Synthesis and Biological Evaluation of 1-(2-(6-Methoxynaphthalen-2-yl)-6-methylnicotinoyl)-4-Substituted Semicarbazides/Thiosemicarbazides as Anti-Tumor Nur77 Modulators
Abstract
:1. Introduction
2. Results
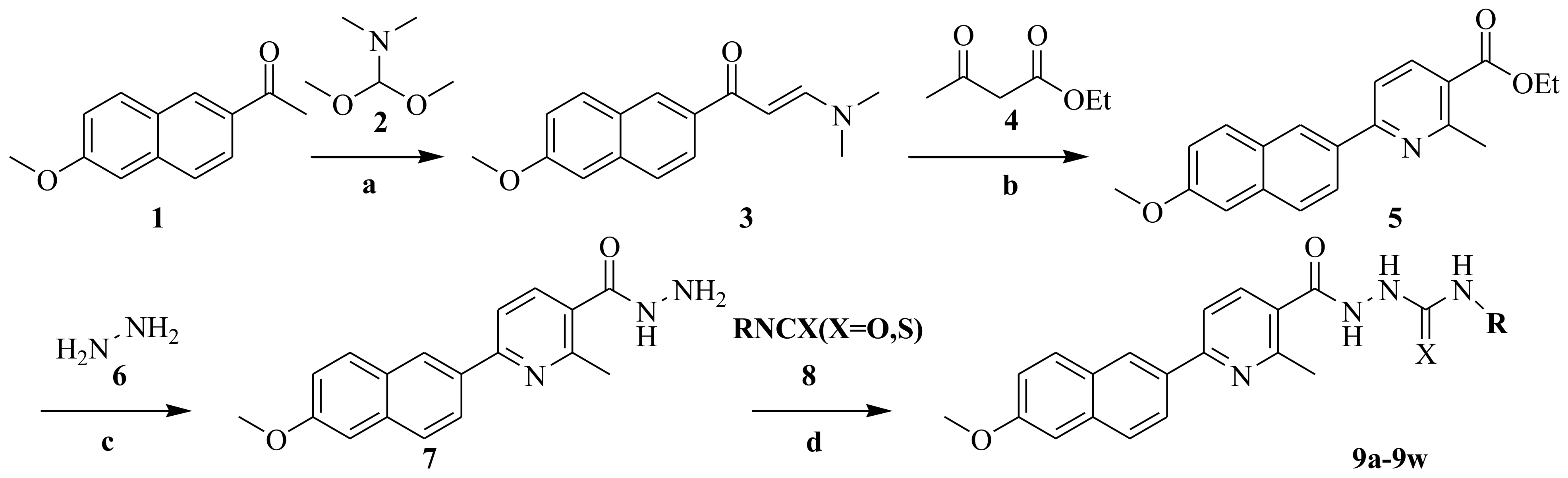
2.1. Chemistry
2.2. Biological Evaluation
2.2.1. Antiproliferative Screening In Vitro
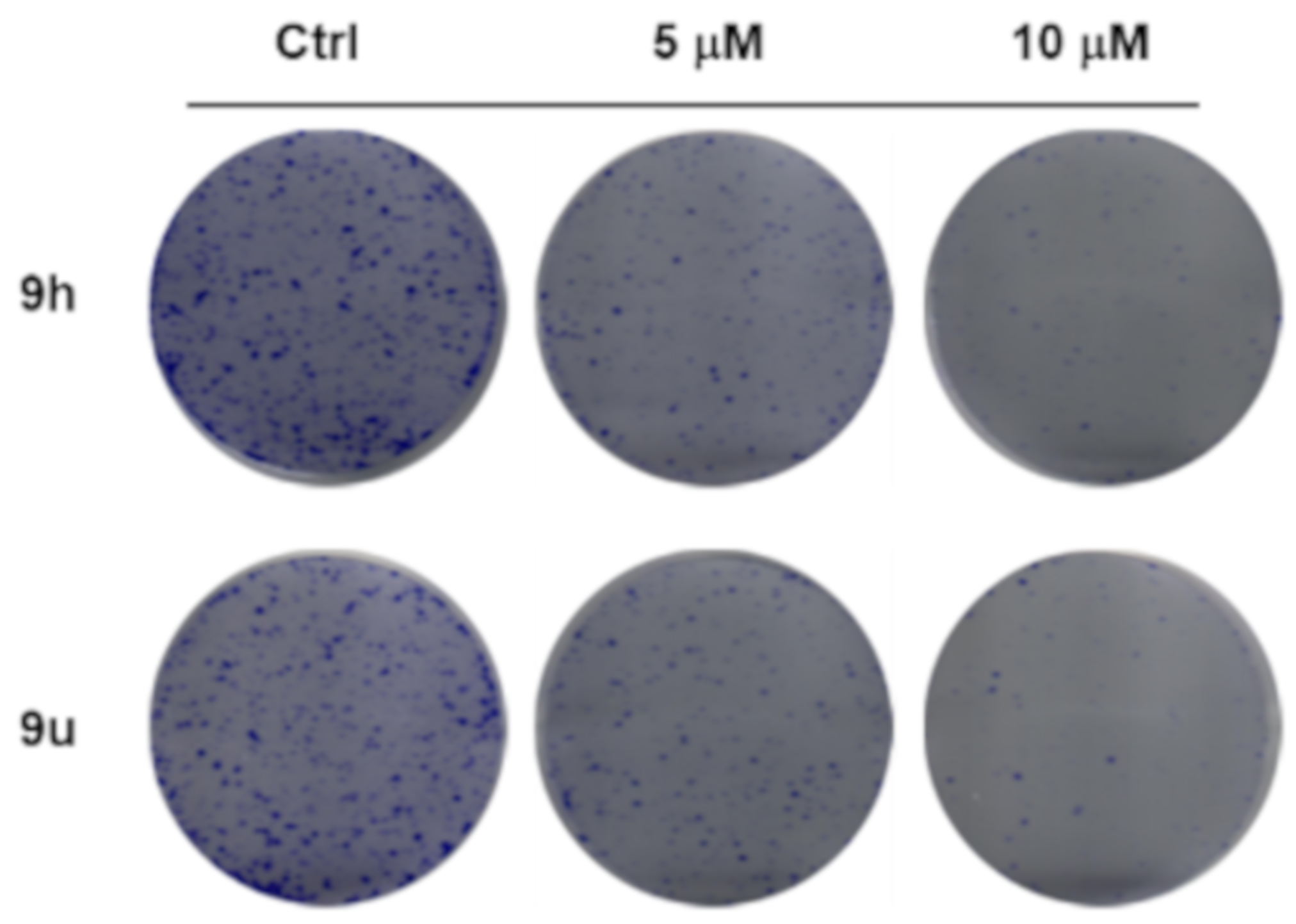
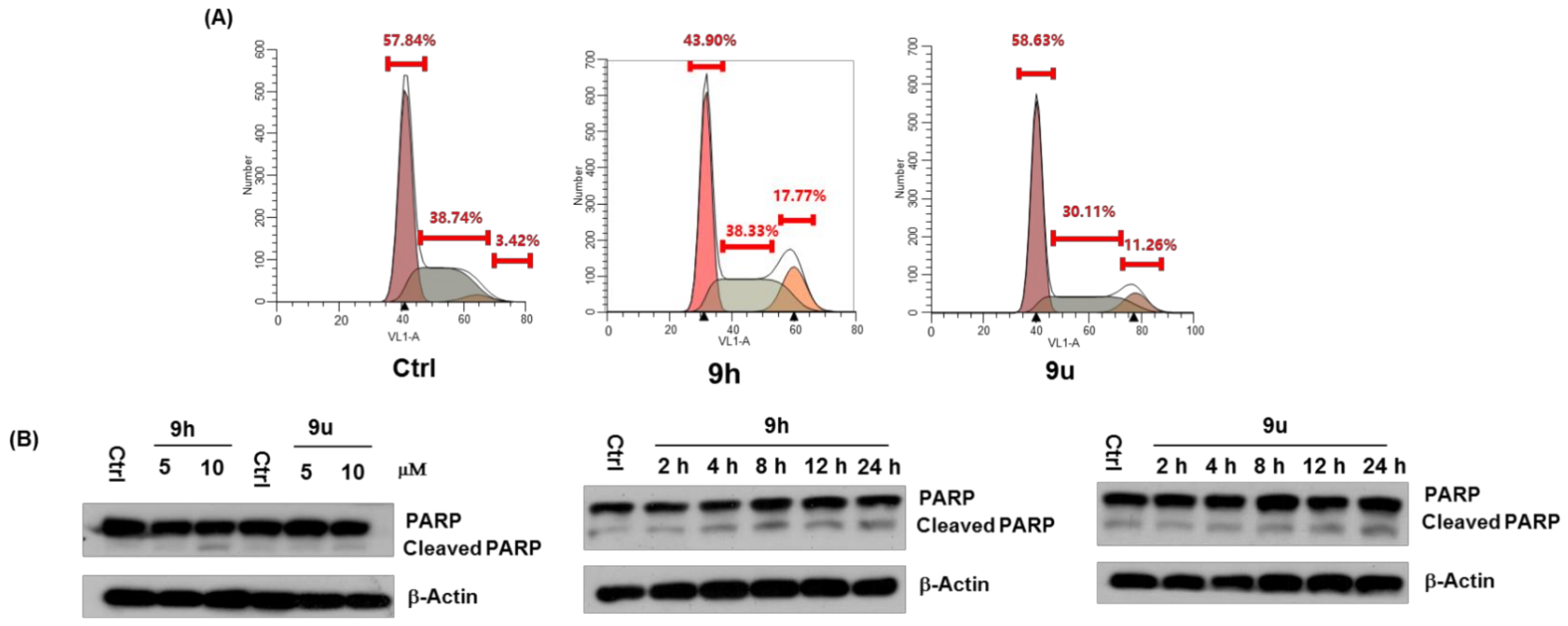
2.2.2. Effect of Compound 9h and 9u on Cell Growth, Cell Cycle and Apoptosis
2.2.3. 9h Induces Expression and Nuclear Export of Nur77, Which May Mediate 9h-Induced Apoptosis
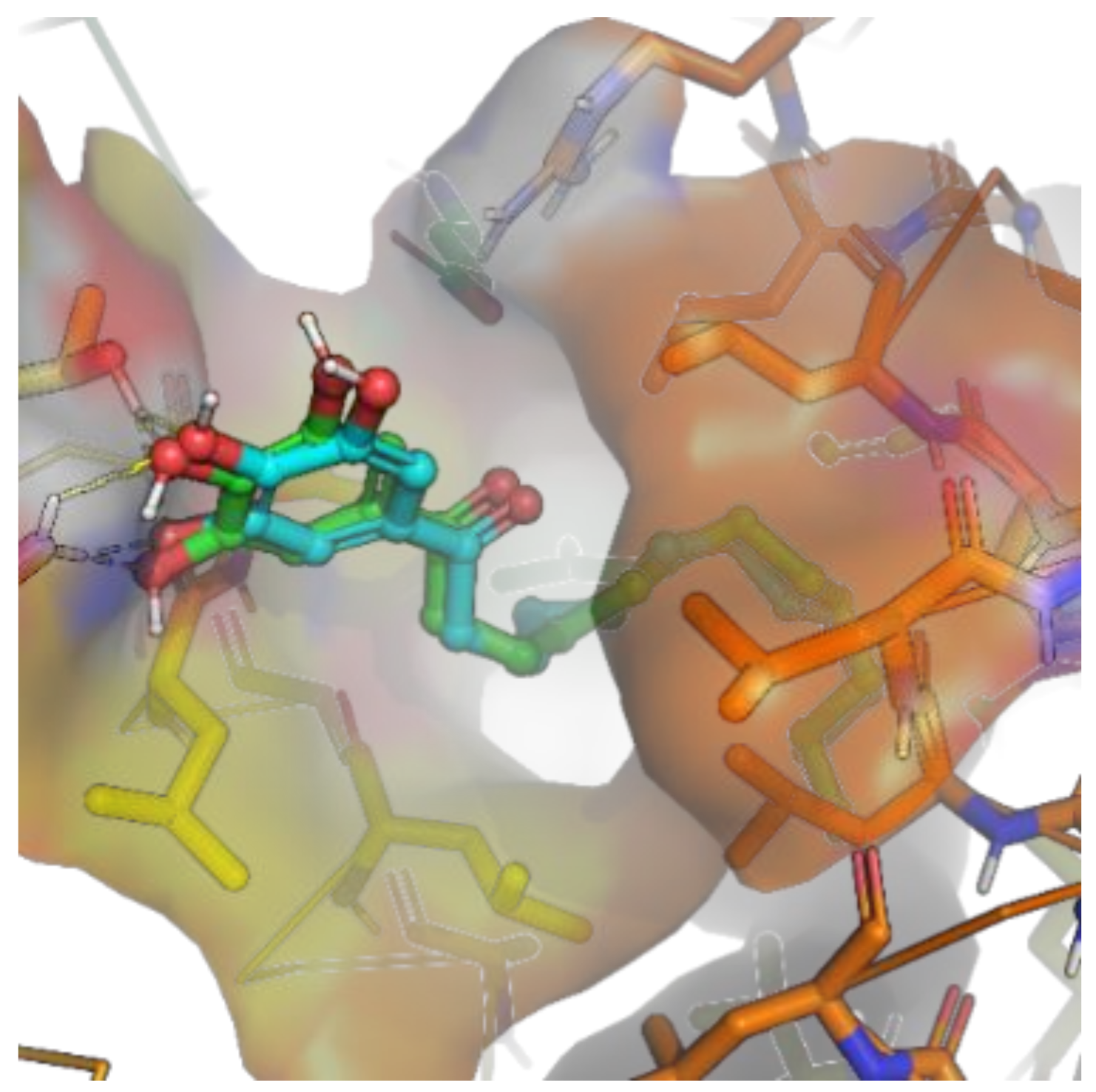
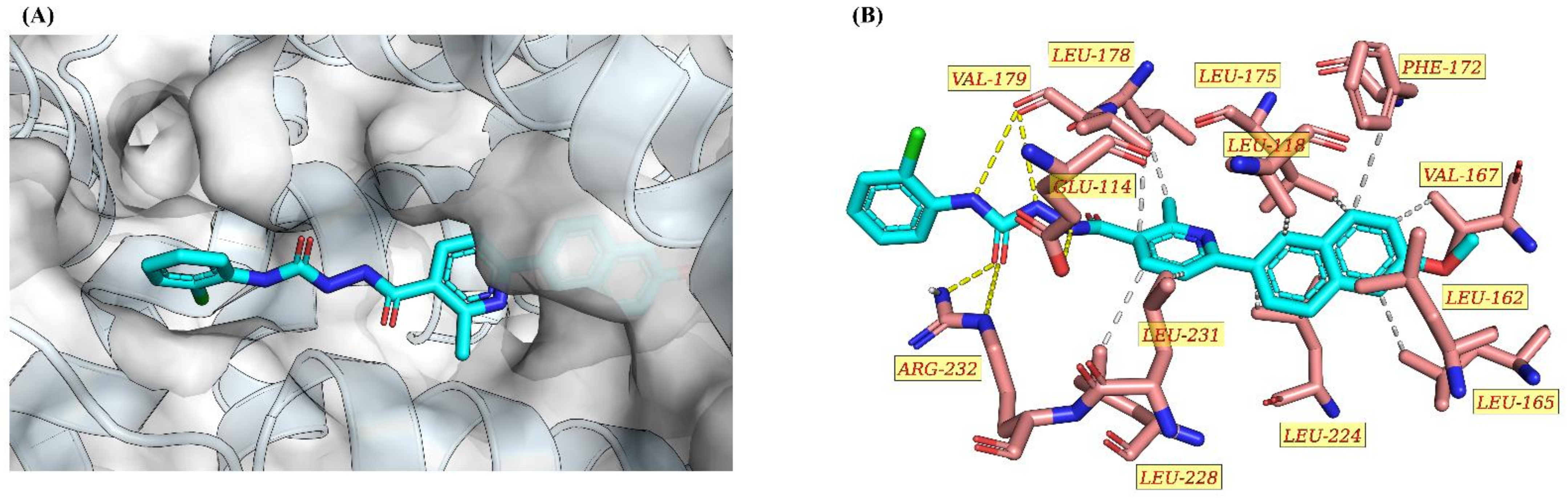
2.2.4. Molecular Docking Study of 9h and Nur77
3. Experimental Section
3.1. General Information
3.2. Synthesis
3.2.1. Procedure for Preparation of the (E)-3-(Dimethylamino)-1-(6-methoxynaphthalen-2-yl)prop-2-en-1-one (3)
3.2.2. Procedure for Preparation of the Ethyl-6-(6-methoxynaphthalen-2-yl)-2-methylnicotinate (5)
3.2.3. Procedure for Preparation of 6-(6-Methoxynaphthalen-2-yl)-2-methylnicotinohydrazide (7)
3.2.4. Procedure for the Preparation of 9a–9w
3.3. Cell Culture
3.4. Cytotoxicity against Cell Lines
3.5. Western Blot Analysis
3.6. Flow Cytometry Assay
3.7. Colony Formation Assay
3.8. Nuclear-Cytosol Fractionation Assay
3.9. Immunofluorescence
3.10. The Docking Experiments
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Al-Lazikani, B.; Hersey, A.; Oprea, T.I. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017, 16, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, S.; Wu, J.; Peng, Y.; Bai, J.; Ning, B.; Wang, X.; Fang, Y.; Han, D.; Ren, S.; et al. The orphan nuclear receptor Nur77 plays a vital role in BPA-induced PC12 cell apoptosis. Ecotoxicol. Environ. Saf. 2021, 213, 112026. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Luo, Q.; Alitongbieke, G.; Chong, S.; Xu, C.; Xie, L.; Chen, X.; Zhang, D.; Zhou, Y.; Wang, Z. Celastrol-Induced Nur77 Interaction with TRAF2 Alleviates Inflammation by Promoting Mitochondrial Ubiquitination and Autophagy. Mol. Cell 2017, 66, 141–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q. The orphan nuclear receptor Nur77 regulates hepatic cholesterol metabolism through the suppression of LDLR and HMGCR expression. Mol. Med. Rep. 2012, 5, 1541–1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lith, S.C.; van Os, B.W.; Seijkens, T.T.P.; de Vries, C.J.M. ‘Nur’turing tumor T cell tolerance and exhaustion: Novel function for Nuclear Receptor Nur77 in immunity. Eur. J. Immunol. 2020, 50, 1643–1652. [Google Scholar] [CrossRef]
- Cortez-Toledo, O.; Schnair, C.; Sangngern, P.; Metzger, D.; Cha, L.C. Nur77 deletion impairs muscle growth during developmental myogenesis and muscle regeneration in mice. PLoS ONE 2017, 12, e0171268. [Google Scholar] [CrossRef] [Green Version]
- Roecken, C. Molecular classification of gastric cancer. Expert Rev. Mol. Diagn. 2017, 17, 293–301. [Google Scholar] [CrossRef]
- Wan, Y.-J. Accelerated Partial Hepatectomy-Induced Liver Cell Proliferation Is Associated with Liver Injury in Nur77 Knockout Mice. Am. J. Pathol. Off. Publ. Am. Assoc. Pathol. 2016, 186, 210. [Google Scholar]
- Wu, Q.; Su, L.; Xiao-Feng, Y.; Zhi-Wei, H.; Wen-Jin, S. Dual roles of Nur77 in selective regulation of apoptosis and cell cycle by TPA and ATRA in gastric cancer cells. Carcinogenesis 2002, 23, 1583–1592. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.H.; Park, J.-S.; Moon, B.; Kim, M.C.; Kim, J.-K.; Lee, S.; Suh, H.; Kim, N.D.; Kim, J.-M.; Park, Y.C.; et al. Orphan Nuclear Receptor Nur77 Translocates to Mitochondria in the Early Phase of Apoptosis Induced by Synthetic Chenodeoxycholic Acid Derivatives in Human Stomach Cancer Cell Line SNU-1. Ann. N. Y. Acad. Sci. 2003, 1010, 171–177. [Google Scholar] [CrossRef]
- To, S.K.; Zeng, J.Z.; Wong, A.S. Nur77: A potential therapeutic target in cancer. Expert Opin. Ther. Targets 2012, 16, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Li, X.X.; Wang, Z.J.; Zheng, Y.; Guan, Y.F.; Yang, P.B.; Chen, X.; Peng, C.; He, J.P.; Ai, Y.L.; Wu, S.F. Nuclear Receptor Nur77 Facilitates Melanoma Cell Survival under Metabolic Stress by Protecting Fatty Acid Oxidation. Mol. Cell 2018, 69, 480–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, X.P.; Zhan, Y.P.; Liu, J.J.; Chen, H.Z.; Ym, W.Q.S. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat. Chem. Biol. 2008, 4, 548–556. [Google Scholar]
- Sun, Z.; Cao, X.; Jiang, M.; Qiu, Y.; Zhou, H.; Chen, L.; Qin, B. Inhibition of β-catenin signaling by nongenomic action of orphan nuclear receptor Nur77. Oncogene 2012, 31, 2653–2667. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; He, H.; Dou, G.; Li, J.; Zhang, X.; Jiang, M.; Li, P.; Huang, X.; Chen, H.; Li, L. Ginsenoside 20(S)-Rh2 Induces Apoptosis and Differentiation of Acute Myeloid Leukemia Cells: Role of Orphan Nuclear Receptor Nur77. J. Agric. Food Chem. 2017, 65, 7687–7697. [Google Scholar] [CrossRef]
- Lee, S.O.; Jin, U.H.; Kang, J.H.; Kim, S.B.; Guthrie, A.S.; Sreevalsan, S.; Lee, J.S.; Safe, S. The Orphan Nuclear Receptor NR4A1 (Nur77) Regulates Oxidative and Endoplasmic Reticulum Stress in Pancreatic Cancer Cells. Mol. Cancer Res. 2014, 12, 527–538. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Zhou, W.; Li, S.-S.; Sun, Z.; Lin, B.; Lang, Y.-Y.; He, J.-Y.; Cao, X.; Yan, T.; Wang, L.; et al. Modulation of orphan nuclear receptor Nur77-mediated apoptotic pathway by acetylshikonin and analogues. Cancer Res. 2008, 68, 8871–8880. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Lin, C.; Ao, M.; Ji, Y.; Tang, B.; Zhou, X.; Fang, M.; Zeng, J.; Wu, Z. Synthesis and biological evaluation of 1-(2-(adamantane-1-yl)-1H-indol-5-yl)-3-substituted urea/thiourea derivatives as anticancer agents. RSC Adv. 2017, 7, 51640–51651. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Yao, J.; Guo, K.; He, F.; Chen, K.; Lin, Z.; Liu, S.; Huang, J.; Wu, Q.; Fang, M.; et al. Design, synthesis, and biological evaluation of 5-((8-methoxy-2-methylquinolin-4-yl)amino)-1 H -indole-2-carbohydrazide derivatives as novel Nur77 modulators. Eur. J. Med. Chem. 2020, 204, 112608. [Google Scholar] [CrossRef]
- Hu, H.; Wu, J.; Ao, M.; Wang, H.; Zhou, T.; Xue, Y.; Qiu, Y.; Fang, M.; Wu, Z. Synthesis, structure–activity relationship studies and biological evaluation of novel 2,5-disubstituted indole derivatives as anticancer agents. Chem. Biol. Drug Des. 2016, 21, 530–541. [Google Scholar] [CrossRef]
- Bo, Z.; Hangzi, C.; Qiao, W. Research progress in orphan nuclear receptor Nur77 on the mechanisms of its multi-biological functions and drug targets. J. Xia Men Univ. 2021, 60, 277–289. [Google Scholar]
- Hu, H.; Wu, J.; Yuan, J.; Wang, Z.; Li, C.; Yan, X.; Fang, M.; Zhao, S. Synthesis and Biological Activity Research of 4-Substitued-1-(2-methyl-6-(pyridin-3-yl)-nicotinoyl) Semicarbazides. Chin. J. Org. Chem. 2019, 39, 2507–2514. [Google Scholar] [CrossRef]
- Hu, H.; Zhang, W.; Wu, J.; Zhang, X.; Sheng, H.; Zhao, S. Synthesisy and Antitumor Activities of Phenylpyridine Substuted Semicarbazides. Chin. Pharmaceut. J. 2019, 54, 747–752. [Google Scholar]
- El Sayed, M.T.; El-Sharief, M.A.; Zarie, E.S.; Morsy, N.; Elsheakh, A.R.; Nayel, M.; Voronkov, A.; Berishvili, V.; Sabry, N.M.; Hassan, G.S.; et al. Design, synthesis, anti-inflammatory antitumor activities, molecular modeling and molecular dynamics simulations of potential naprosyn® analogs as COX-1 and/or COX-2 inhibitors. Bioorgan. Chem. 2018, 76, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Roos, K.; Wu, C.; Damm, W.; Reboul, M.; Stevenson, J.M.; Lu, C.; Dahlgren, M.K.; Mondal, S.; Chen, W.; Wang, L.; et al. OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. J. Chem. Theory Comput. 2019, 15, 1863–1874. [Google Scholar] [CrossRef]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel Procedure for Modeling Ligand/Receptor Induced Fit Effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef]
- Sherman, W.; Beard, H.S.; Farid, R. Use of an Induced Fit Receptor Structure in Virtual Screening. Chem. Biol. Drug Des. 2006, 67, 83–84. [Google Scholar] [CrossRef]
- Farid, R.; Day, T.; Friesner, R.A.; Pearlstein, R.A. New insights about HERG blockade obtained from protein modeling, potential energy mapping, and docking studies. Bioorgan. Med. Chem. 2006, 14, 3160–3173. [Google Scholar] [CrossRef]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef]
- Ouyang, L.; Shi, Z.; Zhao, S.; Wang, F.T.; Zhou, T.T.; Liu, B.; Bao, J.K. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012, 45, 487–498. [Google Scholar] [CrossRef]
- Wong, R.S.Y. Apoptosis in cancer: From pathogenesis to treatment. J. Exp. Clin. Cancer Res. 2011, 30, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]


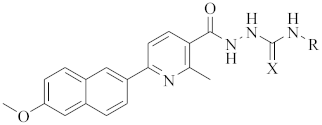 | ||||
|---|---|---|---|---|
| Compound | Structure/R | MTT | ||
| HeLa | HGC-27 | MCF-7 | ||
| 9a |  | 88.39 ± 0.22 | 38.39 ± 0.32 | 60.26 ± 0.11 |
| 9b |  | 30.54 ± 0.40 | 50.26 ± 0.20 | 41.46 ± 0.78 |
| 9c |  | 25.33 ± 0.45 | 23.16 ± 0.20 | 21.13 ± 0.52 |
| 9d | 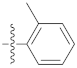 | 20.65 ± 0.26 | 39.12 ± 0.72 | 35.20 ± 0.60 |
| 9e | 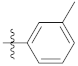 | 24.65 ± 0.24 | 49.24 ± 0.26 | 33.34 ± 0.12 |
| 9f | 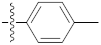 | 24.15 ± 0.61 | 29.44 ± 0.31 | 23.30 ± 0.10 |
| 9g | 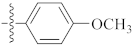 | 20.22 ± 0.31 | 19.40 ± 0.63 | 18.38 ± 0.21 |
| 9h | 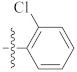 | 4.12 ± 0.08 | 1.40 ± 0.11 | 3.30 ± 0.21 |
| 9i | 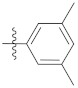 | 20.10 ± 0.44 | 19.67 ± 0.31 | 21.38 ± 0.25 |
| 9j | 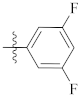 | 12.10 ± 0.21 | 16.56 ± 0.44 | 16.30 ± 0.34 |
| 9k | 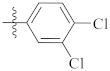 | 10.12 ± 0.69 | 12.40 ± 0.50 | 11.56 ± 0.45 |
| 9l | 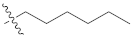 | 45.40 ± 0.11 | 38.10 ± 0.21 | 40.20 ± 0.33 |
| 9m | 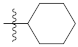 | 34.54 ± 0.37 | 30.20 ± 0.61 | 35.44 ± 0.41 |
| 9n | 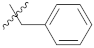 | 25.33 ± 0.10 | 23.16 ± 0.07 | 21.13 ± 0.32 |
| 9o | 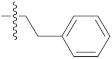 | 20.65 ± 0.22 | 39.12 ± 0.21 | 35.20 ± 0.09 |
| 9p |  | 14.65 ± 0.31 | 19.24 ± 0.09 | 23.34 ± 0.21 |
| 9q | 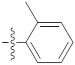 | 14.15 ± 0.22 | 19.44 ± 0.69 | 23.30 ± 0.21 |
| 9r |  | 20.22 ± 0.24 | 19.40 ± 0.11 | 18.38 ± 0.41 |
| 9s | 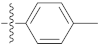 | 16.12 ± 0.07 | 10.40 ± 0.19 | 19.30 ± 0.30 |
| 9t | 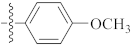 | 10.10 ± 0.61 | 16.67 | 11.38 |
| 9u | 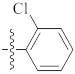 | 6.10 ± 0.50 | 4.56 ± 0.09 | 6.44 ± 0.07 |
| 9v | 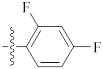 | 10.12 ± 0.11 | 12.40 ± 0.25 | 11.56 ± 0.56 |
| 9w |  | 10.66 ± 0.34 | 16.43 ± 0.31 | 10.90 ± 0.42 |
| Cisplatin | 12.45 ± 0.11 | 8.90 ± 0.20 | 10.54 ± 0.31 | |
| Compound | Cancer Cells (IC50, μM) | Normal Cells (IC50, μM) | |||||
|---|---|---|---|---|---|---|---|
| HepG2 | A549 | MDA-MB231 | H460 | A875 | MRC-5 | LO2 | |
| 9h | 5.1 ± 0.01 | 7.3 ± 0.21 | 4.3 ± 0.33 | 9.2 ± 0.24 | >50 | >50 | >50 |
| 9u | 7.2 ± 0.11 | 11.2 ± 0.46 | 21.0 ± 0.01 | 14.0 ± 0.38 | 15.7 ± 0.11 | 22.1 ± 0.11 | 41.0 ± 0.70 |
| Cisplatin | 15.2 ± 0.29 | 9.3 ± 0.11 | 20.2 ± 0.25 | 16.6 ± 0.09 | 21.2 ± 0.08 | >50 | >50 |
| RESNR | RESTYPE | DIST | RESNR | RESTYPE | DIST |
|---|---|---|---|---|---|
| 118 | LEU | 3.87 | 179 | VAL | 3.47 |
| 162 | LEU | 3.36 | 224 | LEU | 3.52 |
| 165 | LEU | 3.57 | 224 | LEU | 3.61 |
| 167 | VAL | 3.5 | 224 | LEU | 3.41 |
| 172 | PHE | 3.57 | 228 | LEU | 3.45 |
| 175 | LEU | 3.89 | 231 | LEU | 3.46 |
| 178 | LEU | 3.75 |
| RESNR | RESTYPE | SIDECHAIN | DIST_H-A | DIST_D-A | DON_ANGLE | PROTISDON |
|---|---|---|---|---|---|---|
| (Å) | (Å) | (°) | ||||
| 114 | GLU | TRUE | 2.36 | 3.17 | 136.65 | FALSE |
| 179 | VAL | FALSE | 2.54 | 3.49 | 155.72 | FALSE |
| 179 | VAL | FALSE | 2.53 | 3.44 | 149.64 | FALSE |
| 232 | ARG | TRUE | 1.85 | 2.8 | 153.65 | TRUE |
| 232 | ARG | TRUE | 3.39 | 3.96 | 116.77 | TRUE |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, H.; Huang, J.; Cao, Y.; Zhang, Z.; He, F.; Lin, X.; Wu, Q.; Zhao, S. Synthesis and Biological Evaluation of 1-(2-(6-Methoxynaphthalen-2-yl)-6-methylnicotinoyl)-4-Substituted Semicarbazides/Thiosemicarbazides as Anti-Tumor Nur77 Modulators. Molecules 2022, 27, 1698. https://doi.org/10.3390/molecules27051698
Hu H, Huang J, Cao Y, Zhang Z, He F, Lin X, Wu Q, Zhao S. Synthesis and Biological Evaluation of 1-(2-(6-Methoxynaphthalen-2-yl)-6-methylnicotinoyl)-4-Substituted Semicarbazides/Thiosemicarbazides as Anti-Tumor Nur77 Modulators. Molecules. 2022; 27(5):1698. https://doi.org/10.3390/molecules27051698
Chicago/Turabian StyleHu, Hongyu, Jiangang Huang, Yin Cao, Zhaolin Zhang, Fengming He, Xianfu Lin, Qi Wu, and Shengxian Zhao. 2022. "Synthesis and Biological Evaluation of 1-(2-(6-Methoxynaphthalen-2-yl)-6-methylnicotinoyl)-4-Substituted Semicarbazides/Thiosemicarbazides as Anti-Tumor Nur77 Modulators" Molecules 27, no. 5: 1698. https://doi.org/10.3390/molecules27051698






