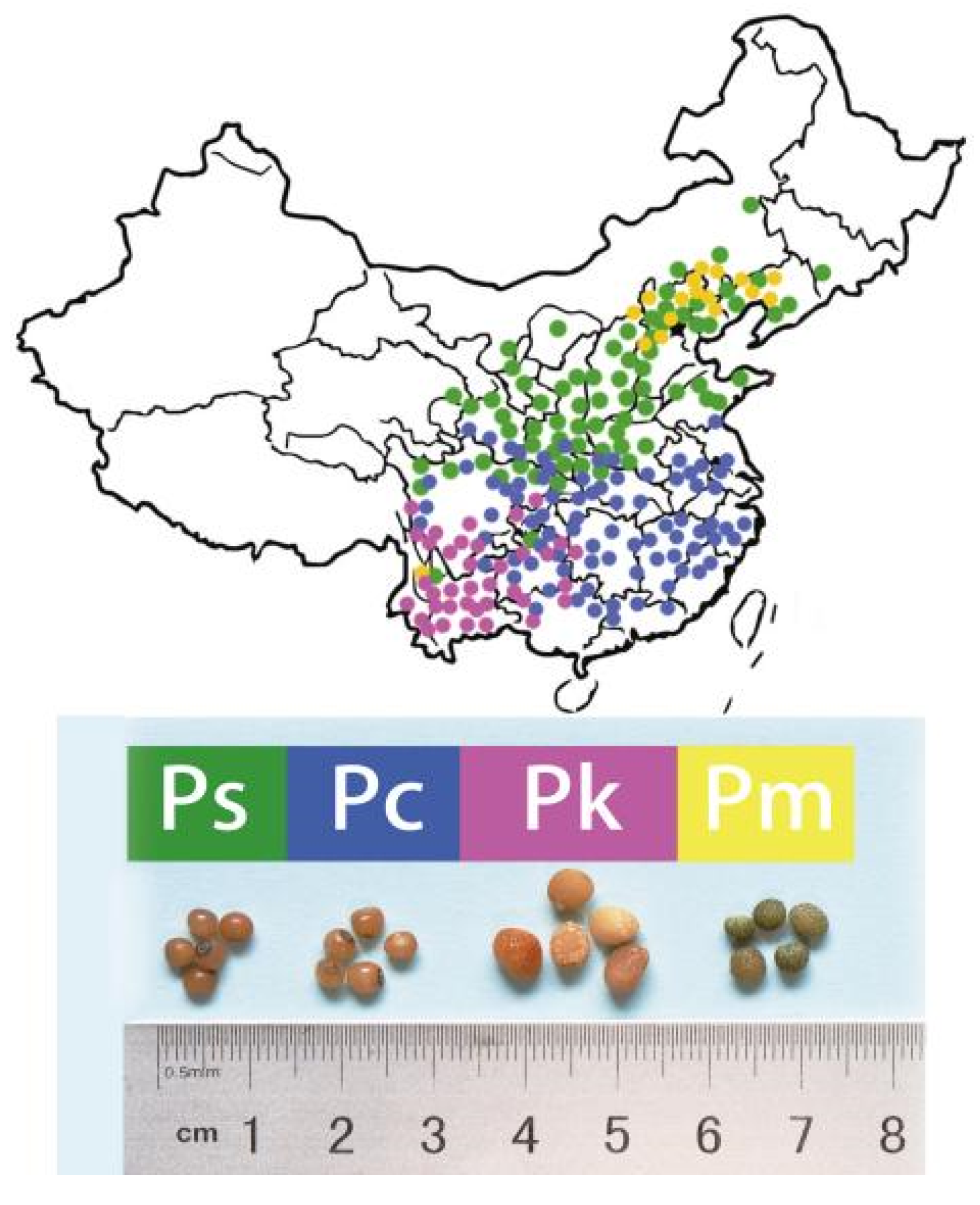

| Ps1 | Ps2 | Pc | Pk | Pm | Total Classes | |
|---|---|---|---|---|---|---|
| Flavonoids | 160 | 160 | 167 | 159 | 149 | 185 |
| Lipids | 116 | 117 | 122 | 124 | 123 | 127 |
| Phenolic acids | 93 | 98 | 100 | 101 | 97 | 105 |
| Amino acids and derivatives | 82 | 86 | 88 | 88 | 85 | 88 |
| Organic acids | 66 | 66 | 72 | 71 | 69 | 74 |
| Alkaloids | 50 | 56 | 54 | 53 | 54 | 57 |
| Nucleotides and derivatives | 45 | 46 | 46 | 46 | 46 | 46 |
| Steroids | 26 | 19 | 25 | 24 | 21 | 36 |
| Terpenoids | 22 | 22 | 21 | 27 | 22 | 31 |
| Lignans and coumarins | 22 | 21 | 22 | 20 | 21 | 24 |
| Tannins | 7 | 7 | 8 | 5 | 8 | 8 |
| Quinones | 1 | 1 | 3 | 3 | 3 | 4 |
| Others | 85 | 85 | 88 | 84 | 86 | 88 |
| Total metabolites | 775 | 782 | 816 | 805 | 784 | 873 |
| Index | Q1 (Da) | Q3 (Da) | Molecular Weight (Da) | Compounds | Ps1 | Ps2 | Pc | Pk | Pm | Ps1 vs. Pc | Ps2 vs. Pc | Ps1 vs. Pk | Ps2 vs. Pk | Ps1 vs. Pm | Ps2 vs. Pm | Pk vs. Pc | Ps1 vs. Ps2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Jmyn6479 | 3.29E + 02 | 1.39E + 02 | 3.30E + 02 | Polygonatone C | + | + | + | + | + | insig | 8.27 | insig | 9.08 | insig | 4.81 | insig | −5.22 |
| Zmcp010827 | 4.15E + 02 | 2.71E + 02 | 4.14E + 02 | Diosgenin | + | + | + | + | + | insig | insig | 5.6 | 8.69 | −3.03 | insig | −6.63 | −3.09 |
| mws1620 | 4.31E + 02 | 2.87E + 02 | 4.30E + 02 | Ruscogenin | + | + | + | − | + | 8.05 | 9.58 | −11.09 | −9.56 | 4.55 | 6.08 | 19.14 | −1.53 |
| pmp001034 | 4.33E + 02 | 2.89E + 02 | 4.32E + 02 | Markogenin | − | − | + | − | − | 12.17 | 12.17 | 12.17 | |||||
| Zmcp008957 | 4.47E + 02 | 4.29E + 02 | 4.46E + 02 | 24-Hydroxypennogenin | − | − | + | − | − | 14.86 | 14.86 | 14.86 | |||||
| Jmyp5224 | 5.93E + 02 | 4.13E + 02 | 5.92E + 02 | 3β,26-diol-25(R)-Δ5,20(22)-diene-furostan-26-O-β-d-glucopyranoside | − | − | + | + | − | 11.39 | 11.39 | 11.5 | 11.5 | insig | |||
| Zmcp006626 | 5.93E + 02 | 4.13E + 02 | 5.92E + 02 | Pennogenin-3-O-glucoside | + | + | + | + | + | −3.18 | insig | 4.68 | 5.08 | insig | insig | −7.86 | insig |
| pmp000750 | 7.09E + 02 | 5.65E + 02 | 7.08E + 02 | Ruscogenin-1-O-xylosyl(1,2)fucoside | − | − | + | − | − | 15.90 | 15.9 | −9.92 | 15.9 | ||||
| pmp000748 | 7.09E + 02 | 5.65E + 02 | 7.08E + 02 | Ruscogenin-1-O-xylosyl(1,3)fucoside | − | − | + | + | − | 19.45 | 19.45 | 8.69 | 8.69 | 10.76 | |||
| pmp000753 | 7.23E + 02 | 5.79E + 02 | 7.22E + 02 | Diosgenin-3-O-rhamnosyl(1,2)glcoside | + | − | + | − | − | 6.71 | 17.1 | −10.4 | −10.4 | 17.1 | −10.4 | ||
| pmp000752 | 7.23E + 02 | 5.79E + 02 | 7.22E + 02 | Ruscogenin-1-O-rhamnosyl(1,2)fucoside | + | − | + | − | − | 6.73 | 16.65 | −9.92 | −9.92 | 16.65 | −9.92 | ||
| Cmdp004421 | 7.37E + 02 | 5.75E + 02 | 7.36E + 02 | kingianoside B | + | + | + | − | − | insig | 4.37 | −17.24 | −12.51 | −17.24 | −12.51 | 16.88 | −4.74 |
| Jmyp4233 | 7.39E + 02 | 5.77E + 02 | 7.38E + 02 | (25S)-spirost-5-en-3β-ol-3-O-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside | + | + | − | + | + | −19.11 | −17.63 | insig | 1.89 | −1.29 | insig | −19.52 | −1.48 |
| pmp000754 | 7.39E + 02 | 5.95E + 02 | 7.38E + 02 | HydroxyDiosgenin-hamnosyl(1,2)glucoside | − | − | + | + | 20.26 | 20.26 | 9.43 | 9.43 | 10.55 | 10.55 | 10.83 | ||
| pmp000755 | 7.39E + 02 | 5.95E + 02 | 7.38E + 02 | HydroxyYamogenin-rhamnosyl(1,2)glucoside | − | − | + | + | + | 20.35 | 20.32 | 9.63 | 9.63 | 10.74 | 10.74 | 10.7 | |
| Hmdp003064 | 7.69E + 02 | 6.07E + 02 | 7.68E + 02 | Kingianoside H | − | − | − | + | − | 14.52 | 14.52 | −14.52 | |||||
| pmp000756 | 7.81E + 02 | 6.19E + 02 | 7.80E + 02 | Ruscogenin-1-O-carboxyglucosyl(1,2)rhamnoside | + | + | + | − | − | −19.11 | −17.63 | insig | 1.89 | −1.29 | insig | −19.52 | −1.48 |
| Hmdp003524 | 8.85E + 02 | 7.23E + 02 | 8.84E + 02 | Gracillin | + | + | − | + | + | 20.26 | 20.26 | 9.43 | 9.43 | 10.55 | 10.55 | 10.83 | |
| Zmcp005230 | 8.85E + 02 | 4.14E + 02 | 8.84E + 02 | Pennogenin-3-O-rhamnosyl(1→4)rhamnosyl(1→2)glucoside | + | − | + | − | − | 20.35 | 20.32 | 9.63 | 9.63 | 10.74 | 10.74 | 10.7 | |
| Zmcp005184 | 8.87E + 02 | 7.07E + 02 | 8.86E + 02 | 24-Hydroxypennogenin-3-O-rhamnosyl(1→2)xyloyl(1→4)glucoside | + | + | − | + | + | 14.52 | 14.52 | −14.52 | |||||
| Hmdp003423 | 9.01E + 02 | 7.39E + 02 | 9.00E + 02 | Neosibiricoside D | − | − | + | + | − | 16.25 | 16.25 | 11.21 | 11.21 | 5.04 | |||
| pmp000762 | 9.01E + 02 | 7.39E + 02 | 9.00E + 02 | Ruscogenin-1-O-rhamnosyl(1,2)glucosyl(1,2)glucoside | − | + | + | − | + | 20.40 | 8.35 | −2.04 | 17.7 | 5.65 | 20.4 | 12.04 | |
| Hmdp003403 | 9.01E + 02 | 7.39E + 02 | 9.00E + 02 | Polygonatoside D | + | + | + | + | + | 2.06 | 4.65 | insig | insig | −2.33 | insig | 1.22 | −2.59 |
| pmp001041 | 9.03E + 02 | 7.41E + 02 | 9.02E + 02 | Timosaponin B-III | + | − | − | −8.75 | 4.56 | 13.31 | −3.17 | insig | −13.31 | −8.75 | |||
| Hmdp002882 | 9.15E + 02 | 4.29E + 02 | 9.14E + 02 | (25S)-Pratioside D1 | + | − | + | − | − | 3.62 | 15.96 | −12.34 | −12.34 | 15.96 | −12.34 | ||
| pmp001042 | 9.19E + 02 | 7.57E + 02 | 9.18E + 02 | Timosaponin D | + | + | − | − | + | −9.83 | insig | −9.83 | insig | 3.49 | 4.68 | −1.18 | |
| Cmdp005440 | 9.31E + 02 | 4.27E + 02 | 9.30E + 02 | kingianoside G | + | − | − | + | − | −9.84 | insig | 9.06 | −9.84 | −9.06 | −9.84 | ||
| Cmdn006559 | 9.29E + 02 | 7.67E + 02 | 9.30E + 02 | kingianoside I | + | − | − | + | + | −9.68 | insig | 11.85 | −1.84 | 7.84 | −11.85 | −9.68 | |
| Zmcp005839 | 1.02E + 03 | 4.15E + 02 | 1.02E + 03 | Parisyunnanoside B | + | + | − | + | + | −16.57 | −16.35 | 3.13 | 3.35 | insig | insig | −19.69 | insig |
| Zmcp005801 | 1.03E + 03 | 4.15E + 02 | 1.03E + 03 | Diosgenin-3-O-glcosyl(1→4)rhamnosyl(1→4)rhamnosyl(1→2)glcoside | + | + | − | + | + | −15.90 | insig | insig | 10.87 | −8.08 | insig | −17.87 | −8.9 |
| Cmlp005341 | 1.03E + 03 | 8.69E + 02 | 1.03E + 03 | Pseudoprotodioscin | + | − | − | + | − | −16.65 | insig | 18.47 | −16.65 | −18.47 | −16.65 | ||
| Jmyp4441 | 1.05E + 03 | 4.15E + 02 | 1.05E + 03 | (3β,14α)-3-O-β-d-glucopyranosyl-(1→2)-[β-d-xylopyranosyl-(1→3)]-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside-yamogenin | + | + | + | + | + | −6.62 | −6.32 | −2.25 | −1.95 | insig | insig | −4.37 | insig |
| Hmqp005575 | 1.05E + 03 | 4.15E + 02 | 1.05E + 03 | Aculeatiside A | + | + | + | + | + | −6.58 | −6.59 | −2.01 | insig | insig | insig | −4.57 | insig |
| Jmyp4236 | 1.06E + 03 | 4.15E + 02 | 1.06E + 03 | polygonatumoside F | + | + | + | + | + | insig | insig | 2.87 | 3.62 | −1.06 | insig | −3.23 | insig |
| Jmyp3897 | 1.08E + 03 | 4.13E + 02 | 1.08E + 03 | (25s)-spirost-5en-3β,14α-diol-3-O-β-d-glucopyranosyl-(1→2)-[β-d-glucopyranosyl-(1→3)]-β-d-glucopyranosyl-(1→4)-β-d-galactopyranoside | + | + | − | + | + | −11.51 | −12.14 | insig | insig | 5.28 | 4.65 | −10.03 | insig |
| Jmyp3798 | 1.21E + 03 | 5.91E + 02 | 1.21E + 03 | Polygonatumoside A | + | + | + | + | + | 3.92 | 5.37 | insig | 2.05 | insig | 2.43 | 3.33 | −1.45 |
| Group Name | R2X | R2Y | Q2 |
|---|---|---|---|
| Ps1_vs._Pc | 0.806 | 1 | 0.966 |
| Ps1_vs._Pk | 0.834 | 1 | 0.990 |
| Ps1_vs._Pm | 0.717 | 1 | 0.965 |
| Ps2_vs._Pc | 0.829 | 1 | 0.989 |
| Ps2_vs._Pk | 0.790 | 0.999 | 0.984 |
| Ps2_vs._Pm | 0.759 | 1 | 0.941 |
| Ps1_vs._Ps2 | 0.650 | 1 | 0.868 |
| Pk_vs._Pc | 0.809 | 1 | 0.966 |
| Group Name | Number of Insig Diff | Number of Sig Diff | Down | Up | Sig Diff % |
|---|---|---|---|---|---|
| Ps1_vs._Pc | 329 | 522 | 90 | 432 | 61.34% |
| Ps1_vs._Pk | 377 | 483 | 99 | 384 | 56.16% |
| Ps1_vs._Pm | 486 | 344 | 122 | 222 | 41.45% |
| Ps2_vs._Pc | 365 | 480 | 98 | 382 | 56.80% |
| Ps2_vs._Pk | 410 | 440 | 101 | 339 | 51.76% |
| Ps2_vs._Pm | 511 | 315 | 143 | 172 | 38.27% |
| Ps1_vs._Ps2 | 591 | 221 | 65 | 156 | 27.22% |
| Pk_vs._Pc | 462 | 401 | 179 | 222 | 46.47% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, J.; Wei, J.; Liao, D.; Ding, Z.; Yao, X.; Sun, P.; Li, X. Untargeted Metabolomics Analysis Revealed the Major Metabolites in the Seeds of four Polygonatum Species. Molecules 2022, 27, 1445. https://doi.org/10.3390/molecules27041445
Qi J, Wei J, Liao D, Ding Z, Yao X, Sun P, Li X. Untargeted Metabolomics Analysis Revealed the Major Metabolites in the Seeds of four Polygonatum Species. Molecules. 2022; 27(4):1445. https://doi.org/10.3390/molecules27041445
Chicago/Turabian StyleQi, Jianjun, Jianhe Wei, Dengqun Liao, Zimian Ding, Xia Yao, Peng Sun, and Xianen Li. 2022. "Untargeted Metabolomics Analysis Revealed the Major Metabolites in the Seeds of four Polygonatum Species" Molecules 27, no. 4: 1445. https://doi.org/10.3390/molecules27041445
APA StyleQi, J., Wei, J., Liao, D., Ding, Z., Yao, X., Sun, P., & Li, X. (2022). Untargeted Metabolomics Analysis Revealed the Major Metabolites in the Seeds of four Polygonatum Species. Molecules, 27(4), 1445. https://doi.org/10.3390/molecules27041445






