Ruthenium(II) Polypyridyl Complexes and Their Use as Probes and Photoreactive Agents for G-quadruplexes Labelling
Abstract
:1. Introduction
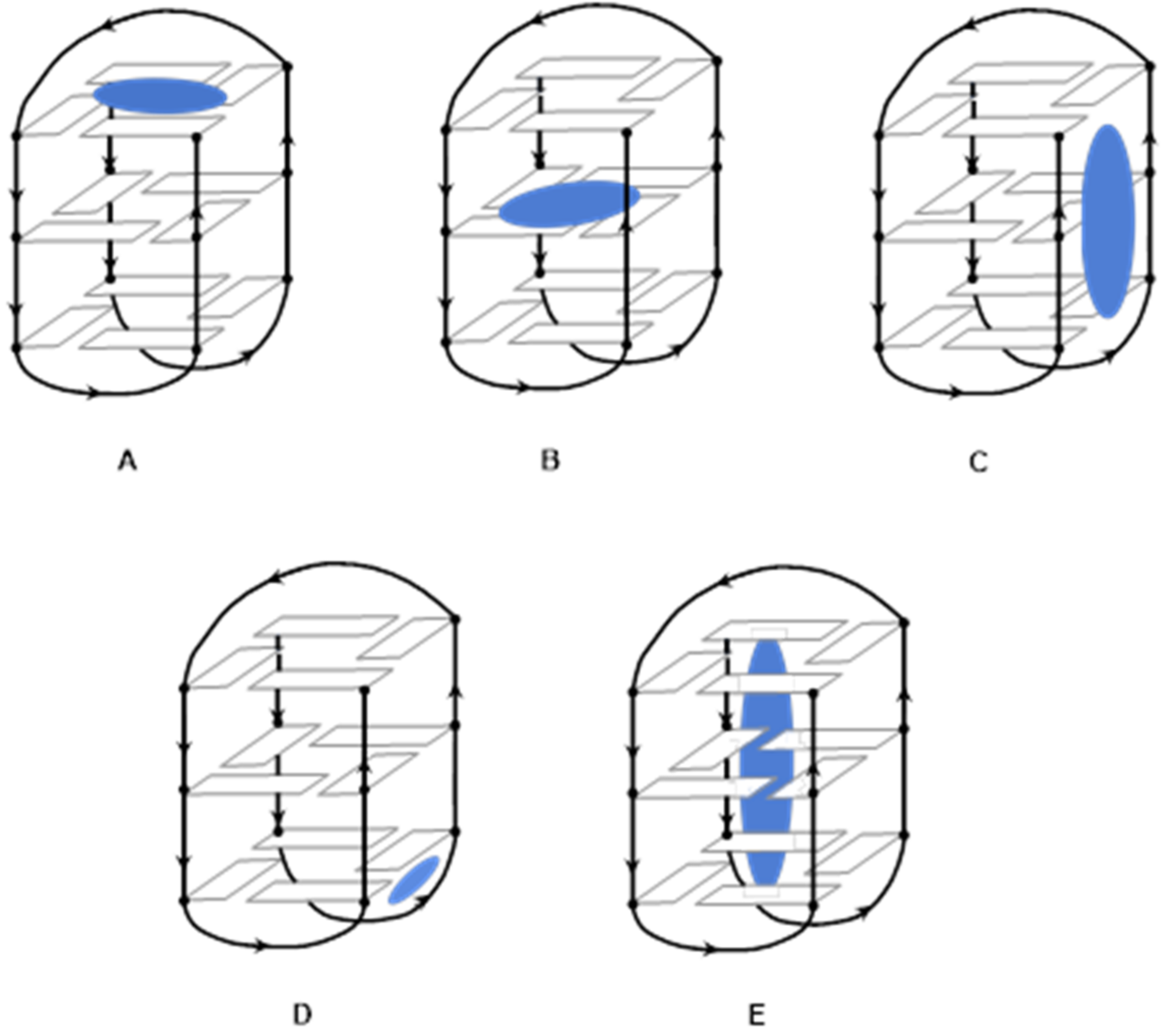
1.1. G-quadruplex Structures
1.2. G-quadruplexes in Human Genome
1.2.1. Telomeres and Their Functions
1.2.2. Cellular Replication
2. Classification and Characteristics of G4 Ligands
2.1. Organic G4 Ligands
2.1.1. Stacking on the Terminal Tetrads
In Situ Protonated Ligands
N-methylated Ligands
2.1.2. Intercalation between Guanine Tetrads
2.1.3. Interaction with Grooves and Loops
2.1.4. Central Insertion
2.2. Metal Complexes-Based G4 Ligands
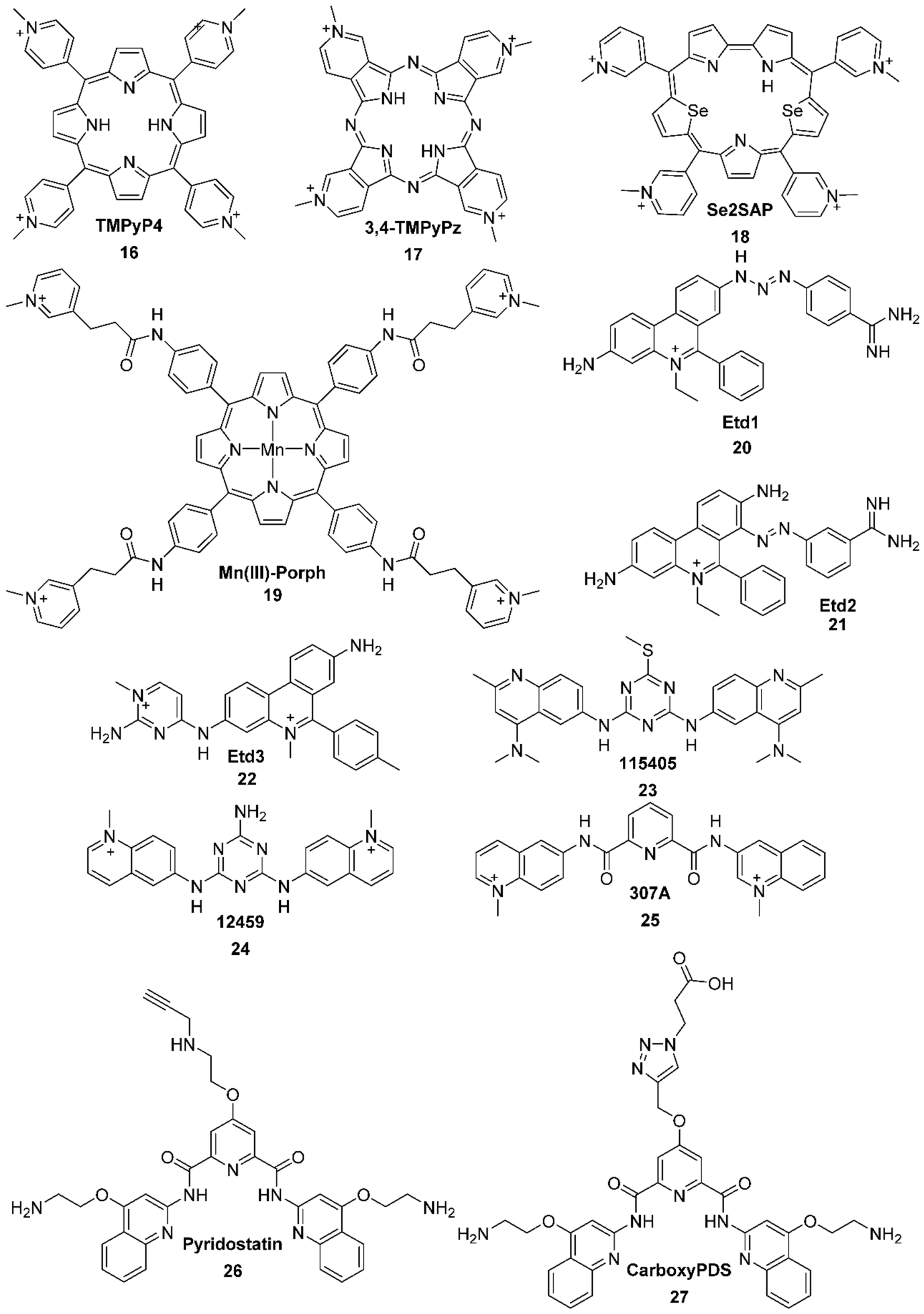
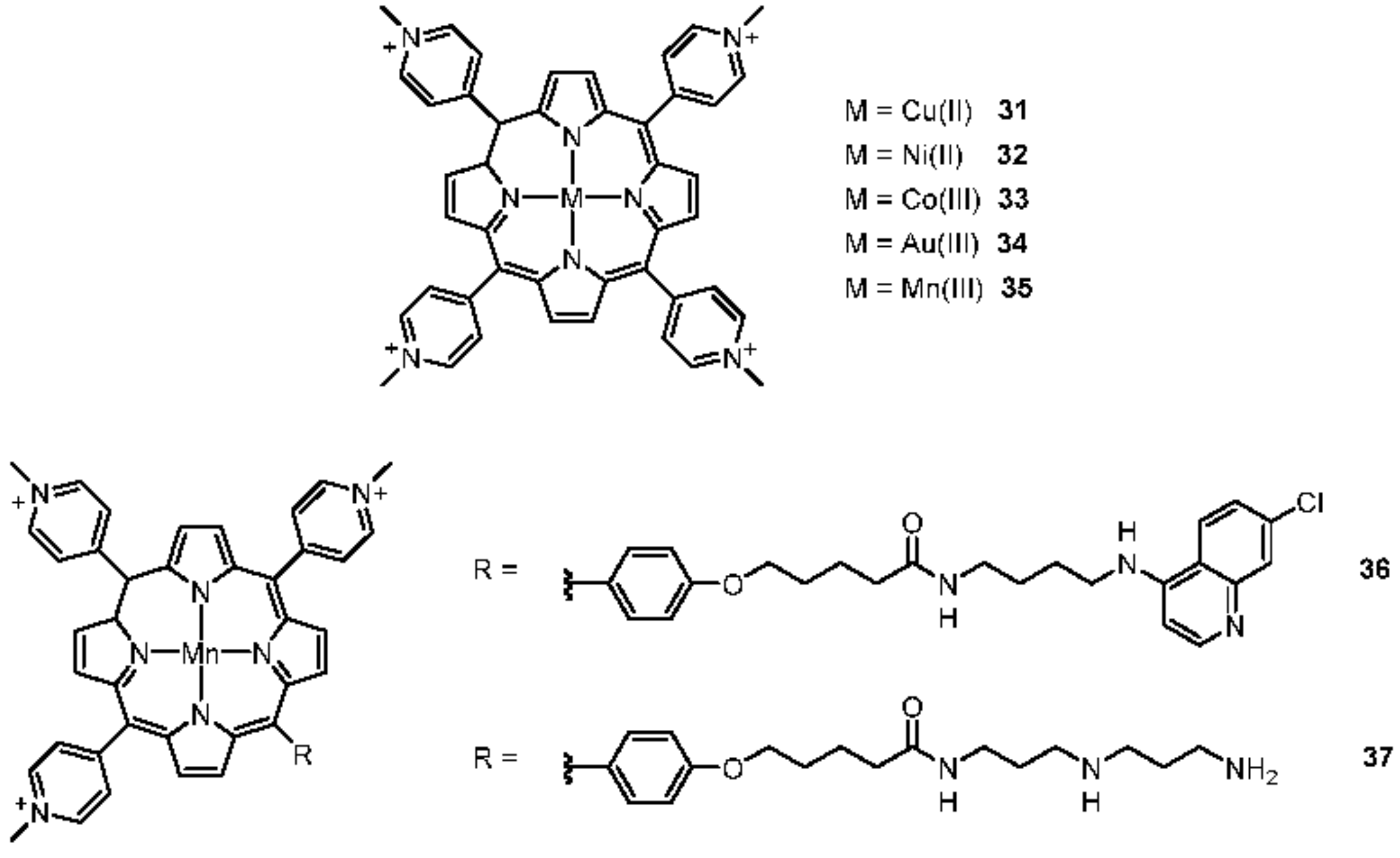
2.2.1. Porphyrines
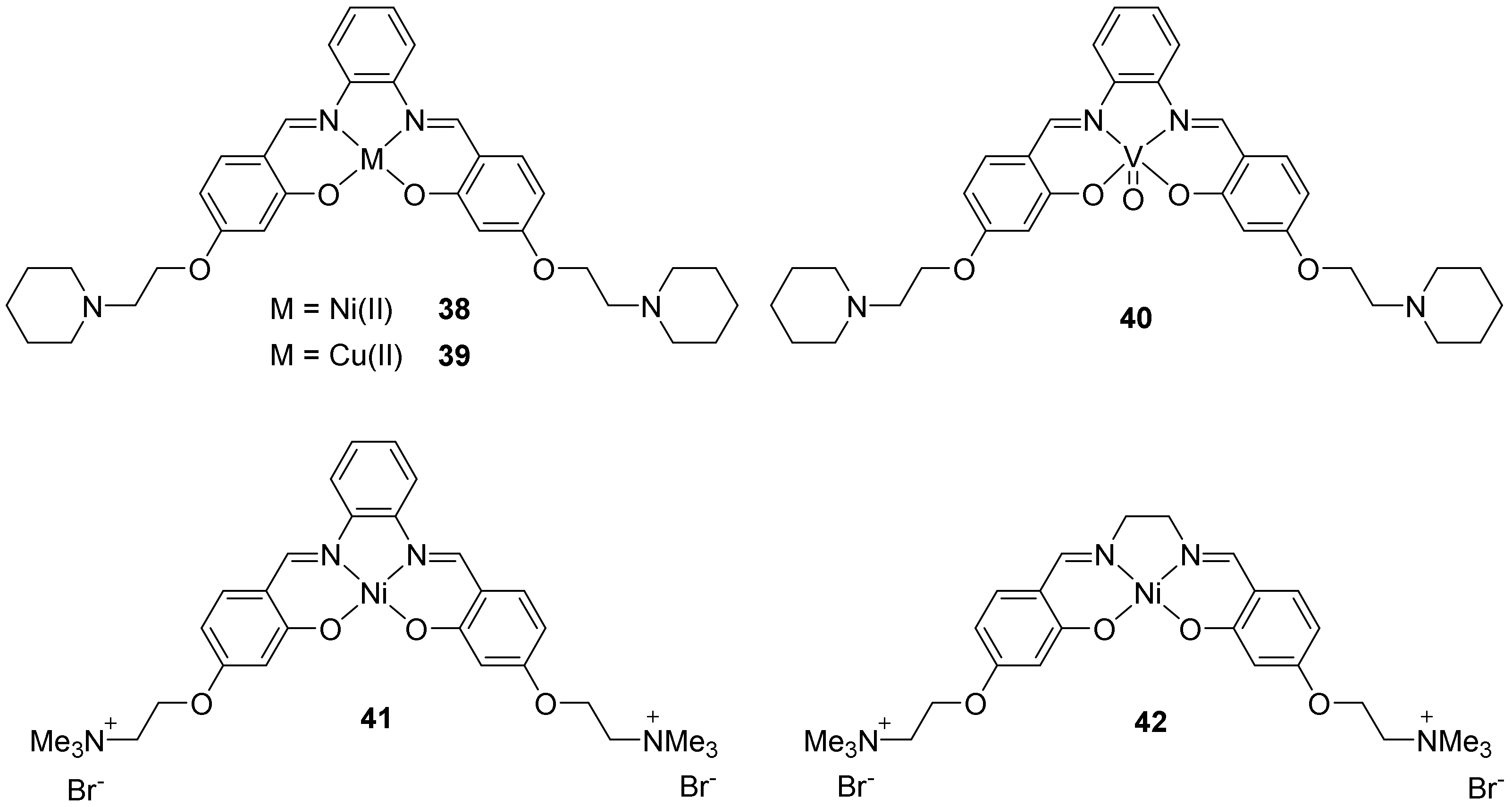
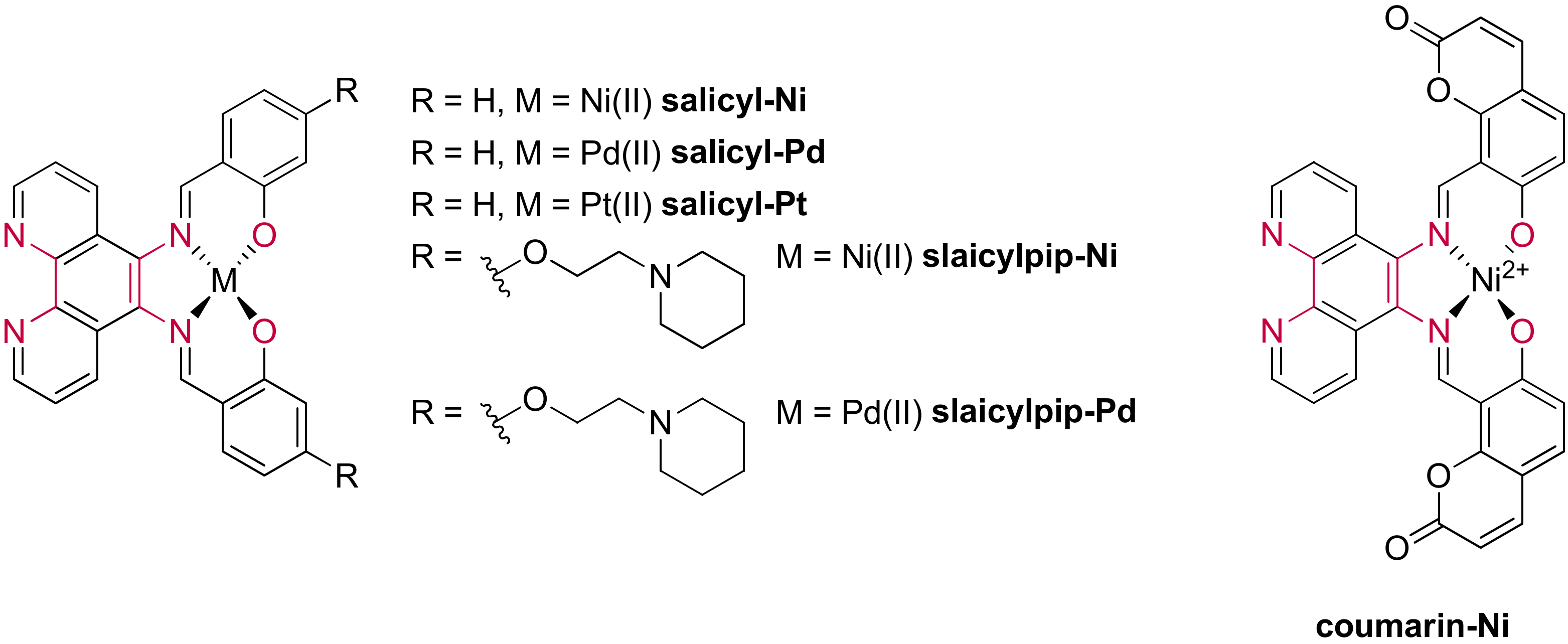
2.2.2. Salphens
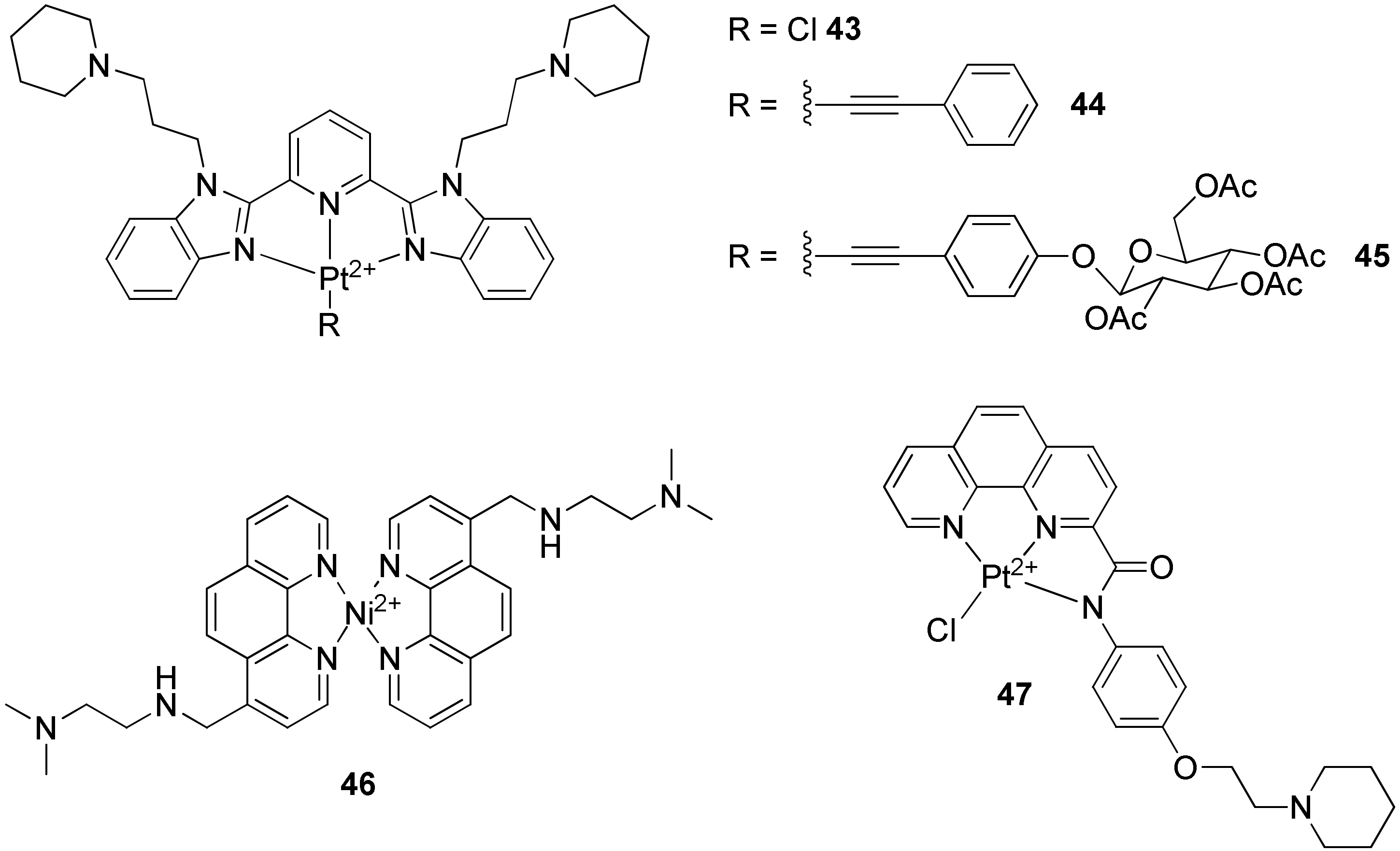
2.2.3. Terpyridines and Other Square Planar Complexes
3. Interactions of Ruthenium(II) Complexes with G-quadruplexes
3.1. Monometallic Ruthenium(II) Complexes
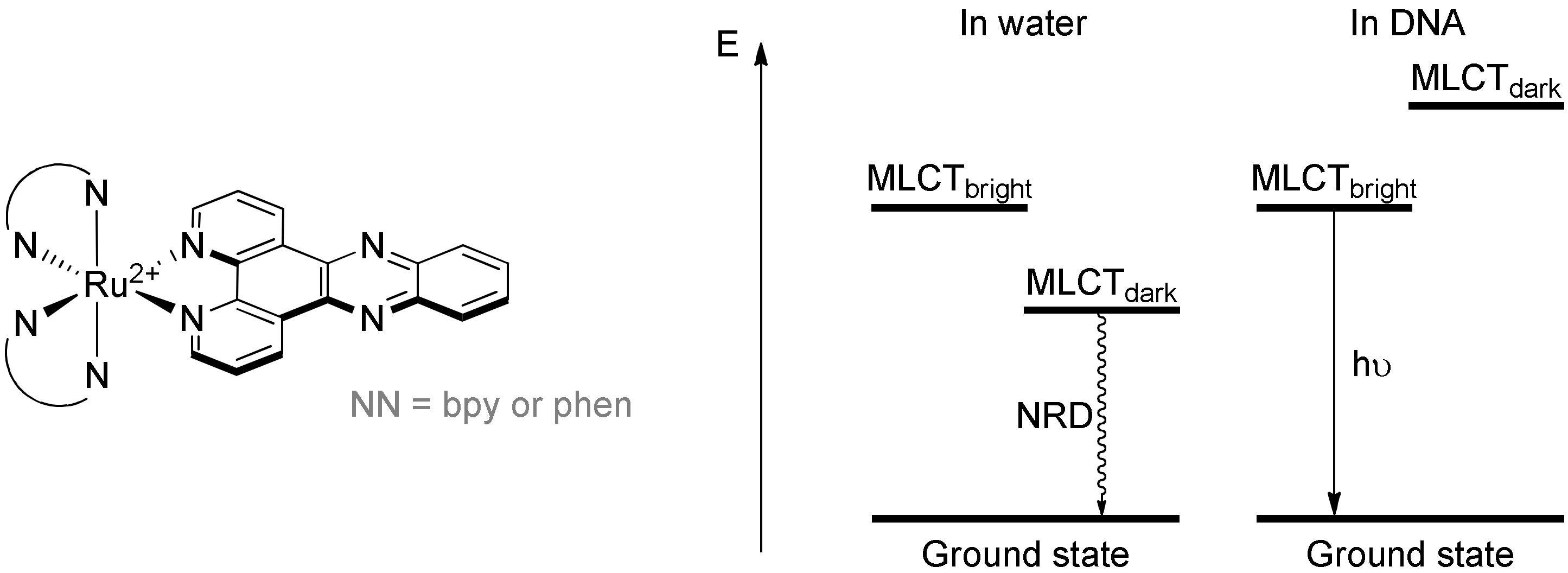
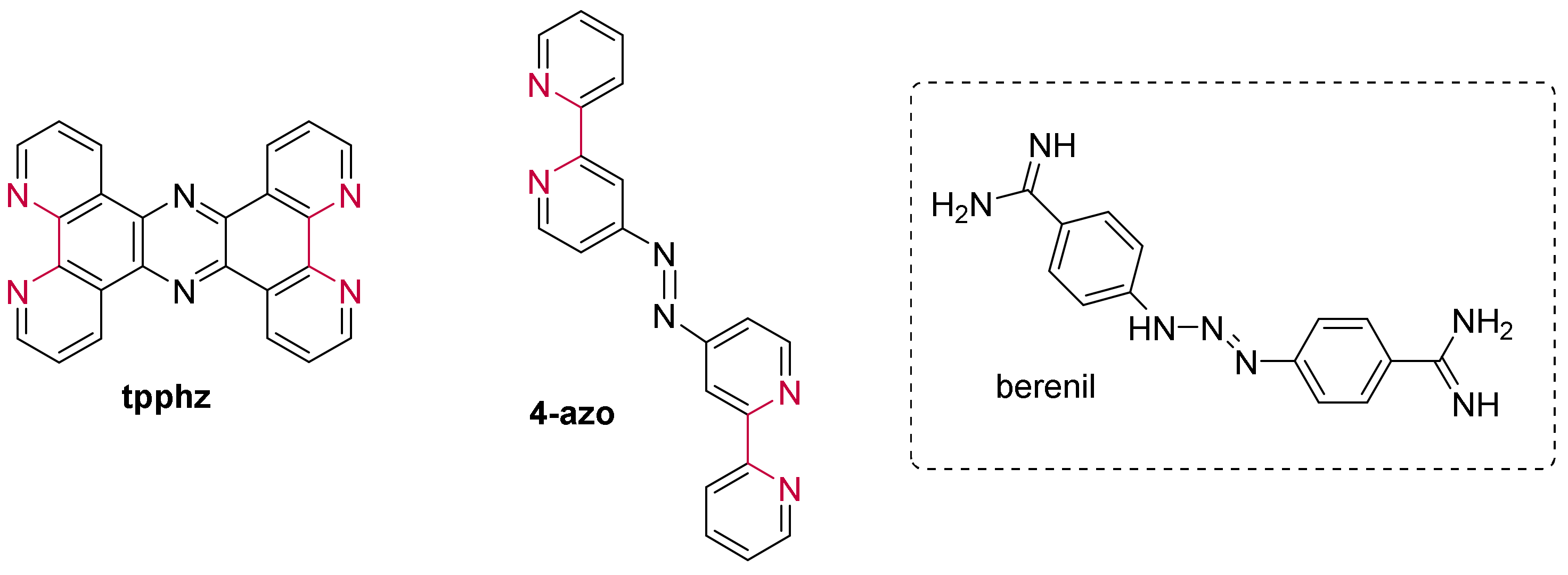
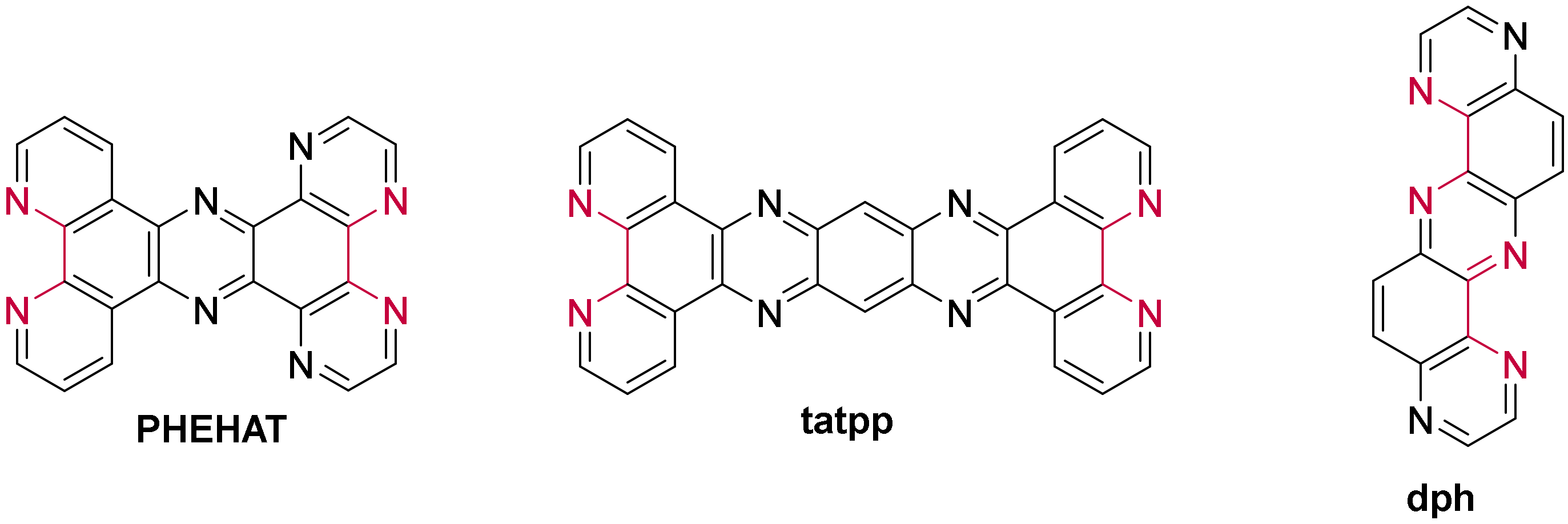
3.1.1. Ruthenium(II) dppz and dppz Derivative Complexes
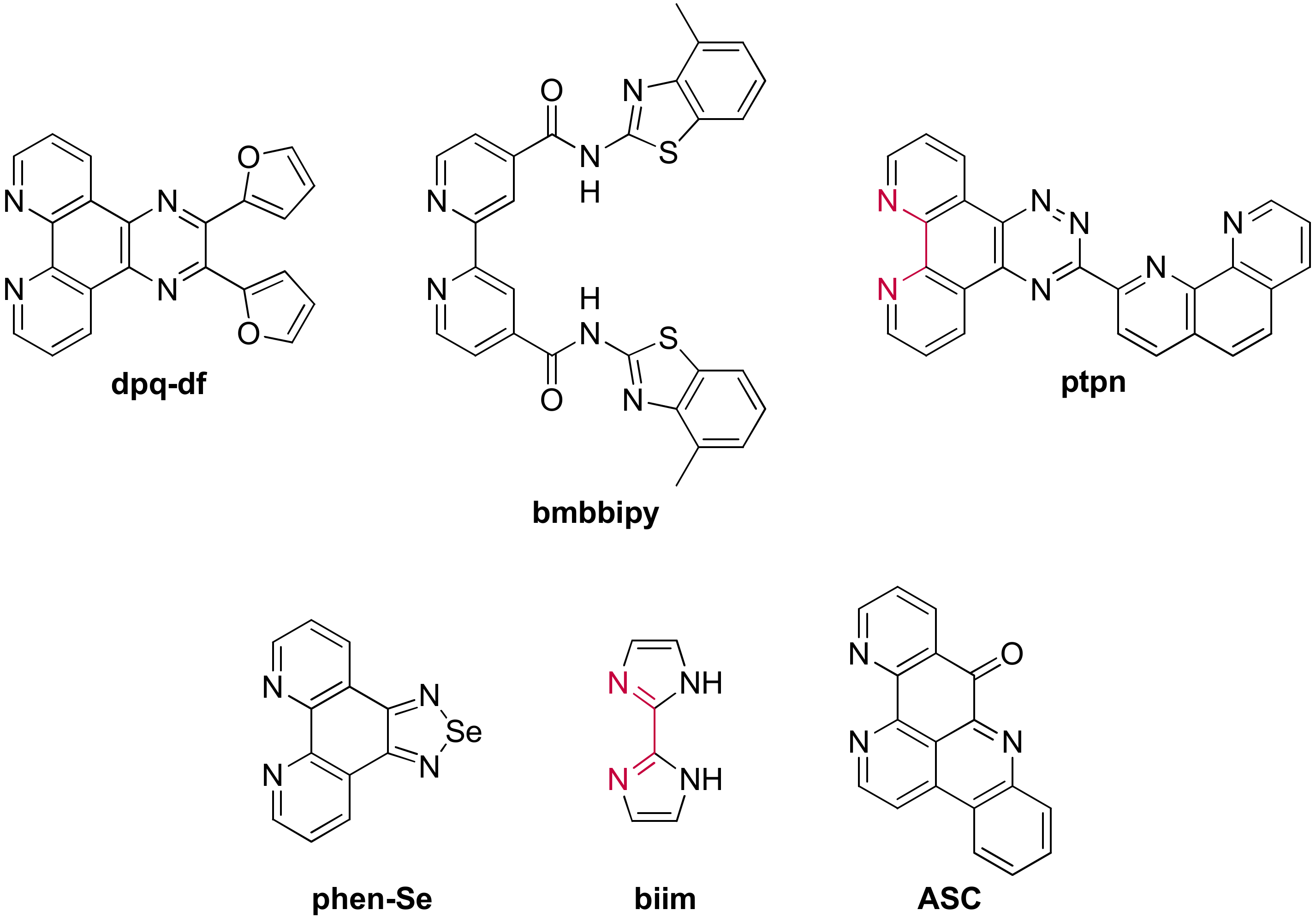
3.1.2. Ruthenium(II) Complexes Bearing a π-Extended Ligand
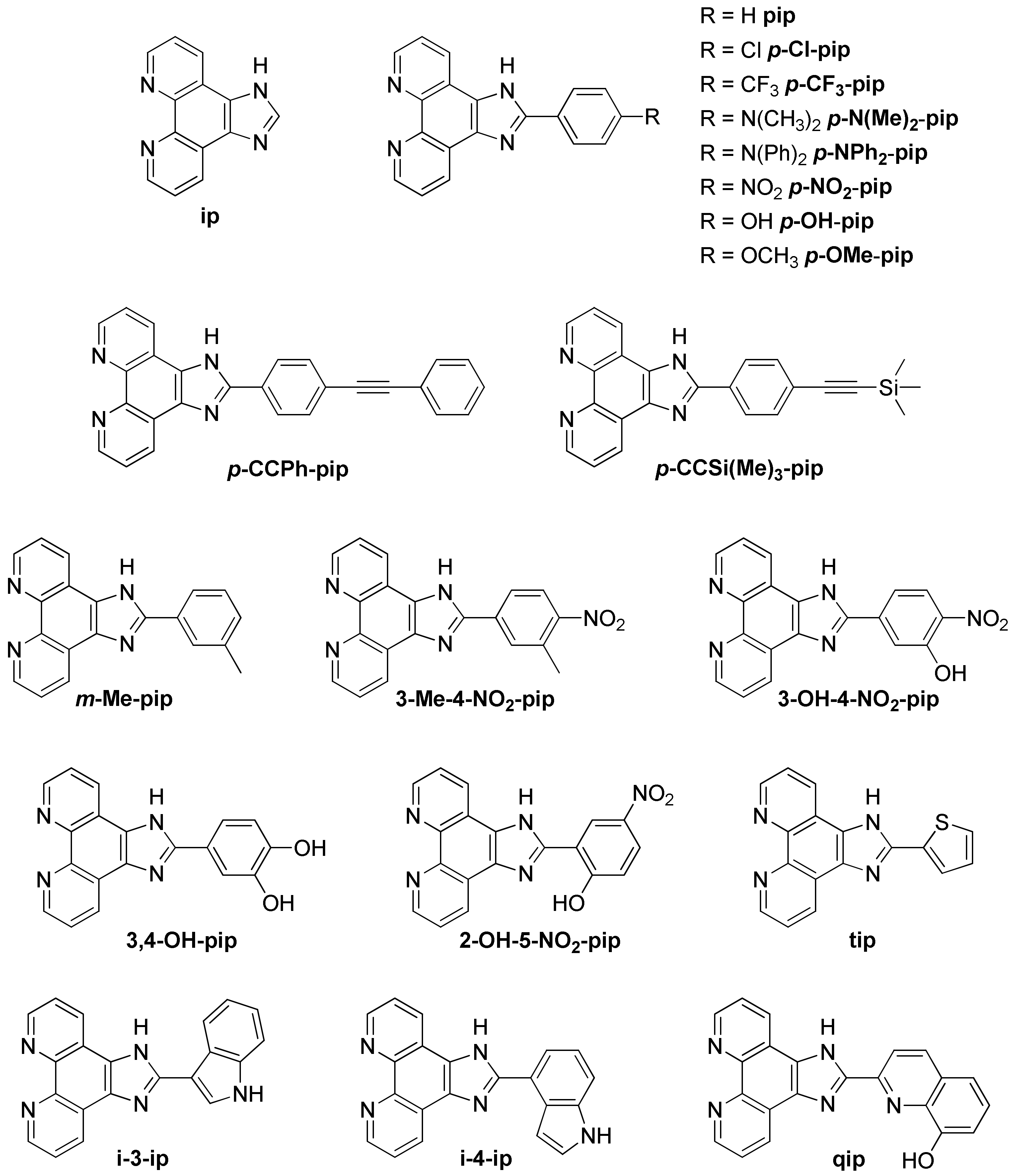
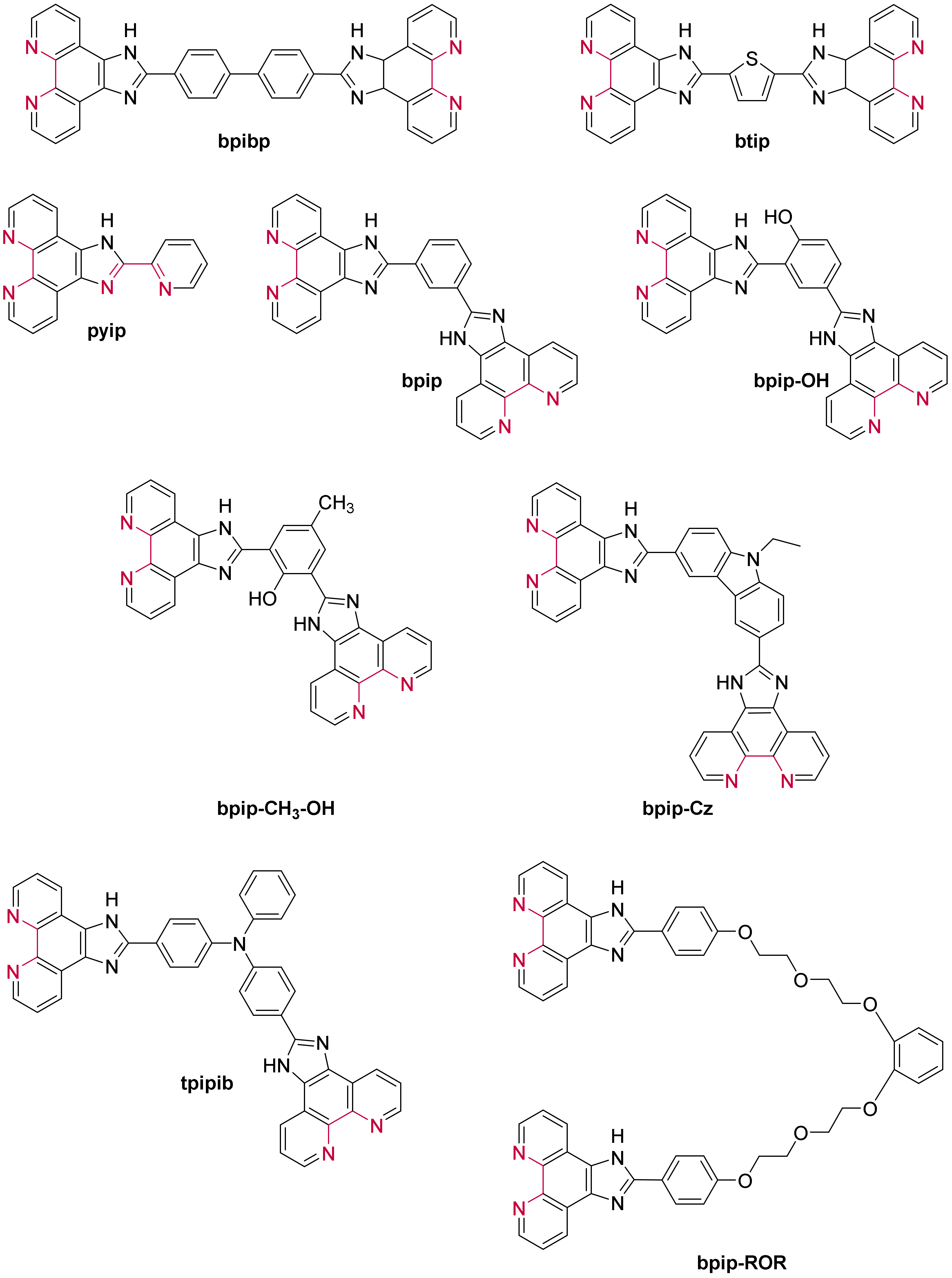
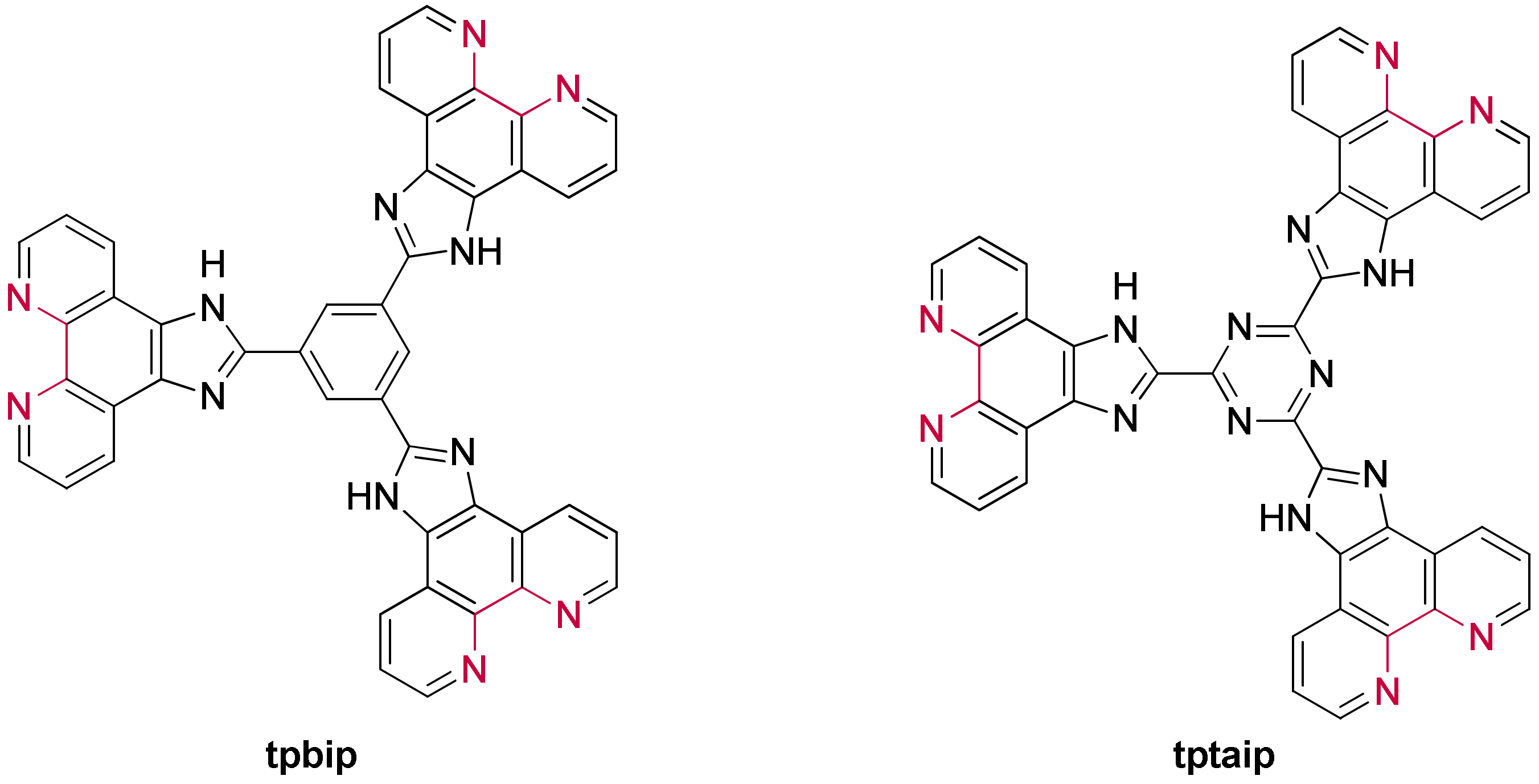
3.1.3. Ruthenium(II) Complexes Bearing Imidazo-Phenanthroline Ligands
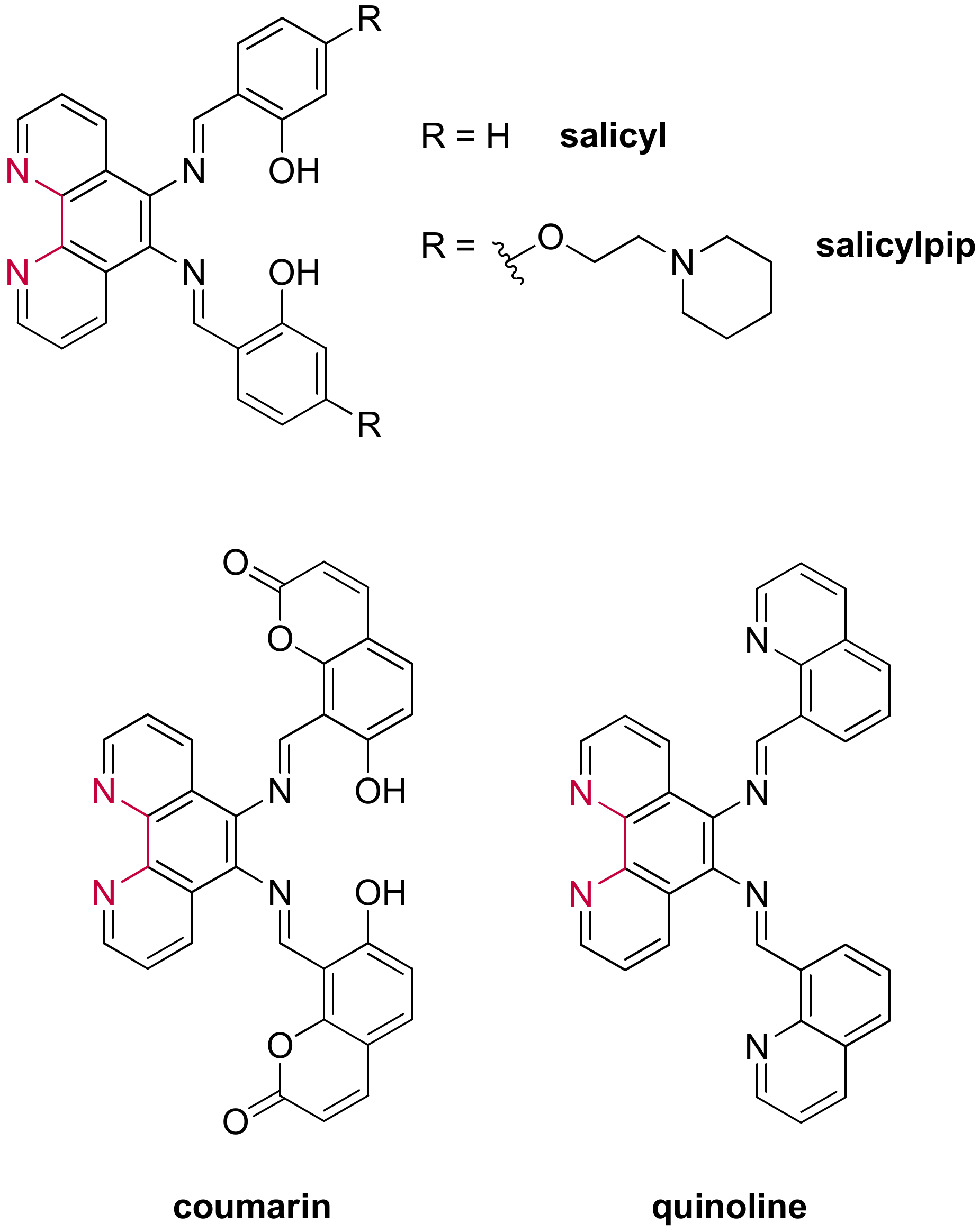
3.1.4. Ruthenium(II) Complexes Bearing Schiff Base-Type Ligands
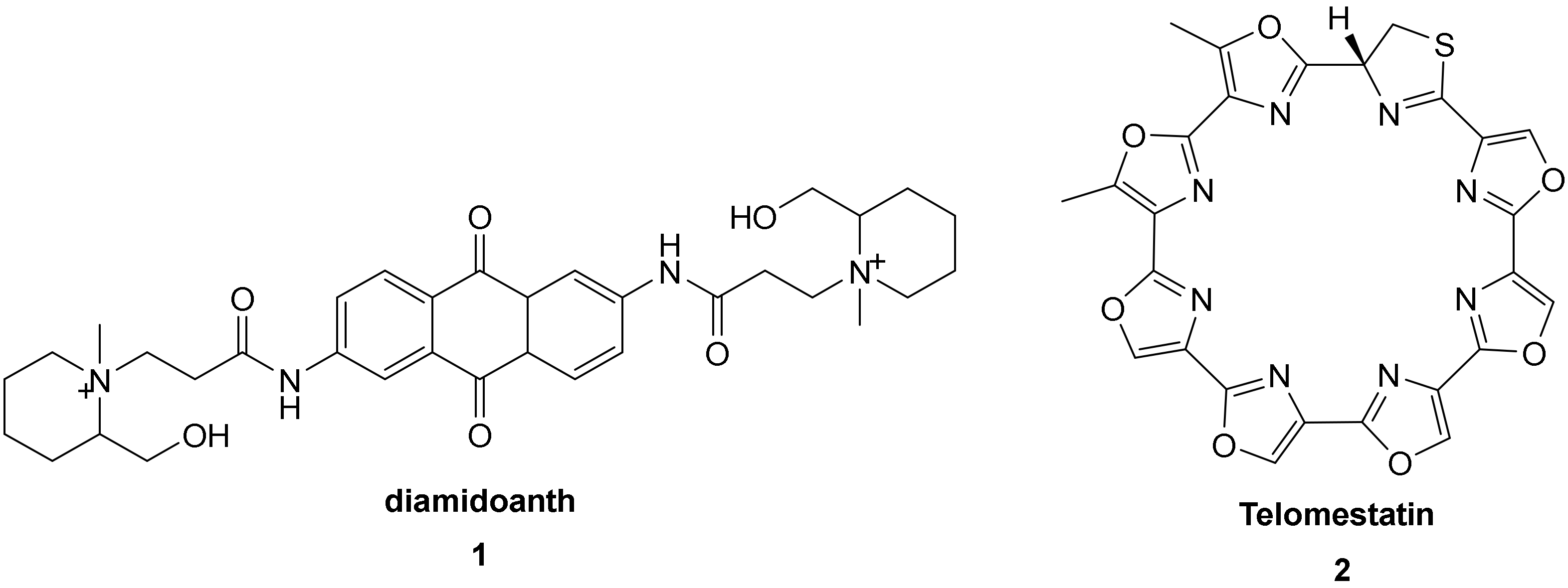
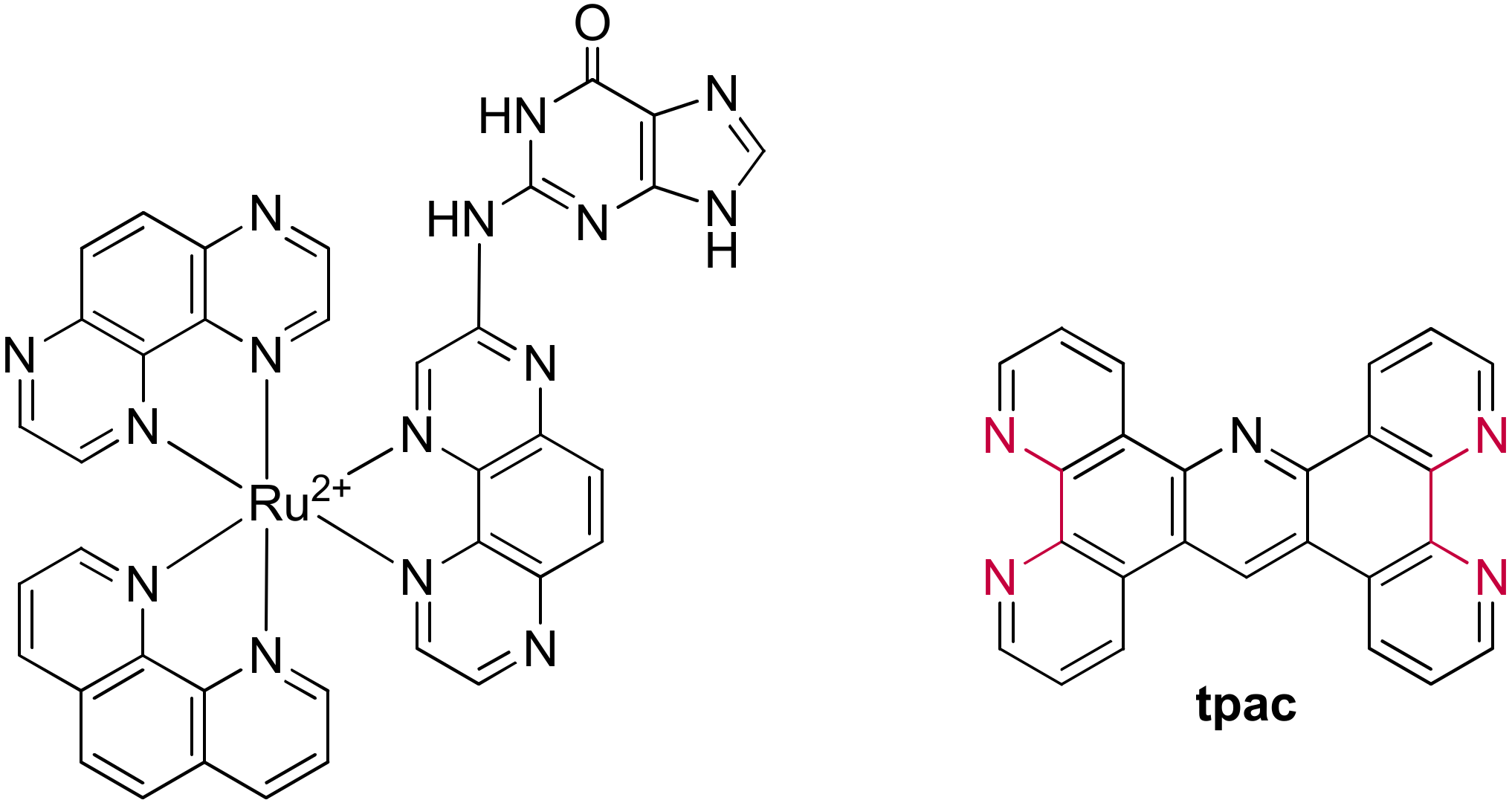
3.2. Bimetallic and Trimetallic Ruthenium(II) Complexes
The First G-quadruplex “Light-Switch” Ruthenium(II) Complexes
4. Photoreaction of Ruthenium(II) Complexes with G-quadruplexes
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Watson, J.D.; Crick, F.H.C. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Gehring, K.; Leroy, J.-L.; Guéron, M. A tetrameric DNA structure with protonated cytosine-cytosine base pairs. Nature 1993, 363, 561–565. [Google Scholar] [CrossRef]
- Phan, A.T.; Mergny, J.-L. Human telomeric DNA: G-quadruplexes, i-motif, and Watson-Crick double helix. Nucleic Acids Res. 2002, 30, 4618–4625. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Lombardía, M.; González, A.; Eritja, R.; Aymamí, J.; Azorin, F.; Coll, M. Crystal structure of a DNA Holliday junction. Nat. Genet. 1999, 6, 913–917. [Google Scholar] [CrossRef] [Green Version]
- Van Rixel, V.H.S.; Busemann, A.; Wissingh, M.F.; Hopkins, S.L.; Siewert, B.; Van de Griend, C.; Siegler, M.A.; Marzo, T.; Papi, F.; Ferraroni, M.; et al. Induction of a Four-Way Junction Structure in the DNA Palindromic Hexanucleotide 5″-d(CGTACG)-3″ by a Mononuclear Platinum Complex. Angew. Chem. Int. Ed. 2019, 58, 9378–9382. [Google Scholar] [CrossRef] [PubMed]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmerman, S.B.; Cohen, G.H.; Davies, D.R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J. Mol. Biol. 1975, 92, 181–192. [Google Scholar] [CrossRef]
- Arnott, S.; Chandrasekaran, R.; Marttila, C.M. Structures for polyinosinic acid and polyguanylic acid. Biochem. J. 1974, 141, 537–543. [Google Scholar] [CrossRef] [Green Version]
- Howard, F.B.; Frazier, J.; Miles, H.T. Stable and metastable forms of poly(G). Biopolymers 1977, 16, 791–809. [Google Scholar] [CrossRef]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [Green Version]
- Kendrick, S.; Hurley, L.H. The Role of G-quadruplex/i-Motif Secondary Structures as Cis-Acting Regulatory Elements. Pure Appl. Chem. 2010, 82, 1609–1621. [Google Scholar] [CrossRef]
- Wright, W.E.; Tesmer, V.M.; Huffman, K.E.; Levene, S.D.; Shay, J.W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997, 11, 2801–2809. [Google Scholar] [CrossRef] [Green Version]
- Sfeir, A.J.; Chai, W.; Shay, J.W.; Wright, W.E. Telomere-End Processing the Terminal Nucleotides of Human Chromosomes. Mol. Cell 2005, 18, 131–138. [Google Scholar] [CrossRef]
- Monchaud, D.; Teulade-Fichou, M.-P. A hitchhiker’s guide to G-quadruplex ligands. Org. Biomol. Chem. 2007, 6, 627–636. [Google Scholar] [CrossRef]
- De Cian, A.; Lacroix, L.; Douarre, C.; Temime-Smaali, N.; Trentesaux, C.; Riou, J.-F.; Mergny, J.-L. Targeting telomeres and telomerase. Biochimie 2008, 90, 131–155. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, Y.; Zhang, W. G-quadruplex Structures and Their Interaction Diversity with Ligands. ChemMedChem 2014, 9, 899–911. [Google Scholar] [CrossRef]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef] [Green Version]
- Huppert, J.L.; Balasubramanian, S. Prevalence of Quadruplexes in the Human Genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef] [Green Version]
- Todd, A.K.; Johnston, M.; Neidle, S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [Google Scholar] [CrossRef] [Green Version]
- Ou, T.-M.; Lu, Y.-J.; Tan, J.-H.; Huang, Z.-S.; Wong, K.-Y.; Gu, L.-Q. G-quadruplexes: Targets in Anticancer Drug Design. ChemMedChem 2008, 3, 690–713. [Google Scholar] [CrossRef]
- Davis, J.T. G-Quartets 40 Years Later: From 5′-GMP to Molecular Biology and Supramolecular Chemistry. Angew. Chem. Int. Ed. 2004, 43, 668–698. [Google Scholar] [CrossRef] [PubMed]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, D.J.; Phan, A.T.; Kuryavyi, V. Human telomere, oncogenic promoter and 5’-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007, 35, 7429–7455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkinson, G.N.; Lee, M.P.H.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Patel, D.J. Two-Repeat Human Telomeric d(TAGGGTTAGGGT) Sequence Forms Interconverting Parallel and Antiparallel G-quadruplexes in Solution: Distinct Topologies, Thermodynamic Properties, and Folding/Unfolding Kinetics. J. Am. Chem. Soc. 2003, 125, 15021–15027. [Google Scholar] [CrossRef] [Green Version]
- Phan, A.T.; Modi, Y.S.; Patel, D.J. Two-repeat Tetrahymena Telomeric d(TGGGGTTGGGGT) Sequence Interconverts Between Asymmetric Dimeric G-quadruplexes in Solution. J. Mol. Biol. 2004, 338, 93–102. [Google Scholar] [CrossRef] [Green Version]
- Kettani, A.; Bouaziz, S.; Gorin, A.; Zhao, H.; Jones, R.A.; Patel, D.J. Solution structure of a Na cation stabilized DNA quadruplex containing G·G·G·G and G·C·G·C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA. J. Mol. Biol. 1998, 282, 619–636. [Google Scholar] [CrossRef] [PubMed]
- Hazel, P.; Parkinson, G.N.; Neidle, S. Topology Variation and Loop Structural Homology in Crystal and Simulated Structures of a Bimolecular DNA Quadruplex. J. Am. Chem. Soc. 2006, 128, 5480–5487. [Google Scholar] [CrossRef]
- Gill, M.L.; Strobel, S.A.; Loria, J.P. 205Tl NMR Methods for the Characterization of Monovalent Cation Binding to Nucleic Acids. J. Am. Chem. Soc. 2005, 127, 16723–16732. [Google Scholar] [CrossRef]
- Haider, S.; Parkinson, G.N.; Neidle, S. Crystal Structure of the Potassium Form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 2002, 320, 189–200. [Google Scholar] [CrossRef]
- Schultze, P.; Hud, N.V.; Smith, F.W.; Feigon, J. The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G4T4G4). Nucleic Acids Res. 1999, 27, 3018–3028. [Google Scholar] [CrossRef] [Green Version]
- Schultze, P.; Smith, F.W.; Feigon, J. Refined solution structure of the dimeric quadruplex formed from the Oxytricha telomeric oligonucleotide d(GGGGTTTTGGGG). Structure 1994, 2, 221–233. [Google Scholar] [CrossRef] [Green Version]
- Smith, F.W.; Feigon, J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature 1992, 356, 164–168. [Google Scholar] [CrossRef]
- Phan, A.T.; Kuryavyi, V.; Burge, S.; Neidle, S.; Patel, D.J. Structure of an Unprecedented G-quadruplex Scaffold in the Human c-kit Promoter. J. Am. Chem. Soc. 2007, 129, 4386–4392. [Google Scholar] [CrossRef] [Green Version]
- Olsen, C.M.; Gmeiner, W.H.; Marky, L.A. Unfolding of G-quadruplexes: Energetic, and Ion and Water Contributions of G-Quartet Stacking. J. Phys. Chem. B 2006, 110, 6962–6969. [Google Scholar] [CrossRef]
- Marathias, V.M.; Bolton, P.H. Determinants of DNA Quadruplex Structural Type: Sequence and Potassium Binding. Biochemistry 1999, 38, 4355–4364. [Google Scholar] [CrossRef]
- Risitano, A.; Fox, K.R. Inosine substitutions demonstrate that intramolecular DNA quadruplexes adopt different conformations in the presence of sodium and potassium. Bioorg. Med. Chem. Lett. 2005, 15, 2047–2050. [Google Scholar] [CrossRef]
- Rujan, I.N.; Meleney, J.C.; Bolton, P.H. Vertebrate Telomere Repeat DNAs Favor External Loop Propeller Quadruplex Structures in the Presence of High Concentrations of Potassium. Nucleic Acids Res. 2005, 33, 2022–2031. [Google Scholar] [CrossRef] [Green Version]
- Ying, L.; Green, J.J.; Li, H.; Klenerman, D.; Balasubramanian, S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 2003, 100, 14629–14634. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.; Okumus, B.; Kim, D.S.; Ha, T. Extreme conformational diversity in human telomeric DNA. Proc. Natl. Acad. Sci. USA 2005, 102, 18938–18943. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Phan, A.T.; Patel, D.J. (3 + 1) Assembly of Three Human Telomeric Repeats into an Asymmetric Dimeric G-quadruplex. J. Am. Chem. Soc. 2005, 127, 17277–17285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Patel, D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1993, 1, 263–282. [Google Scholar] [CrossRef]
- Balagurumoorthy, P.; Brahmachari, S. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994, 269, 21858–21869. [Google Scholar] [CrossRef]
- Li, J.; Correia, J.J.; Wang, L.; Trent, J.O.; Chaires, J.B. Not so Crystal Clear: The Structure of the Human Telomere G-quadruplex in Solution Differs from That Present in a Crystal. Nucleic Acids Res. 2005, 33, 4649–4659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.-C.; Chu, J.-F.; Kao, F.-J.; Chiu, Y.-C.; Lou, P.-J.; Chen, H.-C.; Chang, T.-C. Verification of Antiparallel G-quadruplex Structure in Human Telomeres by Using Two-Photon Excitation Fluorescence Lifetime Imaging Microscopy of the 3,6-Bis(1-methyl-4-vinylpyridinium)carbazole Diiodide Molecule. Anal. Chem. 2006, 78, 2810–2815. [Google Scholar] [CrossRef] [PubMed]
- Hazel, P.; Huppert, J.; Balasubramanian, S.; Neidle, S. Loop-Length-Dependent Folding of G-quadruplexes. J. Am. Chem. Soc. 2004, 126, 16405–16415. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, E.H. Structure and function of telomeres. Nature 1991, 350, 569–573. [Google Scholar] [CrossRef]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.D.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef]
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626. [Google Scholar] [CrossRef] [Green Version]
- Blackburn, E.H. Switching and Signaling at the Telomere. Cell 2001, 106, 661–673. [Google Scholar] [CrossRef] [Green Version]
- Stewart, S.A.; Weinberg, R.A. Telomeres: Cancer to Human Aging. Annu. Rev. Cell Dev. Biol. 2006, 22, 531–557. [Google Scholar] [CrossRef] [Green Version]
- D’Adda Di Fagagna, F.; Teo, S.H.; Jackson, S.P. Functional Links between Telomeres and Proteins of the DNA-Damage Response. Genes Dev. 2004, 18, 1781–1799. [Google Scholar] [CrossRef] [Green Version]
- Zaug, A.J.; Podell, E.R.; Cech, T.R. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. USA 2005, 102, 10864–10869. [Google Scholar] [CrossRef] [Green Version]
- Denchi, E.L.; De Lange, T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 2007, 448, 1068–1071. [Google Scholar] [CrossRef]
- Miyoshi, T.; Kanoh, J.; Saito, M.; Ishikawa, F. Fission Yeast Pot1-Tpp1 Protects Telomeres and Regulates Telomere Length. Science 2008, 320, 1341–1344. [Google Scholar] [CrossRef]
- Gomez, D.; O’Donohue, M.-F.; Wenner, T.; Douarre, C.; Macadré, J.; Koebel, P.; Giraud-Panis, M.-J.; Kaplan, H.; Kolkes, A.; Shin-Ya, K.; et al. The G-quadruplex Ligand Telomestatin Inhibits POT1 Binding to Telomeric Sequences In vitro and Induces GFP-POT1 Dissociation from Telomeres in Human Cells. Cancer Res. 2006, 66, 6908–6912. [Google Scholar] [CrossRef] [Green Version]
- Gomez, D.; Wenner, T.; Brassart, B.; Douarre, C.; O’Donohue, M.-F.; El Khoury, V.; Shin-Ya, K.; Morjani, H.; Trentesaux, C.; Riou, J.-F. Telomestatin-induced Telomere Uncapping Is Modulated by POT1 through G-overhang Extension in HT1080 Human Tumor Cells. J. Biol. Chem. 2006, 281, 38721–38729. [Google Scholar] [CrossRef] [Green Version]
- Gunaratnam, M.; Greciano, O.; Martins, C.; Reszka, A.P.; Schultes, C.M.; Morjani, H.; Riou, J.-F.; Neidle, S. Mechanism of acridine-based telomerase inhibition and telomere shortening. Biochem. Pharmacol. 2007, 74, 679–689. [Google Scholar] [CrossRef]
- D’Adda Di Fagagna, F.; Reaper, P.M.; Clay-Farrace, L.; Fiegler, H.; Carr, P.; Von Zglinicki, T.; Saretzki, G.; Carter, N.P.; Jackson, S.P. A DNA Damage Checkpoint Response in Telomere-Initiated Senescence. Nature 2003, 426, 194–198. [Google Scholar] [CrossRef]
- Tauchi, T.; Shin-Ya, K.; Sashida, G.; Sumi, M.; Nakajima, A.; Shimamoto, T.; Ohyashiki, J.H.; Ohyashiki, K. Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: Involvement of ATM-dependent DNA damage response pathways. Oncogene 2003, 22, 5338–5347. [Google Scholar] [CrossRef] [Green Version]
- Salvati, E.; Leonetti, C.; Rizzo, A.; Scarsella, M.; Mottolese, M.; Galati, R.; Sperduti, I.; Stevens, M.F.; D’Incalci, M.; Blasco, M.A.; et al. Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. J. Clin. Investig. 2007, 117, 3236–3247. [Google Scholar] [CrossRef] [Green Version]
- Qi, H.; Lin, C.-P.; Fu, X.; Wood, L.M.; Liu, A.A.; Tsai, Y.-C.; Chen, Y.; Barbieri, C.M.; Pilch, D.; Liu, L. G-quadruplexes Induce Apoptosis in Tumor Cells. Cancer Res. 2006, 66, 11808–11816. [Google Scholar] [CrossRef] [Green Version]
- Kelland, L. Targeting the Limitless Replicative Potential of Cancer: The Telomerase/Telomere Pathway: Figure 1. Clin. Cancer Res. 2007, 13, 4960–4963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oganesian, L.; Bryan, T.M. Physiological relevance of telomeric G-quadruplex formation: A potential drug target. BioEssays 2007, 29, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in Gene Promoters: A Novel Anticancer Strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murat, P.; Balasubramanian, S. Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 2014, 25, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Sun, D.; Thompson, B.; Cathers, B.E.; Salazar, M.; Kerwin, S.M.; Trent, J.O.; Jenkins, T.C.; Neidle, S.; Hurley, L.H. Inhibition of Human Telomerase by a G-quadruplex-Interactive Compound. J. Med. Chem. 1997, 40, 2113–2116. [Google Scholar] [CrossRef]
- Fedoroff, O.Y.; Salazar, M.; Han, H.; Chemeris, V.V.; Kerwin, S.M.; Hurley, L.H. NMR-Based Model of a Telomerase-Inhibiting Compound Bound to G-quadruplex DNA. Biochemistry 1998, 37, 12367–12374. [Google Scholar] [CrossRef]
- Haider, S.; Parkinson, G.N.; Neidle, S. Structure of a G-quadruplex–Ligand Complex. J. Mol. Biol. 2003, 326, 117–125. [Google Scholar] [CrossRef]
- Gavathiotis, E.; Heald, R.A.; Stevens, M.F.; Searle, M.S. Drug Recognition and Stabilisation of the Parallel-stranded DNA Quadruplex d(TTAGGGT)4 Containing the Human Telomeric Repeat. J. Mol. Biol. 2003, 334, 25–36. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Ghosh, R.; Neidle, S. Structural Basis for Binding of Porphyrin to Human Telomeres. Biochemistry 2007, 46, 2390–2397. [Google Scholar] [CrossRef]
- Campbell, N.H.; Parkinson, G.N.; Reszka, A.P.; Neidle, S. Structural Basis of DNA Quadruplex Recognition by an Acridine Drug. J. Am. Chem. Soc. 2008, 130, 6722–6724. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Cuenca, F.; Neidle, S. Topology Conservation and Loop Flexibility in Quadruplex–Drug Recognition: Crystal Structures of Inter- and Intramolecular Telomeric DNA Quadruplex–Drug Complexes. J. Mol. Biol. 2008, 381, 1145–1156. [Google Scholar] [CrossRef]
- Kim, M.-Y.; Vankayalapati, H.; Shin-Ya, K.; Wierzba, K.; Hurley, L.H. Telomestatin, a Potent Telomerase Inhibitor That Interacts Quite Specifically with the Human Telomeric Intramolecular G-quadruplex. J. Am. Chem. Soc. 2002, 124, 2098–2099. [Google Scholar] [CrossRef]
- Rodriguez, R.; Müller, S.; Yeoman, J.A.; Trentesaux, C.; Riou, J.-F.; Balasubramanian, S. A Novel Small Molecule That Alters Shelterin Integrity and Triggers a DNA-Damage Response at Telomeres. J. Am. Chem. Soc. 2008, 130, 15758–15759. [Google Scholar] [CrossRef] [Green Version]
- Moorhouse, A.D.; Santos, A.M.; Gunaratnam, M.; Moore, M.; Neidle, S.; Moses, J.E. Stabilization of G-quadruplex DNA by Highly Selective Ligands via Click Chemistry. J. Am. Chem. Soc. 2006, 128, 15972–15973. [Google Scholar] [CrossRef]
- Drewe, W.C.; Nanjunda, R.; Gunaratnam, M.; Beltran, M.; Parkinson, G.N.; Reszka, A.P.; Wilson, W.D.; Neidle, S. Rational Design of Substituted Diarylureas: A Scaffold for Binding to G-quadruplex Motifs. J. Med. Chem. 2008, 51, 7751–7767. [Google Scholar] [CrossRef] [Green Version]
- Delaney, S.; Barton, J.K. Charge Transport in DNA Duplex/Quadruplex Conjugates. Biochemistry 2003, 42, 14159–14165. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Kuntz, I.D.; Shafer, R.H. Spectroscopic recognition of guanine dimeric hairpin quadruplexes by a carbocyanine dye. Proc. Natl. Acad. Sci. USA 1996, 93, 2635–2639. [Google Scholar] [CrossRef] [Green Version]
- Harrison, R.J.; Gowan, S.M.; Kelland, L.R.; Neidle, S. Human telomerase inhibition by substituted acridine derivatives. Bioorganic Med. Chem. Lett. 1999, 9, 2463–2468. [Google Scholar] [CrossRef]
- Read, M.A.; Wood, A.A.; Harrison, J.R.; Gowan, S.M.; Kelland, L.R.; Dosanjh, H.S.; Neidle, S. Molecular Modeling Studies on G-quadruplex Complexes of Telomerase Inhibitors: Structure−Activity Relationships. J. Med. Chem. 1999, 42, 4538–4546. [Google Scholar] [CrossRef] [PubMed]
- Campbell, N.; Patel, M.; Tofa, A.B.; Ghosh, R.; Parkinson, G.N.; Neidle, S. Selectivity in Ligand Recognition of G-quadruplex Loops. Biochemistry 2009, 48, 1675–1680. [Google Scholar] [CrossRef]
- Incles, C.M.; Schultes, C.M.; Kempski, H.; Koehler, H.; Kelland, L.R.; Neidle, S. A G-quadruplex telomere targeting agent produces p16-associated senescence and chromosomal fusions in human prostate cancer cells. Mol. Cancer Ther. 2004, 3, 1201–1207. [Google Scholar] [PubMed]
- Moore, M.J.B.; Schultes, C.M.; Cuesta, J.; Cuenca, F.; Gunaratnam, M.; Tanious, F.A.; Wilson, A.W.D.; Neidle, S. Trisubstituted Acridines as G-quadruplex Telomere Targeting Agents. Effects of Extensions of the 3,6- and 9-Side Chains on Quadruplex Binding, Telomerase Activity, and Cell Proliferation. J. Med. Chem. 2005, 49, 582–599. [Google Scholar] [CrossRef]
- Mergny, J.-L.; Maurizot, J.-C. Fluorescence Resonance Energy Transfer as a Probe for G-Quartet Formation by a Telomeric Repeat. ChemBioChem 2001, 2, 124–132. [Google Scholar] [CrossRef]
- De Cian, A.; Guittat, L.; Kaiser, M.; Sacca, B.; Amrane, S.; Bourdoncle, A.; Alberti, P.; Teulade-Fichou, M.-P.; Lacroix, L.; Mergny, J.-L. Fluorescence-based melting assays for studying quadruplex ligands. Methods 2007, 42, 183–195. [Google Scholar] [CrossRef]
- Burger, A.M.; Dai, F.; Schultes, C.M.; Reszka, A.P.; Moore, M.J.; Double, J.A.; Neidle, S. The G-quadruplex-Interactive Molecule BRACO-19 Inhibits Tumor Growth, Consistent with Telomere Targeting and Interference with Telomerase Function. Cancer Res. 2005, 65, 1489–1496. [Google Scholar] [CrossRef] [Green Version]
- Gowan, S.M.; Harrison, J.R.; Patterson, L.; Valenti, M.; Read, M.A.; Neidle, S.; Kelland, L.R. A G-quadruplex-Interactive Potent Small-Molecule Inhibitor of Telomerase Exhibiting in Vitro and in Vivo Antitumor Activity. Mol. Pharmacol. 2002, 61, 1154–1162. [Google Scholar] [CrossRef]
- Incles, C.M.; Schultes, C.M.; Kelland, L.R.; Neidle, S. Acquired Cellular Resistance to Flavopiridol in a Human Colon Carcinoma Cell Line Involves Up-Regulation of the Telomerase Catalytic Subunit and Telomere Elongation. Sensitivity of Resistant Cells to Combination Treatment with a Telomerase Inhibitor. Mol. Pharmacol. 2003, 64, 1101–1108. [Google Scholar] [CrossRef] [Green Version]
- Tuntiwechapikul, W.; Lee, J.T.; Salazar, M. Design and Synthesis of the G-quadruplex-Specific Cleaving Reagent Perylene-EDTA·Iron(II). J. Am. Chem. Soc. 2001, 123, 5606–5607. [Google Scholar] [CrossRef]
- Duan, W.; Rangan, A.; Vankayalapati, H.; Kim, M.Y.; Zeng, Q.; Sun, D.; Han, H.; Fedoroff, O.Y.; Nishioka, D.; Rha, S.Y.; et al. Design and synthesis of fluoroquinophenoxazines that interact with human telomeric G-quadruplexes and their biological effects. Mol. Cancer Ther. 2001, 1, 103–120. [Google Scholar]
- Mehta, A.K.; Shayo, Y.; Vankayalapati, H.; Hurley, L.H.; Schaefer, J. Structure of a Quinobenzoxazine−G-quadruplex Complex by REDOR NMR. Biochemistry 2004, 43, 11953–11958. [Google Scholar] [CrossRef]
- Lichtman, M.A. A historical perspective on the development of the cytarabine (7 days) and daunorubicin (3 days) treatment regimen for acute myelogenous leukemia: 2013 the 40th anniversary of 7 + 3. Blood Cells Mol. Dis. 2013, 50, 119–130. [Google Scholar] [CrossRef]
- Yang, F.; Teves, S.S.; Kemp, C.J.; Henikoff, S. Doxorubicin, DNA Torsion, and Chromatin Dynamics. Biochim. Biophys. Acta-Rev. Cancer 2014, 1845, 84–89. [Google Scholar] [CrossRef] [Green Version]
- Clark, G.R.; Pytel, P.D.; Squire, C.J.; Neidle, S. Structure of the First Parallel DNA Quadruplex-Drug Complex. J. Am. Chem. Soc. 2003, 125, 4066–4067. [Google Scholar] [CrossRef]
- Barrett, M.P.; Gemmell, C.G.; Suckling, C.J. Minor groove binders as anti-infective agents. Pharmacol. Ther. 2013, 139, 12–23. [Google Scholar] [CrossRef]
- Kopka, M.L.; Yoon, C.; Goodsell, D.; Pjura, P.; Dickerson, R.E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc. Natl. Acad. Sci. USA 1985, 82, 1376–1380. [Google Scholar] [CrossRef] [Green Version]
- Pagano, B.; Fotticchia, I.; De Tito, S.; Mattia, C.A.; Mayol, L.; Novellino, E.; Randazzo, A.; Giancola, C. Selective Binding of Distamycin A Derivative to G-quadruplex Structure [d(TGGGGT)]4. J. Nucleic Acids 2010, 2010, 247137. [Google Scholar] [CrossRef] [Green Version]
- Randazzo, A.; Galeone, A.; Mayol, L. 1H-NMR Study of the Interaction of Distamycin A and Netropsin with the Parallel Stranded Tetraplex [d(TGGGT)]4. Chem. Commun. 2001, 11, 1030–1031. [Google Scholar] [CrossRef]
- Cocco, M.J.; Hanakahi, L.A.; Huber, M.D.; Maizels, N. Specific Interactions of Distamycin with G-quadruplex DNA. Nucleic Acids Res. 2003, 31, 2944–2951. [Google Scholar] [CrossRef] [PubMed]
- Martino, L.; Virno, A.; Pagano, B.; Virgilio, A.; Di Micco, S.; Galeone, A.; Giancola, C.; Bifulco, G.; Mayol, L.; Randazzo, A. Structural and Thermodynamic Studies of the Interaction of Distamycin A with the Parallel Quadruplex Structure [d(TGGGGT)]4. J. Am. Chem. Soc. 2007, 129, 16048–16056. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.-L.; Lacroix, L.; Teulade-Fichou, M.-P.; Hounsou, C.; Guittat, L.; Hoarau, M.; Arimondo, P.B.; Vigneron, J.-P.; Lehn, J.-M.; Riou, J.-F.; et al. Telomerase inhibitors based on quadruplex ligands selected by a fluorescence assay. Proc. Natl. Acad. Sci. USA 2001, 98, 3062–3067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hounsou, C.; Guittat, L.; Monchaud, D.; Jourdan, M.; Saettel, N.; Mergny, J.-L.; Teulade-Fichou, M.-P. G-quadruplex Recognition by Quinacridines: A SAR, NMR, and Biological Study. ChemMedChem 2007, 2, 655–666. [Google Scholar] [CrossRef]
- Bertrand, H.; Bombard, S.; Monchaud, D.; Teulade-Fichou, M.-P. A platinum–quinacridine hybrid as a G-quadruplex ligand. JBIC J. Biol. Inorg. Chem. 2007, 12, 1003–1014. [Google Scholar] [CrossRef]
- Teulade-Fichou, M.-P.; Carrasco, C.; Guittat, L.; Bailly, C.; Alberti, P.; Mergny, J.-L.; David, A.; Lehn, J.-M.; Wilson, W.D. Selective Recognition of G-quadruplex Telomeric DNA by a Bis(quinacridine) Macrocycle. J. Am. Chem. Soc. 2003, 125, 4732–4740. [Google Scholar] [CrossRef]
- Allain, C.; Monchaud, D.; Teulade-Fichou, M.-P. FRET Templated by G-quadruplex DNA: A Specific Ternary Interaction Using an Original Pair of Donor/Acceptor Partners. J. Am. Chem. Soc. 2006, 128, 11890–11893. [Google Scholar] [CrossRef]
- Gabelica, V.; Baker, E.S.; Teulade-Fichou, M.-P.; De Pauw, E.; Bowers, M.T. Stabilization and Structure of Telomeric and c-myc Region Intramolecular G-quadruplexes: The Role of Central Cations and Small Planar Ligands. J. Am. Chem. Soc. 2007, 129, 895–904. [Google Scholar] [CrossRef]
- Haudecoeur, R.; Stefan, L.; Denat, F.; Monchaud, D. A Model of Smart G-quadruplex Ligand. J. Am. Chem. Soc. 2012, 135, 550–553. [Google Scholar] [CrossRef]
- Xu, W.; Tan, J.-H.; Chen, S.-B.; Hou, J.-Q.; Li, D.; Huang, Z.-S.; Gu, L.-Q. Studies on the binding of 5-N-methylated quindoline derivative to human telomeric G-quadruplex. Biochem. Biophys. Res. Commun. 2011, 406, 454–458. [Google Scholar] [CrossRef]
- Tan, J.-H.; Ou, T.-M.; Hou, J.-Q.; Lu, Y.-J.; Huang, S.-L.; Luo, H.-B.; Wu, J.-Y.; Huang, Z.-S.; Wong, K.-Y.; Gu, L.-Q. Isaindigotone Derivatives: A New Class of Highly Selective Ligands for Telomeric G-quadruplex DNA. J. Med. Chem. 2009, 52, 2825–2835. [Google Scholar] [CrossRef]
- Han, F.X.; Wheelhouse, R.T.; Hurley, L.H. Interactions of TMPyP4 and TMPyP2 with Quadruplex DNA. Structural Basis for the Differential Effects on Telomerase Inhibition. J. Am. Chem. Soc. 1999, 121, 3561–3570. [Google Scholar] [CrossRef]
- Izbicka, E.; Wheelhouse, R.; Raymond, E.; Davidson, K.K.; Lawrence, R.A.; Sun, D.; Windle, B.E.; Hurley, L.H.; Von Hoff, D.D. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 1999, 59, 639–644. [Google Scholar]
- Shi, D.-F.; Wheelhouse, R.T.; Sun, D.; Hurley, L.H. Quadruplex-Interactive Agents as Telomerase Inhibitors: Synthesis of Porphyrins and Structure−Activity Relationship for the Inhibition of Telomerase. J. Med. Chem. 2001, 44, 4509–4523. [Google Scholar] [CrossRef]
- Kim, M.-Y.; Gleason-Guzman, M.; Izbicka, E.; Nishioka, D.; Hurley, L.H. The different biological effects of telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular G-quadruplex structures. Cancer Res. 2003, 63, 3247–3256. [Google Scholar]
- Pirotton, S.; Muller, C.; Pantoustier, N.; Botteman, F.; Collinet, S.; Grandfils, C.; Dandrifosse, G.; Degée, P.; Dubois, P.; Raes, M. Enhancement of Transfection Efficiency Through Rapid and Noncovalent Post-PEGylation of Poly(Dimethylaminoethyl Methacrylate)/DNA Complexes. Pharm. Res. 2004, 21, 1471–1479. [Google Scholar] [CrossRef]
- Sun, D.; Guo, K.; Rusche, J.J.; Hurley, L.H. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005, 33, 6070–6080. [Google Scholar] [CrossRef]
- Dexheimer, T.S.; Sun, D.; Hurley, L.H. Deconvoluting the Structural and Drug-Recognition Complexity of the G-quadruplex-Forming Region Upstream of the bcl-2 P1 Promoter. J. Am. Chem. Soc. 2006, 128, 5404–5415. [Google Scholar] [CrossRef] [Green Version]
- Guo, K.; Pourpak, A.; Beetz-Rogers, K.; Gokhale, V.; Sun, D.; Hurley, L.H. Formation of Pseudosymmetrical G-quadruplex and i-Motif Structures in the Proximal Promoter Region of the RET Oncogene. J. Am. Chem. Soc. 2007, 129, 10220–10228. [Google Scholar] [CrossRef] [Green Version]
- Freyer, M.W.; Buscaglia, R.; Kaplan, K.; Cashman, D.; Hurley, L.H.; Lewis, E.A. Biophysical Studies of the c-MYC NHE III1 Promoter: Model Quadruplex Interactions with a Cationic Porphyrin. Biophys. J. 2007, 92, 2007–2015. [Google Scholar] [CrossRef] [Green Version]
- Phan, A.T.; Kuryavyi, V.; Gaw, H.Y.; Patel, D.J. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005, 1, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Chaires, J.B. Sequence and Structural Selectivity of Nucleic Acid Binding Ligands. Biochemistry 1999, 38, 16067–16075. [Google Scholar] [CrossRef] [PubMed]
- De Cian, A.; Guittat, L.; Shin-Ya, K.; Riou, J.-F.; Mergny, J.-L. Affinity and selectivity of G4 ligands measured by FRET. Nucleic Acids Symp. Ser. 2005, 49, 235–236. [Google Scholar] [CrossRef]
- De Armond, R.; Wood, S.; Sun, D.; Hurley, L.H.; Ebbinghaus, S.W. Evidence for the Presence of a Guanine Quadruplex Forming Region within a Polypurine Tract of the Hypoxia Inducible Factor 1α Promoter. Biochemistry 2005, 44, 16341–16350. [Google Scholar] [CrossRef]
- Monchaud, D.; Allain, C.; Teulade-Fichou, M.-P. Development of a fluorescent intercalator displacement assay (G4-FID) for establishing quadruplex-DNA affinity and selectivity of putative ligands. Bioorg. Med. Chem. Lett. 2006, 16, 4842–4845. [Google Scholar] [CrossRef]
- Haq, I.; Trent, J.O.; Chowdhry, B.Z.; Jenkins, T.C. Intercalative G-Tetraplex Stabilization of Telomeric DNA by a Cationic Porphyrin1. J. Am. Chem. Soc. 1999, 121, 1768–1779. [Google Scholar] [CrossRef]
- Wei, C.; Jia, G.; Yuan, J.; Feng, Z.; Li, C. A Spectroscopic Study on the Interactions of Porphyrin with G-quadruplex DNAs. Biochemistry 2006, 45, 6681–6691. [Google Scholar] [CrossRef]
- Gonçalves, D.P.N.; Rodriguez, R.; Balasubramanian, S.; Sanders, J.K.M. Tetramethylpyridiniumporphyrazines—a new class of G-quadruplex inducing and stabilising ligands. Chem. Commun. 2006, 4685–4687. [Google Scholar] [CrossRef] [Green Version]
- Dixon, I.M.; Lopez, F.; Tejera, A.M.; Estève, J.-P.; Blasco, M.A.; Pratviel, G.; Meunier, B. A G-quadruplex Ligand with 10000-Fold Selectivity over Duplex DNA. J. Am. Chem. Soc. 2007, 129, 1502–1503. [Google Scholar] [CrossRef] [PubMed]
- Seenisamy, J.; Bashyam, S.; Gokhale, V.; Vankayalapati, H.; Sun, D.; Siddiqui-Jain, A.; Streiner, N.; Shin-Ya, K.; White, E.; Wilson, A.W.D.; et al. Design and Synthesis of an Expanded Porphyrin That Has Selectivity for the c-MYC G-quadruplex Structure. J. Am. Chem. Soc. 2005, 127, 2944–2959. [Google Scholar] [CrossRef]
- Rezler, E.M.; Seenisamy, J.; Bashyam, S.; Kim, M.-Y.; White, E.; Wilson, W.D.; Hurley, L.H. Telomestatin and Diseleno Sapphyrin Bind Selectively to Two Different Forms of the Human Telomeric G-quadruplex Structure. J. Am. Chem. Soc. 2005, 127, 9439–9447. [Google Scholar] [CrossRef]
- Koeppel, F.; Riou, J.F.; Laoui, A.; Mailliet, P.; Arimondo, P.B.; Labit, D.; Petitgenet, O.; Hélène, C.; Mergny, J.L. Ethidium Derivatives Bind to G-Quartets, Inhibit Telomerase and Act as Fluorescent Probes for Quadruplexes. Nucleic Acids Res. 2001, 29, 1087–1096. [Google Scholar] [CrossRef] [Green Version]
- Riou, J.-F.; Guittat, L.; Mailliet, P.; Laoui, A.; Renou, E.; Petitgenet, O.; Megnin-Chanet, F.; Helene, C.; Mergny, J.-L. Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc. Natl. Acad. Sci. USA 2002, 99, 2672–2677. [Google Scholar] [CrossRef] [Green Version]
- Gomez, D.; Aouali, N.; Londoño-Vallejo, A.; Lacroix, L.; Mégnin-Chanet, F.; Lemarteleur, T.; Douarre, C.; Shin-Ya, K.; Mailliet, P.; Trentesaux, C.; et al. Resistance to the Short Term Antiproliferative Activity of the G-quadruplex Ligand 12459 Is Associated with Telomerase Overexpression and Telomere Capping Alteration. J. Biol. Chem. 2003, 278, 50554–50562. [Google Scholar] [CrossRef] [Green Version]
- Gomez, D.; Lemarteleur, T.; Lacroix, L.; Mailliet, P.; Mergny, J.L.; Riou, J.F. Telomerase Downregulation Induced by the G-quadruplex Ligand 12459 in A549 Cells Is Mediated by HTERT RNA Alternative Splicing. Nucleic Acids Res. 2004, 32, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Douarre, C.; Gomez, D.; Morjani, H.; Zahm, J.M.; O’Donohue, M.F.; Eddabra, L.; Mailliet, P.; Riou, J.F.; Trentesaux, C. Overexpression of Bcl-2 Is Associated with Apoptotic Resistance to the G-quadruplex Ligand 12459 but Is Not Sufficient to Confer Resistance to Long-Term Senescence. Nucleic Acids Res. 2005, 33, 2192–2203. [Google Scholar] [CrossRef]
- Lemarteleur, T.; Gomez, D.; Paterski, R.; Mandine, E.; Mailliet, P.; Riou, J.-F. Stabilization of the c-myc gene promoter quadruplex by specific ligands’ inhibitors of telomerase. Biochem. Biophys. Res. Commun. 2004, 323, 802–808. [Google Scholar] [CrossRef]
- Pennarun, G.; Granotier, C.; Gauthier, L.R.; Gomez, D.; Hoffschir, F.; Mandine, E.; Riou, J.-F.; Mergny, J.-L.; Mailliet, P.; Boussin, F.D. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene 2005, 24, 2917–2928. [Google Scholar] [CrossRef]
- Rodriguez, R.; Miller, K.M.; Forment, J.; Bradshaw, C.; Nikan, M.; Britton, S.; Oelschlaegel, T.; Xhemalce, B.; Balasubramanian, S.; Jackson, S.P. Small-molecule–induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012, 8, 301–310. [Google Scholar] [CrossRef] [Green Version]
- Biffi, G.; Di Antonio, M.; Tannahill, D.; Balasubramanian, S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2013, 6, 75–80. [Google Scholar] [CrossRef]
- Lubitz, I.; Borovok, N.; Kotlyar, A. Interaction of Monomolecular G4-DNA Nanowires with TMPyP: Evidence for Intercalation. Biochemistry 2007, 46, 12925–12929. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, C.M.; Welsh, S.J.; Strauss, S.J.; Dale, A.G.; Todd, A.K.; Nanjunda, R.; Wilson, W.D.; Neidle, S. A novel series of G-quadruplex ligands with selectivity for HIF-expressing osteosarcoma and renal cancer cell lines. Bioorg. Med. Chem. Lett. 2012, 22, 5984–5988. [Google Scholar] [CrossRef]
- Anantha, N.V.; Azam, M.; Sheardy, R.D. Porphyrin Binding to Quadruplexed T4G4. Biochemistry 1998, 37, 2709–2714. [Google Scholar] [CrossRef]
- Li, Q.; Xiang, J.; Li, X.; Chen, L.; Xu, X.; Tang, Y.; Zhou, Q.; Li, L.; Zhang, H.; Sun, H.; et al. Stabilizing parallel G-quadruplex DNA by a new class of ligands: Two non-planar alkaloids through interaction in lateral grooves. Biochimie 2009, 91, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.; Pantoş, G.D.; Gonçalves, D.P.N.; Sanders, J.K.M.; Balasubramanian, S. Ligand-Driven G-quadruplex Conformational Switching By Using an Unusual Mode of Interaction. Angew. Chem. 2007, 119, 5501–5503. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.-H.; Shi, S.; Yao, J.-L.; Gao, X.; Huang, H.-L.; Yao, T.-M. Two structurally analogous ruthenium complexes as naked-eye and reversible molecular “light switch” for G-quadruplex DNA. J. Inorg. Biochem. 2014, 140, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-J.; Yu, L.; Hao, Z.-F.; Zhao, Y. Interactions of ruthenium complexes containing indoloquinoline moiety with human telomeric G-quadruplex DNA. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 124, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.E.; Arnal, A.A.; Neidle, S.; Vilar, R. Stabilization of G-quadruplex DNA and Inhibition of Telomerase Activity by Square-Planar Nickel(II) Complexes. J. Am. Chem. Soc. 2006, 128, 5992–5993. [Google Scholar] [CrossRef]
- Keating, L.R.; Szalai, V.A. Parallel-Stranded Guanine Quadruplex Interactions with a Copper Cationic Porphyrin. Biochemistry 2004, 43, 15891–15900. [Google Scholar] [CrossRef]
- Evans, S.E.; Mendez, M.A.; Turner, K.B.; Keating, L.R.; Grimes, R.T.; Melchoir, S.; Szalai, V.A. End-stacking of copper cationic porphyrins on parallel-stranded guanine quadruplexes. JBIC J. Biol. Inorg. Chem. 2007, 12, 1235–1249. [Google Scholar] [CrossRef]
- Romera, C.; Bombarde, O.; Bonnet, R.; Gomez, D.; Dumy, P.; Calsou, P.; Gwan, J.-F.; Lin, J.-H.; Defrancq, E.; Pratviel, G. Improvement of porphyrins for G-quadruplex DNA targeting. Biochimie 2011, 93, 1310–1317. [Google Scholar] [CrossRef]
- Vialas, C.; Pratviel, G.; Meunier, B. Oxidative Damage Generated by an Oxo-Metalloporphyrin onto the Human Telomeric Sequence. Biochemistry 2000, 39, 9514–9522. [Google Scholar] [CrossRef]
- Maraval, A.; Franco, S.; Vialas, C.; Pratviel, G.; Blasco, M.A.; Meunier, B. Porphyrin–aminoquinoline conjugates as telomerase inhibitors. Org. Biomol. Chem. 2003, 1, 921–927. [Google Scholar] [CrossRef]
- Dixon, I.; Lopez, F.; Estève, J.-P.; Tejera, A.M.; Blasco, M.A.; Pratviel, G.; Meunier, B. Porphyrin Derivatives for Telomere Binding and Telomerase Inhibition. ChemBioChem 2004, 6, 123–132. [Google Scholar] [CrossRef]
- Ren, L.; Zhang, A.; Huang, J.; Wang, P.; Weng, X.; Zhang, L.; Liang, F.; Tan, Z.; Zhou, X. Quaternary Ammonium Zinc Phthalocyanine: Inhibiting Telomerase by Stabilizing G quadruplexes and Inducing G-quadruplex Structure Transition and Formation. ChemBioChem 2007, 8, 775–780. [Google Scholar] [CrossRef]
- Mikuma, T.; Ohyama, T.; Terui, N.; Yamamoto, Y.; Hori, H. Coordination Complex between Haemin and Parallel-Quadruplexed d(TTAGGG). Chem. Commun. 2003, 3, 1708–1709. [Google Scholar] [CrossRef]
- Ohyama, T.; Kato, Y.; Mita, H.; Yamamoto, Y. Exogenous Ligand Binding Property of a Heme–DNA Coordination Complex. Chem. Lett. 2006, 35, 126–127. [Google Scholar] [CrossRef]
- Arola-Arnal, A.; Benet-Buchholz, J.; Neidle, S.; Vilar, R. Effects of Metal Coordination Geometry on Stabilization of Human Telomeric Quadruplex DNA by Square-Planar and Square-Pyramidal Metal Complexes. Inorg. Chem. 2008, 47, 11910–11919. [Google Scholar] [CrossRef]
- Campbell, N.H.; Karim, N.H.A.; Parkinson, G.N.; Gunaratnam, M.; Petrucci, V.; Todd, A.K.; Vilar, R.; Neidle, S. Molecular Basis of Structure–Activity Relationships between Salphen Metal Complexes and Human Telomeric DNA Quadruplexes. J. Med. Chem. 2011, 55, 209–222. [Google Scholar] [CrossRef]
- Terenzi, A.; Bonsignore, R.; Spinello, A.; Gentile, C.; Martorana, A.; Ducani, C.; Högberg, B.; Almerico, A.M.; Lauria, A.; Barone, G. Selective G-quadruplex stabilizers: Schiff-base metal complexes with anticancer activity. RSC Adv. 2014, 4, 33245–33256. [Google Scholar] [CrossRef]
- Davis, K.J.; Richardson, C.; Beck, J.L.; Knowles, B.M.; Guédin, A.; Mergny, J.-L.; Willis, A.C.; Ralph, S.F. Synthesis and characterisation of nickel Schiff base complexes containing the meso-1,2-diphenylethylenediamine moiety: Selective interactions with a tetramolecular DNA quadruplex. Dalton Trans. 2015, 44, 3136–3150. [Google Scholar] [CrossRef] [PubMed]
- Terenzi, A.; Lötsch, D.; Van Schoonhoven, S.; Roller, A.; Kowol, C.R.; Berger, W.; Keppler, B.K.; Barone, G. Another step toward DNA selective targeting: Ni(II) and Cu(II) complexes of a Schiff base ligand able to bind gene promoter G-quadruplexes. Dalton Trans. 2016, 45, 7758–7767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, A.; Kamra, M.; Roy, S.; Muniyappa, K.; Bhattacharya, S. Novel Oligopyrrole Carboxamide based Nickel(II) and Palladium(II) Salens, Their Targeting of Human G-quadruplex DNA, and Selective Cancer Cell Toxicity. Chem. Asian J. 2016, 11, 2542–2554. [Google Scholar] [CrossRef]
- Lecarme, L.; Prado, E.; De Rache, A.; Nicolau-Travers, M.-L.; Gellon, G.; Dejeu, J.; Lavergne, T.; Jamet, H.; Gomez, D.; Mergny, J.-L.; et al. Efficient Inhibition of Telomerase by Nickel-Salophen Complexes. ChemMedChem 2016, 11, 1133–1136. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Kamra, M.; Roy, S.; Muniyappa, K.; Bhattacharya, S. Enhanced G-quadruplex DNA Stabilization and Telomerase Inhibition by Novel Fluorescein Derived Salen and Salphen Based Ni(II) and Pd(II) Complexes. Bioconjugate Chem. 2017, 28, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.J.; Assadawi, N.M.O.; Pham, S.Q.T.; Birrento, M.L.; Richardson, C.; Beck, J.L.; Willis, A.C.; Ralph, S.F. Effect of structure variations on the quadruplex DNA binding ability of nickel Schiff base complexes. Dalton Trans. 2018, 47, 13573–13591. [Google Scholar] [CrossRef] [PubMed]
- Farine, G.; Migliore, C.; Terenzi, A.; Lo Celso, F.; Santoro, A.; Bruno, G.; Bonsignore, R.; Barone, G. On the G-quadruplex Binding of a New Class of Nickel(II), Copper(II), and Zinc(II) Salphen-Like Complexes. Eur. J. Inorg. Chem. 2021, 2021, 1332–1336. [Google Scholar] [CrossRef]
- Ruehl, C.L.; Lim, A.H.M.; Kench, T.; Mann, D.J.; Vilar, R. An Octahedral Cobalt(III) Complex with Axial NH 3 Ligands that Templates and Selectively Stabilises G--quadruplex DNA. Chem. Eur. J. 2019, 25, 9691–9700. [Google Scholar] [CrossRef]
- Wu, P.; Ma, D.-L.; Leung, C.-H.; Yan, S.-C.; Zhu, N.; Abagyan, R.; Che, C.-M. Stabilization of G-quadruplex DNA with Platinum(II) Schiff Base Complexes: Luminescent Probe and Down-Regulation of c-myc Oncogene Expression. Chem. Eur. J. 2009, 15, 13008–13021. [Google Scholar] [CrossRef]
- Rakers, V.; Cadinu, P.; Edel, J.B.; Vilar, R. Development of microfluidic platforms for the synthesis of metal complexes and evaluation of their DNA affinity using online FRET melting assays. Chem. Sci. 2018, 9, 3459–3469. [Google Scholar] [CrossRef] [Green Version]
- Abd Karim, N.H.; Mendoza, O.; Shivalingam, A.; Thompson, A.J.; Ghosh, S.; Kuimova, M.K.; Vilar, R. Salphen metal complexes as tunable G-quadruplex binders and optical probes. RSC Adv. 2013, 4, 3355–3363. [Google Scholar] [CrossRef]
- Bertrand, H.; Monchaud, D.; De Cian, A.; Guillot, R.; Mergny, J.-L.; Teulade-Fichou, M.-P. The importance of metal geometry in the recognition of G-quadruplex-DNA by metal-terpyridine complexes. Org. Biomol. Chem. 2007, 5, 2555–2559. [Google Scholar] [CrossRef]
- Bertrand, H.; Bombard, S.; Monchaud, D.; Talbot, E.; Guédin, A.; Mergny, J.-L.; Grünert, R.; Bednarski, P.J.; Teulade-Fichou, M.-P. Exclusive platination of loop adenines in the human telomeric G-quadruplex. Org. Biomol. Chem. 2009, 7, 2864–2871. [Google Scholar] [CrossRef]
- Largy, E.; Hamon, F.; Rosu, F.; Gabelica, V.; De Pauw, E.; Guédin, A.; Mergny, J.-L.; Teulade-Fichou, M.-P. Tridentate N-Donor Palladium(II) Complexes as Efficient Coordinating Quadruplex DNA Binders. Chem. Eur. J. 2011, 17, 13274–13283. [Google Scholar] [CrossRef]
- Trajkovski, M.; Morel, E.; Hamon, F.; Bombard, S.; Teulade-Fichou, M.-P.; Plavec, J. Interactions of Pt-ttpy with G-quadruplexes Originating from Promoter Region of the c-myc Gene Deciphered by NMR and Gel Electrophoresis Analysis. Chem. Eur. J. 2015, 21, 7798–7807. [Google Scholar] [CrossRef]
- Wang, P.; Leung, C.-H.; Ma, D.-L.; Yan, S.-C.; Che, C.-M. Structure-Based Design of Platinum(II) Complexes as c-myc Oncogene Down-Regulators and Luminescent Probes for G-quadruplex DNA. Chem. Eur. J. 2010, 16, 6900–6911. [Google Scholar] [CrossRef]
- Musetti, C.; Krapcho, A.P.; Palumbo, M.; Sissi, C. Effect of G-quadruplex Polymorphism on the Recognition of Telomeric DNA by a Metal Complex. PLoS ONE 2013, 8, e58529. [Google Scholar] [CrossRef] [Green Version]
- Reed, J.E.; Neidle, S.; Vilar, R. Stabilisation of human telomeric quadruplex DNA and inhibition of telomerase by a platinum–phenanthroline complex. Chem. Commun. 2007, 4366–4368. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Khayamian, T.; Hadadzadeh, H.; Jannesari, Z.; Khaksar, G. Spectroscopic, biological, and molecular modeling studies on the interactions of [Fe(III)-meloxicam] with G-quadruplex DNA and investigation of its release from bovine serum albumin (BSA) nanoparticles. J. Biomol. Struct. Dyn. 2015, 33, 2316–2329. [Google Scholar] [CrossRef]
- Castor, K.K.; Metera, K.L.; Tefashe, U.M.; Serpell, C.J.; Mauzeroll, J.; Sleiman, H.F. Cyclometalated Iridium(III) Imidazole Phenanthroline Complexes as Luminescent and Electroluminescent G-quadruplex DNA Binders. Inorg. Chem. 2015, 54, 6958–6967. [Google Scholar] [CrossRef]
- Lu, L.; Wang, M.; Mao, Z.; Kang, T.-S.; Chen, X.-P.; Lu, J.-J.; Leung, C.-H.; Ma, D.-L. A Novel Dinuclear Iridirum(III) Complex as a G-quadruplex Selective Probe for the Luminescent Switch-On Detection of Transcription Factor HIF-1α. Sci. Rep. 2016, 6, 22458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Mao, Z.; Kang, T.-S.; Wong, C.-Y.; Mergny, J.-L.; Leung, C.-H.; Ma, D.-L. Conjugating a groove-binding motif to an Ir(iii) complex for the enhancement of G-quadruplex probe behavior. Chem. Sci. 2016, 7, 2516–2523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.; Lu, L.; Kang, T.-S.; Mergny, J.-L.; Leung, C.-H.; Ma, D.-L. Interaction of an Iridium(III) Complex with G-quadruplex DNA and Its Application in Luminescent Switch-On Detection of Siglec-5. Anal. Chem. 2016, 88, 10290–10295. [Google Scholar] [CrossRef] [PubMed]
- Paris, J.P.; Brandt, W.W. Charge transfer luminescence of a ruthenium(II) chelate. J. Am. Chem. Soc. 1959, 81, 5001–5002. [Google Scholar] [CrossRef]
- Balzani, V.; Juris, A.; Venturi, M.; Campagna, S.; Serroni, S. Luminescent and Redox-Active Polynuclear Transition Metal Complexes. Chem. Rev. 1996, 96, 759–834. [Google Scholar] [CrossRef] [PubMed]
- Campagna, S.; Puntoriero, F.; Nastasi, F.; Bergamini, G.; Balzani, V. Photochemistry and Photophysics of Coordination Compounds: Ruthenium. In Photochemistry and Photophysics of Coordination Compounds I; Balzani, V., Campagna, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 280, pp. 117–214. [Google Scholar]
- Juris, A.; Balzani, V.; Barigelletti, F.; Campagna, S.; Belser, P.; Von Zelewsky, A. Ru(II) polypyridine complexes: Photophysics, photochemistry, eletrochemistry, and chemiluminescence. Coord. Chem. Rev. 1988, 84, 85–277. [Google Scholar] [CrossRef]
- Di Pietro, M.L.; La Ganga, G.; Nastasi, L.; Puntoriero, F. Ru(II)-Dppz Derivatives and Their Interactions with DNA: Thirty Years and Counting. Appl. Sci. 2021, 11, 3038. [Google Scholar] [CrossRef]
- Olson, E.J.C.; Hu, D.; Hörmann, A.; Jonkman, A.M.; Arkin, M.R.; Stemp, E.D.A.; Barton, A.J.K.; Barbara, P.F. First Observation of the Key Intermediate in the “Light-Switch” Mechanism of [Ru(phen)2dppz]2+. J. Am. Chem. Soc. 1997, 119, 11458–11467. [Google Scholar] [CrossRef]
- Olofsson, J.; Önfelt, B.; Lincoln, P. Three-State Light Switch of [Ru(phen)2dppz]2+: Distinct Excited-State Species with Two, One, or No Hydrogen Bonds from Solvent. J. Phys. Chem. A 2004, 108, 4391–4398. [Google Scholar] [CrossRef]
- Szalai, V.A.; Thorp, H.H. Electron Transfer in Tetrads: Adjacent Guanines Are Not Hole Traps in G Quartets. J. Am. Chem. Soc. 2000, 122, 4524–4525. [Google Scholar] [CrossRef]
- Rubio-Magnieto, J.; Kajouj, S.; Di Meo, F.; Fossépré, M.; Trouillas, P.; Norman, P.; Linares, M.; Moucheron, C.; Surin, M. Binding Modes and Selectivity of Ruthenium Complexes to Human Telomeric DNA G-quadruplexes. Chem. Eur. J. 2018, 24, 15577–15588. [Google Scholar] [CrossRef]
- Shi, S.; Geng, X.; Zhao, J.; Yao, T.; Wang, C.; Yang, D.; Zheng, L.; Ji, L. Interaction of [Ru(bpy)2(dppz)]2+ with human telomeric DNA: Preferential binding to G-quadruplexes over i-motif. Biochimie 2010, 92, 370–377. [Google Scholar] [CrossRef]
- Shi, S.; Zhao, J.; Geng, X.; Yao, T.; Huang, H.; Liu, T.; Zheng, L.; Li, Z.; Yang, D.; Ji, L. Molecular “light switch” for G-quadruplexes and i-motif of human telomeric DNA: [Ru(phen)2(dppz)]2+. Dalton Trans. 2010, 39, 2490–2493. [Google Scholar] [CrossRef]
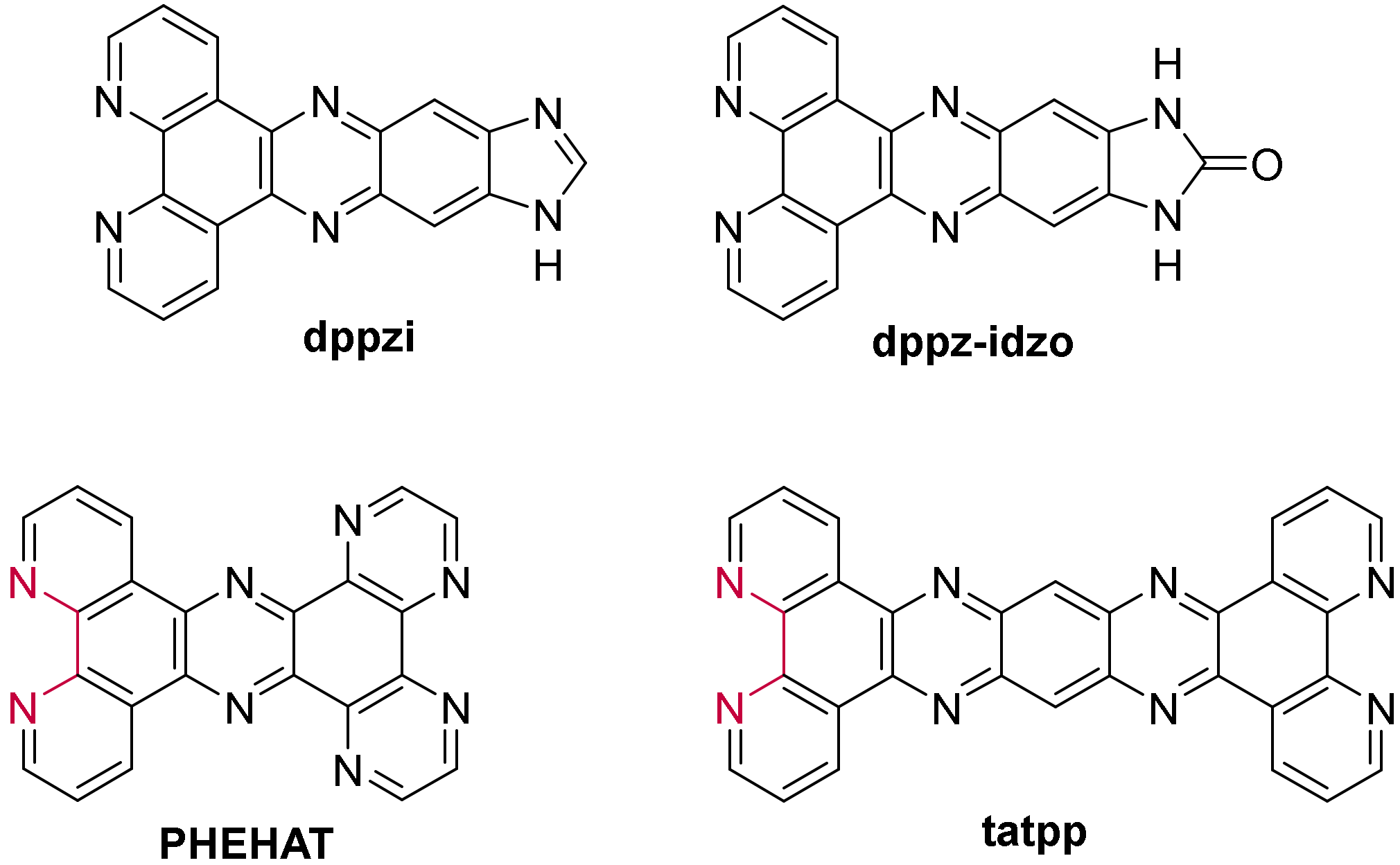
- Shi, S.; Xu, J.-H.; Gao, X.; Huang, H.-L.; Yao, T.-M. Binding Behaviors for Different Types of DNA G-quadruplexes: Enantiomers of [Ru(Bpy)2(L)]2+ (L=dppz, Dppz-Idzo). Chem. Eur. J. 2015, 21, 11435–11445. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, H.S.; Jang, M.D.; Han, S.W.; Kim, S.K.; Lee, Y.A. Enantioselective Light Switch Effect of Δ- and Λ-[Ru(Phenanthroline)2Dipyrido[3,2-a:2′,3′-c]Phenazine]2+ Bound to G-quadruplex DNA. J. Biomol. Struct. Dyn. 2018, 36, 1948–1957. [Google Scholar] [CrossRef]
- Devereux, S.J.; Poynton, F.E.; Baptista, F.; Gunnlaugsson, T.; Cardin, C.J.; Sazanovich, I.V.; Towrie, M.; Kelly, J.M.; Quinn, S.J. Caught in the Loop: Binding of the [Ru(phen)2(dppz)] 2+ Light-Switch Compound to Quadruplex DNA in Solution Informed by Time-Resolved Infrared Spectroscopy. Chem. Eur. J. 2020, 26, 17103–17109. [Google Scholar] [CrossRef]
- Yang, C.; Zhou, Q.; Jiao, Z.; Zhao, H.; Huang, C.-H.; Zhu, B.-Z.; Su, H. Ultrafast excited state dynamics and light-switching of [Ru(phen)2(dppz)]2+ in G-quadruplex DNA. Commun. Chem. 2021, 4, 68. [Google Scholar] [CrossRef]
- McQuaid, K.; Hall, J.P.; Baumgaertner, L.; Cardin, D.J.; Cardin, C.J. Three thymine/adenine binding modes of the ruthenium complex Λ-[Ru(TAP)2(dppz)]2+ to the G-quadruplex forming sequence d(TAGGGTT) shown by X-ray crystallography. Chem. Commun. 2019, 55, 9116–9119. [Google Scholar] [CrossRef]
- McQuaid, K.; Abell, H.; Gurung, S.P.; Allan, D.R.; Winter, G.; Sørensen, T.L.-M.; Cardin, D.J.; Brazier, J.A.; Cardin, C.J.; Hall, J.P. Structural Studies Reveal Enantiospecific Recognition of a DNA G-quadruplex by a Ruthenium Polypyridyl Complex. Angew. Chem. Int. Ed. 2019, 58, 9881–9885. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Lv, C.; Gao, X.; Zhao, J.; Yao, J.; Sun, W.; Huang, H.; Yao, T.; Ji, L. [Ru(bpy)2(bppp)]2+ binds two different forms of the human telomeric G-quadruplex structure. Inorg. Chem. Commun. 2012, 24, 212–215. [Google Scholar] [CrossRef]
- Shi, S.; Huang, H.-L.; Gao, X.; Yao, J.-L.; Lv, C.-Y.; Zhao, J.; Sun, W.-L.; Yao, T.-M.; Ji, L.-N. A comparative study of the interaction of two structurally analogous ruthenium complexes with human telomeric G-quadruplex DNA. J. Inorg. Biochem. 2013, 121, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Wachter, E.; Moyá, D.; Glazer, E.C. Combining a Ru(II) “Building Block” and Rapid Screening Approach to Identify DNA Structure-Selective “Light Switch” Compounds. ACS Comb. Sci. 2016, 19, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Pei, L.; Fan, X.; Shi, S. [Ru(L)2(3-tppp)]2+ (L = bpy, phen) stabilizes two different forms of the human telomeric G-quadruplex DNA. Inorg. Chem. Commun. 2016, 72, 7–12. [Google Scholar] [CrossRef]
- Wachter, E.; Moyá, D.; Parkin, S.; Glazer, E.C. Ruthenium Complex “Light Switches” That Are Selective for Different G-quadruplex Structures. Chem. Eur. J. 2016, 22, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; An, Y.; Zhang, L.; Chen, H.-Y.; Wang, Y.-J.; Mao, Z.-W.; Ji, L.-N. Studies on synthesis, characterization, and G-quadruplex binding of Ru(II) complexes containing two dppz ligands. J. Inorg. Biochem. 2010, 105, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Bouzada, D.; Salvadó, I.; Barka, G.; Rama, G.; Martínez-Costas, J.; Lorca, R.; Somoza, Á.; Melle-Franco, M.; Vázquez, M.E.; López, M.V. Selective G-quadruplex binding by oligoarginine-Ru(dppz) metallopeptides. Chem. Commun. 2017, 54, 658–661. [Google Scholar] [CrossRef] [PubMed]
- Wachter, E.; Howerton, B.S.; Hall, E.C.; Parkin, S.; Glazer, E.C. A new type of DNA “light-switch”: A dual photochemical sensor and metalating agent for duplex and G-quadruplex DNA. Chem. Commun. 2013, 50, 311–313. [Google Scholar] [CrossRef]
- Liao, G.; Chen, X.; Wu, J.; Qian, C.; Wang, H.; Ji, L.; Chao, H. Novel ruthenium(II) polypyridyl complexes as G-quadruplex stabilisers and telomerase inhibitors. Dalton Trans. 2014, 43, 7811–7819. [Google Scholar] [CrossRef]
- Shi, S.; Zhao, J.; Gao, X.; Lv, C.; Yang, L.; Hao, J.; Huang, H.; Yao, J.; Sun, W.; Yao, T.; et al. Molecular “Light Switch” for G-quadruplex DNA: Cycling the Switch on and Off. Dalt. Trans. 2012, 41, 5789–5793. [Google Scholar] [CrossRef]
- Shi, S.; Gao, X.; Huang, H.; Zhao, J.; Yao, T. Effect of the Ancillary Ligands on the Spectral Properties and G-quadruplexes DNA Binding Behavior: A Combined Experimental and Theoretical Study. Chem. Eur. J. 2015, 21, 13390–13400. [Google Scholar] [CrossRef]
- Yao, J.-L.; Gao, X.; Sun, W.; Fan, X.-Z.; Shi, S.; Yao, T.-M. A Naked-Eye On–Off–On Molecular “Light Switch” Based on a Reversible “Conformational Switch” of G-quadruplex DNA. Inorg. Chem. 2012, 51, 12591–12593. [Google Scholar] [CrossRef]
- Yao, J.-L.; Gao, X.; Sun, W.; Shi, S.; Yao, T.-M. [Ru(bpy)2dppz-idzo]2+: A colorimetric molecular “light switch” and powerful stabilizer for G-quadruplex DNA. Dalton Trans. 2013, 42, 5661–5672. [Google Scholar] [CrossRef]
- Hu, X.; Yang, D.; Yao, T.; Gao, R.; Wumaier, M.; Shi, S. Regulation of multi-factors (tail/loop/link/ions) for G-quadruplex enantioselectivity of Δ- and Λ- [Ru(bpy)2(dppz-idzo)]2+. Dalton Trans. 2018, 47, 5422–5430. [Google Scholar] [CrossRef]
- Mikek, C.G.; Machha, V.R.; White, J.C.; Martin, L.R.; West, S.J.; Butrin, A.; Shumaker, C.; Gwin, J.C.; Alatrash, N.; MacDonnell, F.M.; et al. The Thermodynamic Effects of Ligand Structure on the Molecular Recognition of Mono- and Birutheniumium polypyridyl Complexes with G-quadruplex DNA. Eur. J. Inorg. Chem. 2017, 2017, 3953–3960. [Google Scholar] [CrossRef]
- Yu, H.-J.; Zhao, Y.; Mo, W.-J.; Hao, Z.-F.; Yu, L. Ru-indoloquinoline complex as a selective and effective human telomeric G-quadruplex binder. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 132, 84–90. [Google Scholar] [CrossRef]
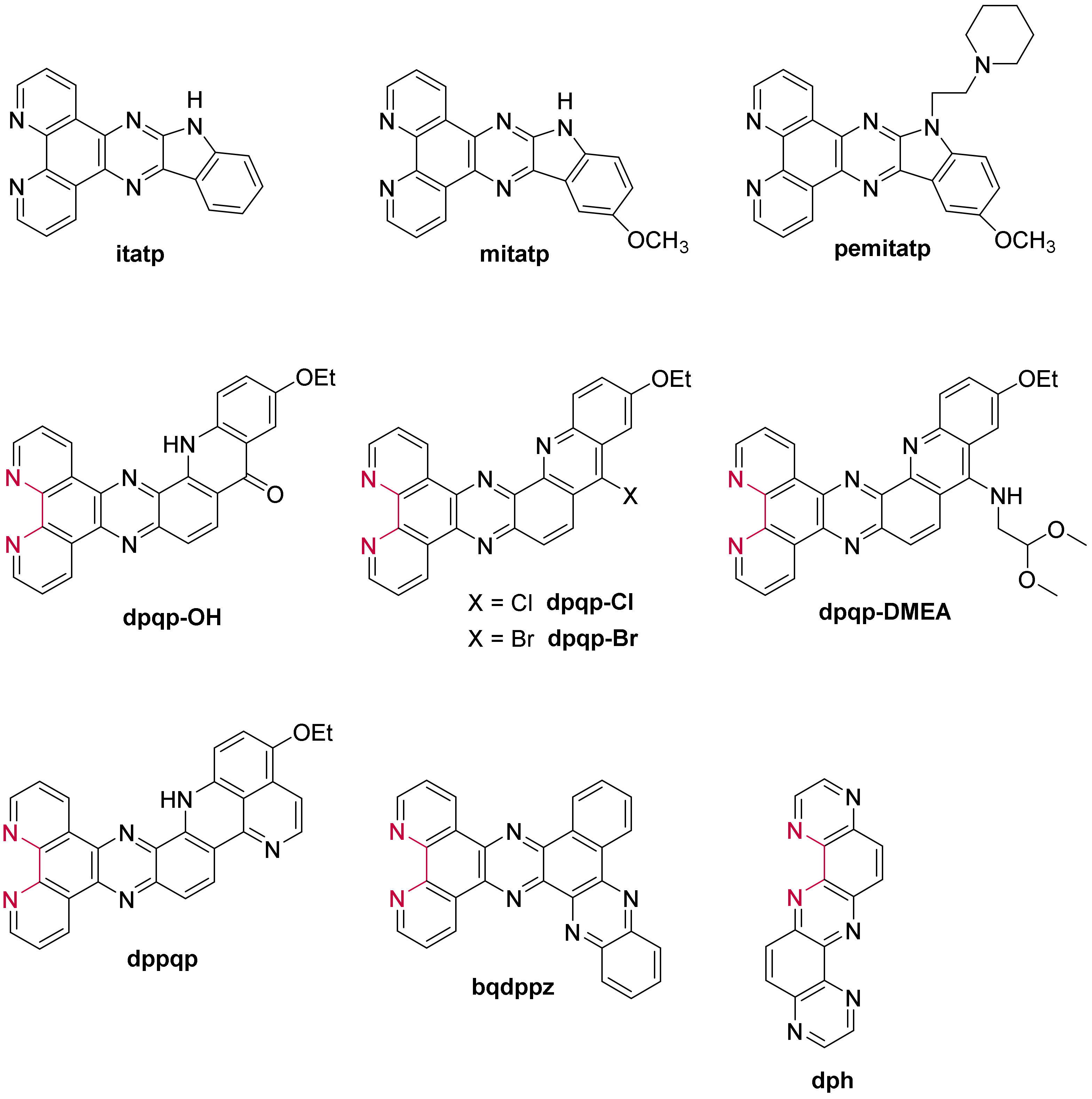
- Lefebvre, J.-F.; Saadallah, D.; Traber, P.; Kupfer, S.; Gräfe, S.; Dietzek, B.; Baussanne, I.; De Winter, J.; Gerbaux, P.; Moucheron, C.; et al. Synthesis of three series of ruthenium tris-diimine complexes containing acridine-based π-extended ligands using an efficient “Chemistry on the Complex” approach. Dalton Trans. 2016, 45, 16298–16308. [Google Scholar] [CrossRef]
- Saadallah, D.; Bellakhal, M.; Amor, S.; Lefebvre, J.-F.; Chavarot-Kerlidou, M.; Baussanne, I.; Moucheron, C.; Demeunynck, M.; Monchaud, D. Selective Luminescent Labeling of DNA and RNA Quadruplexes by π-Extended Ruthenium Light-Up Probes. Chem. Eur. J. 2017, 23, 4967–4972. [Google Scholar] [CrossRef]
- Collie, G.W.; Sparapani, S.; Parkinson, G.N.; Neidle, S. Structural Basis of Telomeric RNA Quadruplex–Acridine Ligand Recognition. J. Am. Chem. Soc. 2011, 133, 2721–2728. [Google Scholar] [CrossRef]
- Neidle, S. Quadruplex Nucleic Acids as Novel Therapeutic Targets. J. Med. Chem. 2016, 59, 5987–6011. [Google Scholar] [CrossRef]
- Laguerre, A.; Stefan, L.; Larrouy, M.; Genest, D.; Novotna, J.; Pirrotta, M.; Monchaud, D. A Twice-As-Smart Synthetic G-Quartet: PyroTASQ Is Both a Smart Quadruplex Ligand and a Smart Fluorescent Probe. J. Am. Chem. Soc. 2014, 136, 12406–12414. [Google Scholar] [CrossRef]
- Yang, S.; Amor, S.; Laguerre, A.; Wong, J.M.; Monchaud, D. Real-time and quantitative fluorescent live-cell imaging with quadruplex-specific red-edge probe (G4-REP). Biochim. Biophys. Acta–Gen. Subj. 2017, 1861, 1312–1320. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.-L.; Chen, X.; Ji, L.-N.; Chao, H. Visual specific luminescent probing of hybrid G-quadruplex DNA by a ruthenium polypyridyl complex. Chem. Commun. 2012, 48, 10781–10783. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, L.; Liu, M.; Qiu, Y.; Wang, M.; Liu, X.; Shen, G.; Yu, R. A novel, label-free fluorescent aptasensor for cocaine detection based on a G-quadruplex and ruthenium polypyridyl complex molecular light switch. Anal. Methods 2016, 8, 3740–3746. [Google Scholar] [CrossRef]
- Piraux, G.; Bar, L.; Abraham, M.; Lavergne, T.; Jamet, H.; Dejeu, J.; Marcélis, L.; Defrancq, E.; Elias, B. New Ruthenium-Based Probes for Selective G-quadruplex Targeting. Chem. Eur. J. 2017, 23, 11872–11880. [Google Scholar] [CrossRef]
- Liu, D.; Liu, Y.; Wang, C.; Shi, S.; Sun, D.; Gao, F.; Zhang, Q.; Liu, J. Polypyridyl Complexes of Ruthenium(II): Stabilization of G-quadruplex DNA and Inhibition of Telomerase Activity. ChemPlusChem 2012, 77, 551–562. [Google Scholar] [CrossRef]
- Sun, D.; Liu, Y.; Liu, D.; Zhang, R.; Yang, X.; Liu, J. Stabilization of G-quadruplex DNA, Inhibition of Telomerase Activity and Live Cell Imaging Studies of Chiral Ruthenium(II) Complexes. Chem.-Eur. J. 2012, 18, 4285–4295. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Liu, Y.; Wang, C.; Sun, D.; Yang, X.; Liu, Y.; Liu, J. Chiral Ruthenium(II) Polypyridyl Complexes: Stabilization of G-quadruplex DNA, Inhibition of Telomerase Activity and Cellular Uptake. PLoS ONE 2012, 7, e50902. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, Q.; Wu, X.-H.; Sun, F.-Y.; Chen, L.-M.; Chen, J.-C.; Yang, S.-L.; Mei, W.-J. Ruthenium(II) complexes as apoptosis inducers by stabilizing c-myc G-quadruplex DNA. Eur. J. Med. Chem. 2014, 80, 316–324. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Y.; Yang, L.; Cao, C.; Zhou, Y.; Liu, J. Stabilization for loop isomers of c-myc G-quadruplex DNA and anticancer activity by ruthenium complexes. MedChemComm 2014, 5, 1724–1728. [Google Scholar] [CrossRef]
- Sun, D.; Liu, Y.; Yu, Q.; Liu, D.; Zhou, Y.; Liu, J. Selective nuclei accumulation of ruthenium(II) complex enantiomers that target G-quadruplex DNA. J. Inorg. Biochem. 2015, 150, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wu, Q.; Zhang, H.; Wang, Q.; Wang, X.; Mei, W.; Wu, X.; Zheng, W. Microwave-assisted synthesis of ruthenium(II) complexes with alkynes as potential inhibitor by selectively recognizing c-myc G-quadruplex DNA. J. Inorg. Biochem. 2017, 176, 113–122. [Google Scholar] [CrossRef]
- Liu, X.-W.; Liu, N.-Y.; Deng, Y.-Q.; Wang, S.; Liu, T.; Tang, Y.-C.; Chen, Y.-D.; Lu, J.-L. A Luminescence Probe for c-myc G-quadruplex by a Triphenylamine-Appended Ruthenium Complex. Appl. Organomet. Chem. 2021, 35, e6143. [Google Scholar] [CrossRef]
- Yu, Q.; Liu, Y.; Zhang, J.; Yang, F.; Sun, D.; Liu, D.; Zhou, Y.; Liu, J. Ruthenium(II) polypyridyl complexes as G-quadruplex inducing and stabilizing ligands in telomeric DNA. Metallomics 2013, 5, 222–231. [Google Scholar] [CrossRef]
- Zhang, Z.; Mei, W.; Wu, X.; Wang, X.; Wang, B.; Chen, S. Synthesis and Characterization of Chiral Ruthenium(II) Complexes Λ/Δ-[Ru(Bpy)2(H2Iip)](ClO4)2 as Stabilizers of c-Myc G-quadruplex DNA. J. Coord. Chem. 2015, 68, 1465–1475. [Google Scholar] [CrossRef]
- Xu, X.; Wang, S.; Mi, Y.; Zhao, H.; Zheng, Z.; Zhao, X. A hydroxyquinoline-appended ruthenium(II)-polypyridyl complex that induces and stabilizes G-quadruplex DNA. J. Coord. Chem. 2019, 72, 201–217. [Google Scholar] [CrossRef]
- Liu, X.-W.; Liu, N.-Y.; Deng, Y.-Q.; Wang, S.; Liu, T. Topoisomerase I Inhibition, DNA Photocleavage Activity, and G-quadruplex DNA ‘Light Switch’ Based on Nitro-Substituted Ruthenium Complexes. Russ. J. Inorg. Chem. 2020, 65, 1186–1195. [Google Scholar]
- Liu, X.; Liu, N.; Deng, Y.; Wang, S.; Liu, T.; Tang, Y.; Chen, Y.; Lu, J. An unexpected fluorescent probe for G-quadruplex DNA based on a nitro-substituted ruthenium (II) complex. Appl. Organomet. Chem. 2020, 34, e5703. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, Q.; Li, Q.; Yang, L.; Chen, L.; Zhou, Y.; Liu, J. A ruthenium(II) complex capable of inducing and stabilizing bcl-2 G-quadruplex formation as a potential cancer inhibitor. J. Inorg. Biochem. 2014, 134, 1–11. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, J.; Yang, L.; Yu, Q.; Chen, Q.; Qin, X.; Le, F.; Zhang, Q.; Liu, J. Stabilization of G-quadruplex DNA and inhibition of telomerase activity studies of ruthenium(II) complexes. J. Inorg. Biochem. 2014, 130, 122–129. [Google Scholar] [CrossRef]
- Liu, X.-W.; Liu, N.-Y.; Deng, Y.-Q.; Wang, S.; Liu, T.; Tang, Y.-C.; Chen, Y.-D.; Lu, J.-L. Nitro-Substituted Ruthenium(II) Complex: A New Strategy for a G-quadruplex DNA Fluorescent Probe. Inorg. Chem. 2019, 58, 16326–16329. [Google Scholar] [CrossRef]
- Weynand, J.; Diman, A.; Abraham, M.; Marcélis, L.; Jamet, H.; Decottignies, A.; Dejeu, J.; Defrancq, E.; Elias, B. Towards the Development of Photo-Reactive Ruthenium(II) Complexes Targeting Telomeric G-quadruplex DNA. Chem. Eur. J. 2018, 24, 19216–19227. [Google Scholar] [CrossRef] [PubMed]
- Sheet, S.K.; Rabha, M.; Sen, B.; Patra, S.K.; Aguan, K.; Khatua, S. Ruthenium(II) Complex-Based G-quadruplex DNA Selective Luminescent ‘Light-up’ Probe for RNase H Activity Detection. ChemBioChem 2021, 22, 2880–2887. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wu, J.-H.; Lai, Y.-W.; Zhao, R.; Chao, H.; Ji, L.-N. Targeting telomeric G-quadruplexes with the ruthenium(II) complexes [Ru(bpy)2(ptpn)]2+ and [Ru(phen)2(ptpn)]2+. Dalton Trans. 2013, 42, 4386–4397. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Sun, N.; Zhou, Y.; Liu, D.; Zhang, Q.; Liu, J. Anticancer activity of novel ruthenium complex with 1,10-phenanthrolineselenazole as potent telomeric G-quadruplex inhibitor. Inorg. Chem. Commun. 2012, 20, 142–146. [Google Scholar] [CrossRef]
- Xia, Y.; Chen, Q.; Qin, X.; Sun, D.; Zhang, J.; Liu, J. Studies of Ruthenium(II)-2,2′-Bisimidazole Complexes on Binding to G-quadruplex DNA and Inducing Apoptosis in HeLa Cells. New J. Chem. 2013, 37, 3706–3715. [Google Scholar] [CrossRef]
- Wumaier, M.; Shi, J.-J.; Yao, T.-M.; Hu, X.-C.; Gao, R.-R.; Shi, S. G-quadruplex and duplex DNA binding studies of novel Ruthenium(II) complexes containing ascididemin ligands. J. Inorg. Biochem. 2019, 196, 110681. [Google Scholar] [CrossRef]
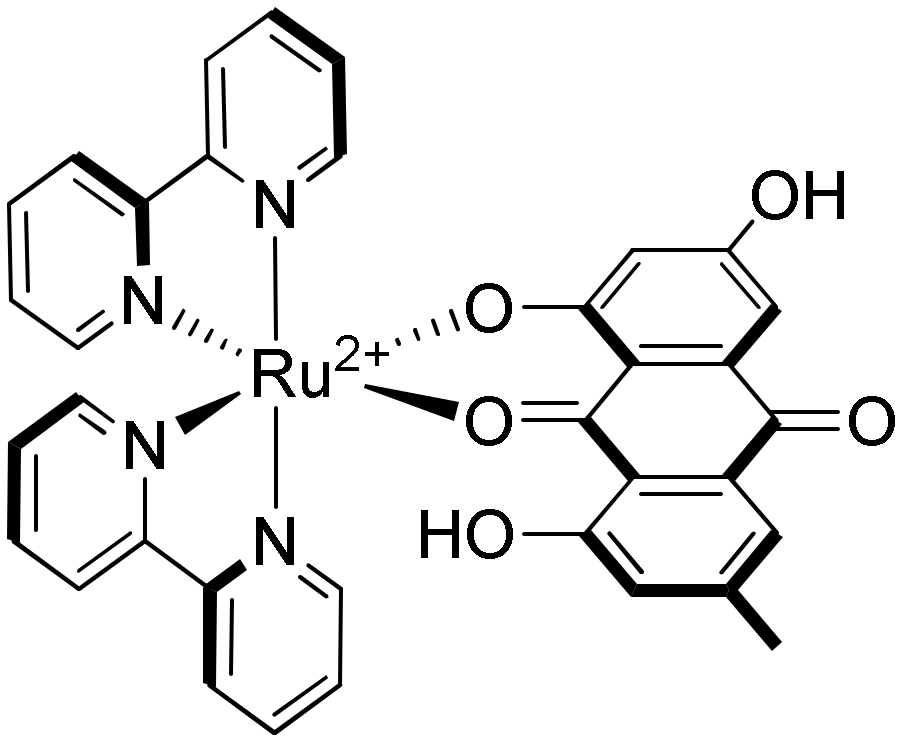
- Zhang, Z.; Wu, X.-H.; Sun, F.-Q.; Shan, F.; Chen, J.-C.; Chen, L.-M.; Zhou, Y.-S.; Mei, W.-J. Synthesis, characterization of ruthenium(II) complex of 1,3,8-trihydroxy-6-methyl-anthraquinone (emodin) and its binding behavior with c-myc G-quadruplex. Inorg. Chim. Acta 2014, 418, 23–29. [Google Scholar] [CrossRef]
- He, L.; Chen, X.; Meng, Z.; Wang, J.; Tian, K.; Li, T.; Shao, F. Octahedral ruthenium complexes selectively stabilize G-quadruplexes. Chem. Commun. 2016, 52, 8095–8098. [Google Scholar] [CrossRef]
- Prasad, B.; Jamroskovic, J.; Bhowmik, S.; Kumar, R.; Romell, T.; Sabouri, N.; Chorell, E. Flexible Versus Rigid G-quadruplex DNA Ligands: Synthesis of Two Series of Bis-indole Derivatives and Comparison of Their Interactions with G-quadruplex DNA. Chem. Eur. J. 2018, 24, 7926–7938. [Google Scholar] [CrossRef]
- Ihmels, H.; Karbasiyoun, M.; Löhl, K.; Stremmel, C. Structural flexibility versus rigidity of the aromatic unit of DNA ligands: Binding of aza- and azoniastilbene derivatives to duplex and quadruplex DNA. Org. Biomol. Chem. 2019, 17, 6404–6413. [Google Scholar] [CrossRef] [Green Version]
- Gillard, M.; Weynand, J.; Bonnet, H.; Loiseau, F.; Decottignies, A.; Dejeu, J.; Defrancq, E.; Elias, B. Flexible Ru(II) Schiff Base Complexes: G-quadruplex DNA Binding and Photo-Induced Cancer Cell Death. Chem. Eur. J. 2020, 26, 13849–13860. [Google Scholar] [CrossRef]
- Friedman, A.E.; Chambron, J.C.; Sauvage, J.P.; Turro, N.J.; Barton, J.K. A molecular light switch for DNA: Ru(bpy)2(dppz)2+. J. Am. Chem. Soc. 1990, 112, 4960–4962. [Google Scholar] [CrossRef]
- Hartshorn, R.M.; Barton, J.K. Novel dipyridophenazine complexes of ruthenium(II): Exploring luminescent reporters of DNA. J. Am. Chem. Soc. 1992, 114, 5919–5925. [Google Scholar] [CrossRef]
- Rajput, C.; Rutkaite, R.; Swanson, L.; Haq, I.; Thomas, J.A. Dinuclear Monointercalating Ru(II) Complexes That Display High Affinity Binding to Duplex and Quadruplex DNA. Chem. Eur. J. 2006, 12, 4611–4619. [Google Scholar] [CrossRef]
- Wilson, T.; Williamson, M.P.; Thomas, J.A. Differentiating quadruplexes: Binding preferences of a luminescent dinuclear ruthenium(II) complex with four-stranded DNA structures. Org. Biomol. Chem. 2010, 8, 2617–2621. [Google Scholar] [CrossRef]
- Wilson, T.; Costa, P.J.; Félix, V.; Williamson, M.P.; Thomas, J.A. Structural Studies on Dinuclear Ruthenium(II) Complexes That Bind Diastereoselectively to an Antiparallel Folded Human Telomere Sequence. J. Med. Chem. 2013, 56, 8674–8683. [Google Scholar] [CrossRef]
- Gill, M.; Garcia-Lara, J.; Foster, S.J.; Smythe, C.; Battaglia, G.; Thomas, J. A ruthenium(II) polypyridyl complex for direct imaging of DNA structure in living cells. Nat. Chem. 2009, 1, 662–667. [Google Scholar] [CrossRef]
- Gonzalez, V.; Wilson, T.; Kurihara, I.; Imai, A.; Thomas, J.A.; Otsuki, J. A dinuclear ruthenium(II) complex that functions as a label-free colorimetric sensor for DNA. Chem. Commun. 2008, 1868–1870. [Google Scholar] [CrossRef]
- Zhao, X.-L.; Zhao, H.-Q.; Xu, X.-X.; Li, Z.-S.; Wang, K.-Z. Inducement and stabilization of G-quadruplex DNA by a thiophene-containing dinuclear ruthenium(II) complex. J. Coord. Chem. 2017, 70, 2094–2112. [Google Scholar] [CrossRef]
- Zhao, H.; Xu, X.; Wang, S.; Mi, Y.; Zheng, Z.; Zhao, X. A dinuclear ruthenium(II) complex as an inducer and potential luminescent switch-on probe for G-quadruplex DNA. Transit. Met. Chem. 2018, 43, 539–548. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, D.; Huang, J.; Deng, M.; Zhang, M.; Zhou, X. High fluorescence selectivity and visual detection of G-quadruplex structures by a novel dinuclear ruthenium complex. Chem. Commun. 2009, 46, 743–745. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Liu, Y.; Liu, Y.; Qin, X.; Zhou, Y.; Liu, J. Dinuclear ruthenium complexes display loop isomer selectivity to c-MYC DNA G-quadruplex and exhibit anti-tumour activity. J. Inorg. Biochem. 2016, 156, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Gao, S.; Cao, T.; Liu, J.; Gao, X.; Hao, J.; Lv, C.; Huang, H.; Xu, J.; Yao, T. Targeting Human Telomeric G-quadruplex DNA and Inhibition of Telomerase Activity With [(dmb)2Ru(obip)Ru(dmb)2]4+. PLoS ONE 2013, 8, e84419. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Chen, X.; Wu, J.; Wang, J.; Ji, L.; Chao, H. Dinuclear Ruthenium(II) Complexes That Induce and Stabilise G-quadruplex DNA. Chem. Eur. J. 2015, 21, 4008–4020. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Liao, G.-L.; Chen, X.; Zhao, C.-Y.; Chao, H.; Ji, L.-N. Trinuclear Ru(II) polypyridyl complexes as human telomeric quadruplex DNA stabilizers. Inorg. Chem. Commun. 2010, 13, 1050–1053. [Google Scholar] [CrossRef]
- Lecomte, J.-P.; Kirsch-De Mesmaeker, A.; Feeney, M.M.; Kelly, J. Ruthenium(II) Complexes with 1,4,5,8,9,12-Hexaazatriphenylene and 1,4,5,8-Tetraazaphenanthrene Ligands: Key Role Played by the Photoelectron Transfer in DNA Cleavage and Adduct Formation. Inorg. Chem. 1995, 34, 6481–6491. [Google Scholar] [CrossRef]
- Blasius, R.; Moucheron, C.; Kirsch-De Mesmaeker, A. Photoadducts of Metallic Compounds with Nucleic Acids—Role Played by the Photoelectron Transfer Process and by the TAP and HAT Ligands in the RuII Complexes. Eur. J. Inorg. Chem. 2004, 2004, 3971–3979. [Google Scholar] [CrossRef]
- Wagenknecht, H.-A.; Stemp, E.D.A.; Barton, J.K. Evidence of Electron Transfer from Peptides to DNA: Oxidation of DNA-Bound Tryptophan Using the Flash-Quench Technique. J. Am. Chem. Soc. 1999, 122, 1–7. [Google Scholar] [CrossRef]
- Moucheron, C. From cisplatin to photoreactive Ru complexes: Targeting DNA for biomedical applications. New J. Chem. 2008, 33, 235–245. [Google Scholar] [CrossRef]
- Keane, P.M.; O’Sullivan, K.; Poynton, F.E.; Poulsen, B.C.; Sazanovich, I.V.; Towrie, M.; Cardin, C.J.; Sun, X.-Z.; George, M.W.; Gunnlaugsson, T.; et al. Understanding the factors controlling the photo-oxidation of natural DNA by enantiomerically pure intercalating ruthenium polypyridyl complexes through TA/TRIR studies with polydeoxynucleotides and mixed sequence oligodeoxynucleotides. Chem. Sci. 2020, 11, 8600–8609. [Google Scholar] [CrossRef]
- Ghizdavu, L.; Pierard, F.; Rickling, S.; Aury, S.; Surin, M.; Beljonne, D.; Lazzaroni, R.; Murat, P.; Defrancq, E.; Moucheron, C.; et al. Oxidizing Ru(II) Complexes as Irreversible and Specific Photo-Cross-Linking Agents of Oligonucleotide Duplexes. Inorg. Chem. 2009, 48, 10988–10994. [Google Scholar] [CrossRef]
- Rickling, S.; Ghisdavu, L.; Pierard, F.; Gerbaux, P.; Surin, M.; Murat, P.; Defrancq, E.; Moucheron, C.; Kirsch-De Mesmaeker, A. A Rigid Dinuclear Ruthenium(II) Complex as an Efficient Photoactive Agent for Bridging Two Guanine Bases of a Duplex or Quadruplex Oligonucleotide. Chem. Eur. J. 2010, 16, 3951–3961. [Google Scholar] [CrossRef]
- Archer, S.A.; Raza, A.; Dröge, F.; Robertson, C.; Auty, A.J.; Chekulaev, D.; Weinstein, J.A.; Keane, T.; Meijer, A.J.H.M.; Haycock, J.W.; et al. A dinuclear ruthenium(II) phototherapeutic that targets duplex and quadruplex DNA. Chem. Sci. 2019, 10, 3502–3513. [Google Scholar] [CrossRef] [Green Version]






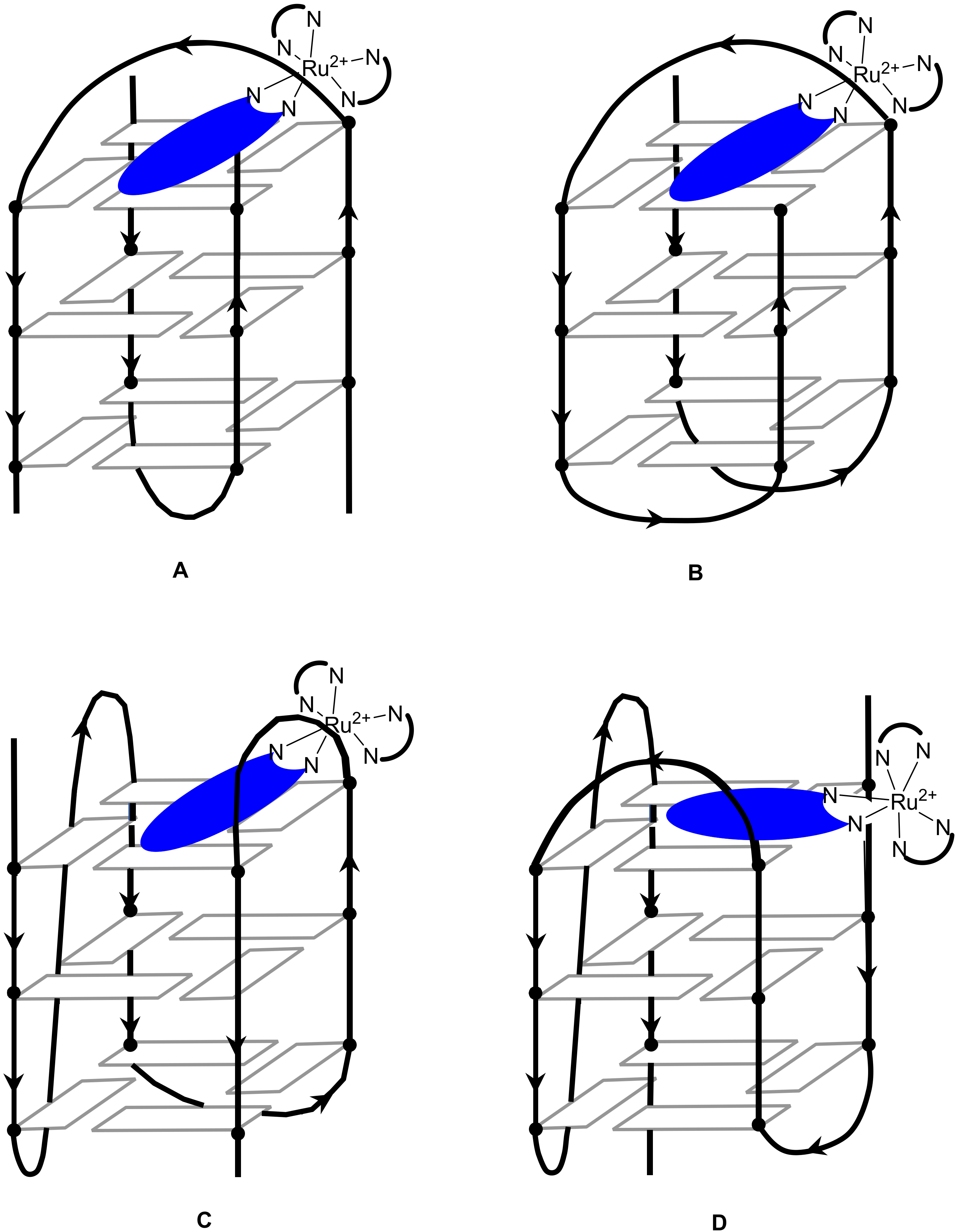
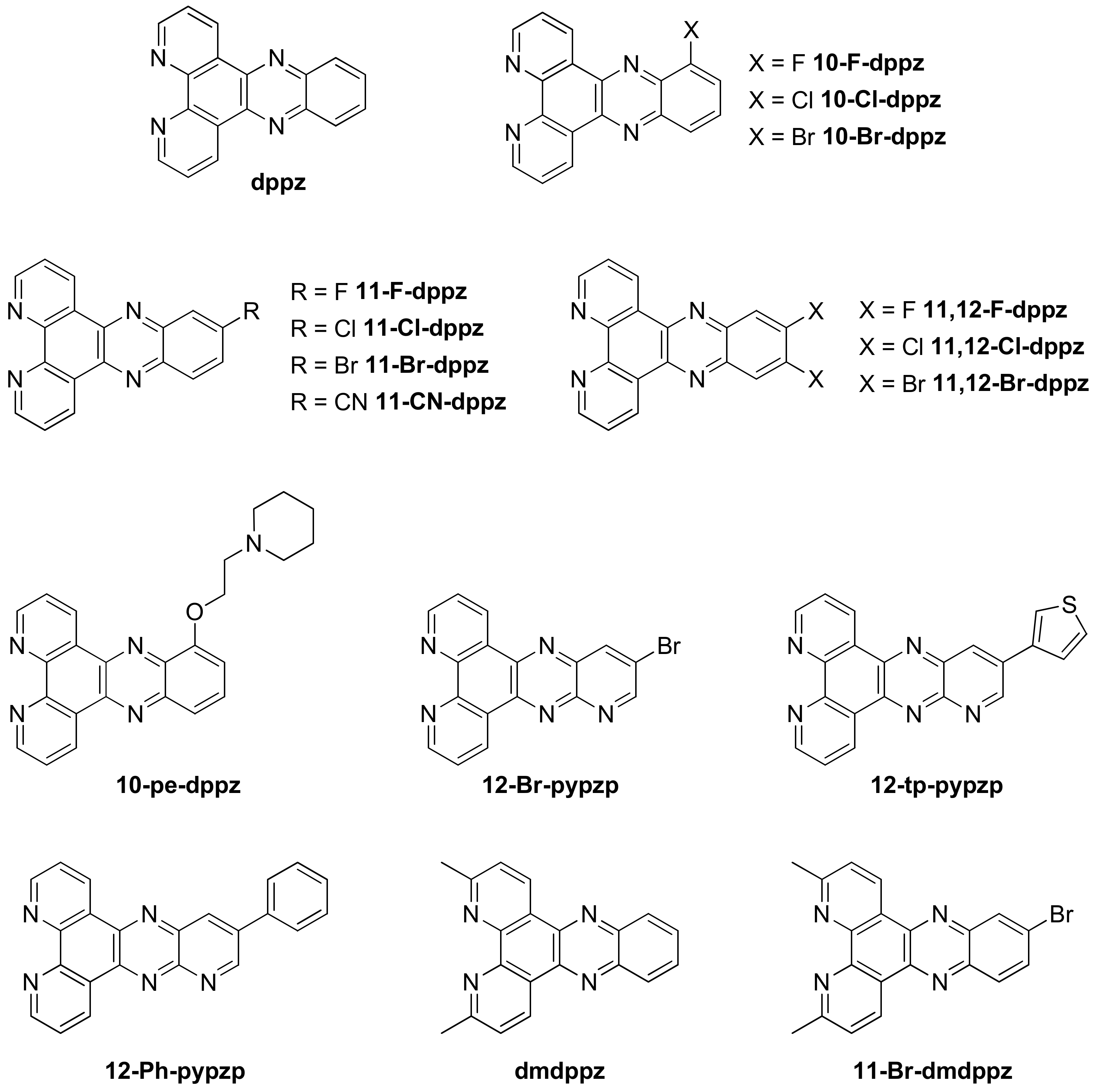
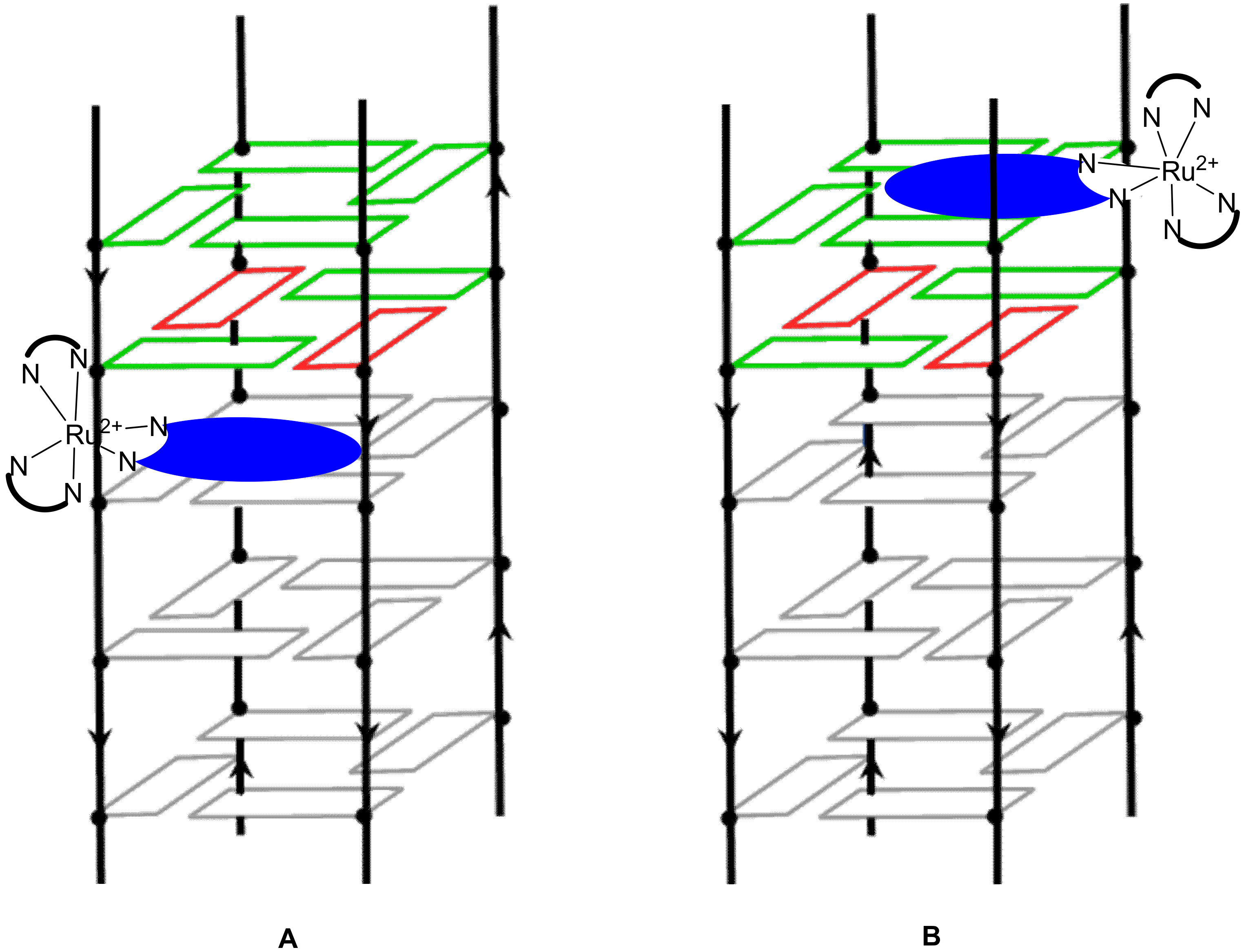
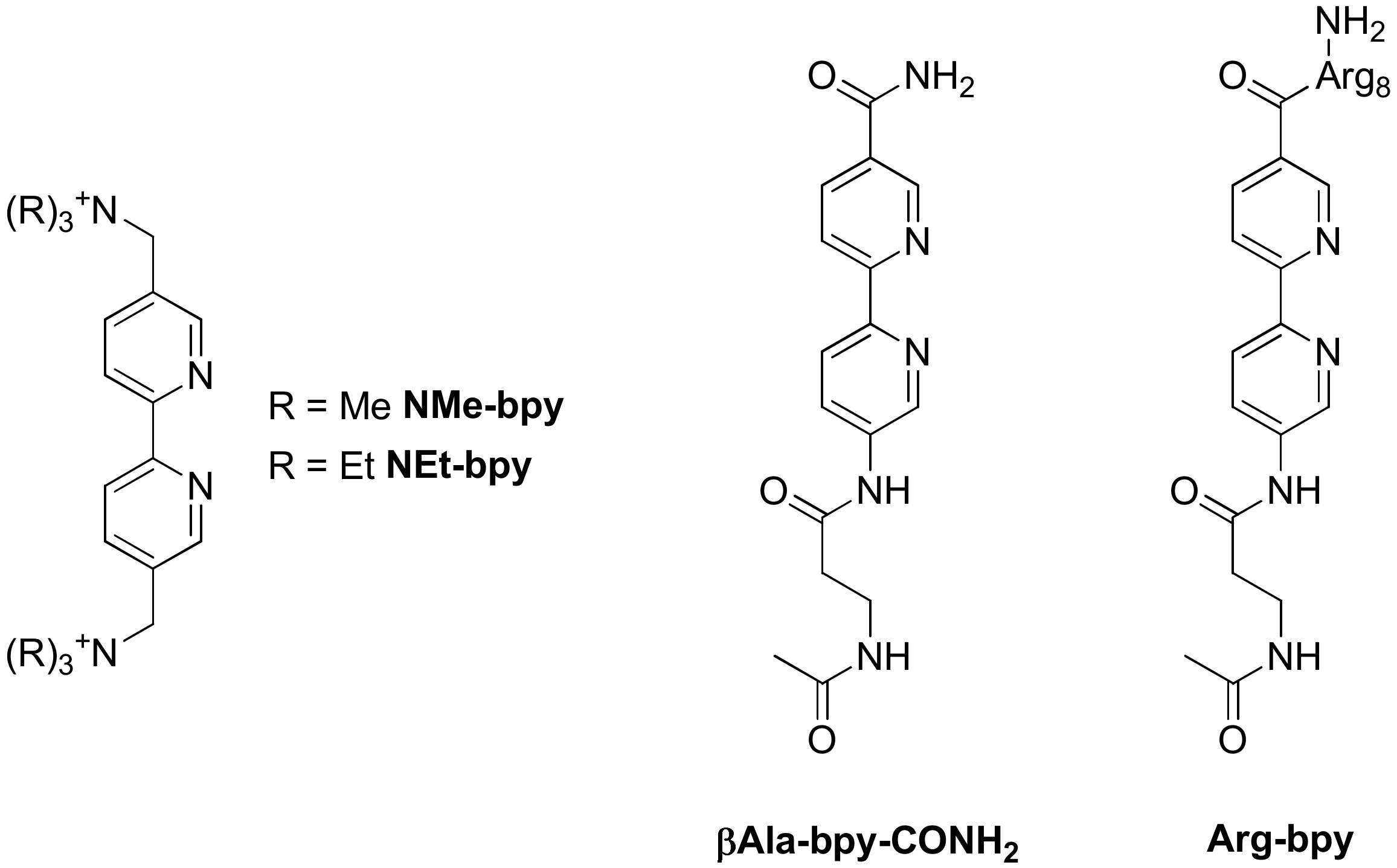
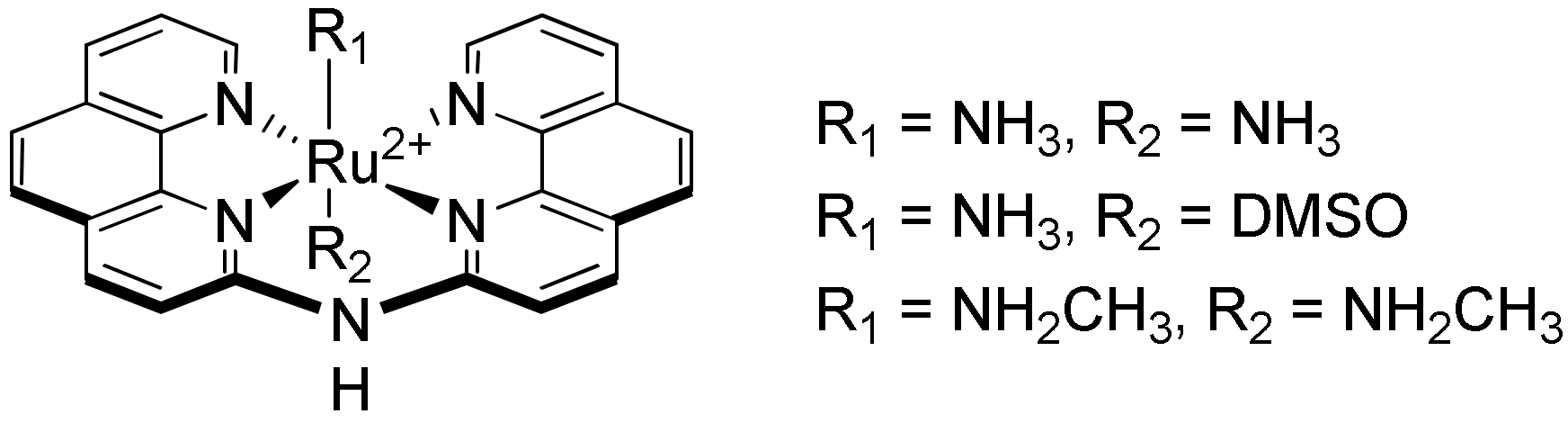
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, J.; Teunens, T.; Tisaun, J.; Denuit, L.; Moucheron, C. Ruthenium(II) Polypyridyl Complexes and Their Use as Probes and Photoreactive Agents for G-quadruplexes Labelling. Molecules 2022, 27, 1541. https://doi.org/10.3390/molecules27051541
Jiang J, Teunens T, Tisaun J, Denuit L, Moucheron C. Ruthenium(II) Polypyridyl Complexes and Their Use as Probes and Photoreactive Agents for G-quadruplexes Labelling. Molecules. 2022; 27(5):1541. https://doi.org/10.3390/molecules27051541
Chicago/Turabian StyleJiang, Julie, Titouan Teunens, Jérôme Tisaun, Laura Denuit, and Cécile Moucheron. 2022. "Ruthenium(II) Polypyridyl Complexes and Their Use as Probes and Photoreactive Agents for G-quadruplexes Labelling" Molecules 27, no. 5: 1541. https://doi.org/10.3390/molecules27051541





