Inhibition of the Quorum Sensing System, Elastase Production and Biofilm Formation in Pseudomonas aeruginosa by Psammaplin A and Bisaprasin
Abstract
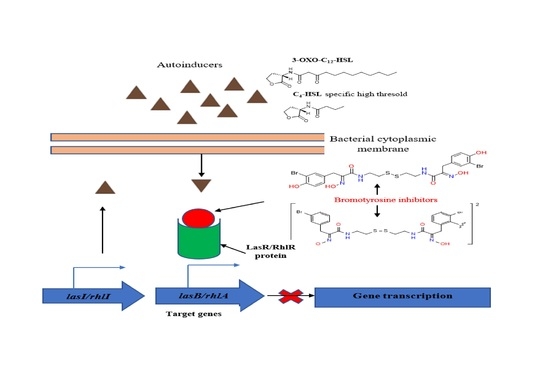
:1. Introduction
2. Results and Discussion
2.1. Isolation and Structure Elucidation
2.2. Inhibition of the Quorum Sensing Systems of P. aeruginosa
2.3. Effects of Psammaplin A and Bisaprasin on Elastase Production in P. aeruginosa
2.4. Evaluation of Psammaplin A and Bisaprasin against Biofilm Formation in P. aeruginosa
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Sponge Collection and Identification
3.3. Extraction and Isolation of Psammaplin A (1) and Bisaprasin (2)
3.4. Bacterial Strains
3.5. P. aeruginosa Quorum Sensing Inhibition Assays
3.6. Elastase Assay
3.7. Biofilm Assay Screening
3.8. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Aminov, R.A. Brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, L. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Yadav, G.; Thakuria, B.; Madan, M.; Agwan, V.; Pandey, A. Linezolid and vancomycin-resistant Enterococci: A therapeutic problem. J. Clin. Diagn. Res. 2017, 11, GC07–GC11. [Google Scholar] [CrossRef] [PubMed]
- Markwart, R.; Willrich, N.; Haller, S.; Noll, I.; Koppe, U.; Werner, G.; Eckmanns, T.; Reuss, A. The rise in vancomycin-resistant Enterococcus faecium in Germany: Data from the German antimicrobial resistance surveillance (ARS). Antimicrob. Resist. Infect. Control 2019, 8, 147. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.; Basuino, L.; Diep, B.; Hamilton, S.; Chatterjee, S.S.; Chambers, H. Ceftobiprole- and ceftaroline-resistant methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2015, 59, 2960–2963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harkins, C.; Pichon, B.; Doumith, M.; Parkhill, J.; Westh, H.; Tomasz, A.; de Lencastre, H.; Bentley, S.; Kearns, A.; Holden, M. Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol. 2017, 18, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabat, A.; Tinelli, M.; Grundmann, H.; Akkerboom, V.; Monaco, M.; Del Grosso, M.; Errico, G.; Pantosti, A.; Friedrich, A. Daptomycin resistant Staphylococcus aureus clinical strain with novel non-synonymous mutations in the mprf and vras genes: A new insight into daptomycin resistance. Front. Microbiol. 2018, 9, 2705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marty, F.; Yeh, W.; Wennersten, C.; Venkataraman, L.; Albano, E.; Alyea, E.; Gold, H.; Baden, L.; Pillai, S. Emergence of a clinical daptomycin-resistant Staphylococcus aureus isolate during treatment of methicillin-resistant Staphylococcus aureus Bacteremia and Osteomyelitis. J. Clin. Microbiol. 2006, 44, 595–597. [Google Scholar] [CrossRef] [Green Version]
- Lombardi, F.; Gaia, P.; Valaperta, R.; Cornetta, M.; Tejada, M.; Di Girolamo, L.; Moroni, A.; Ramundo, F.; Colombo, A.; Valisi, M.; et al. Emergence of carbapenem-resistant Klebsiella pneumoniae: Progressive spread and four-year period of observation in a cardiac surgery division. Biomed. Res. Int. 2015, 2015, 871947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David, S.; Reuter, S.; Harris, S.; Glasner, C.; Feltwell, T.; Argimon, S.; Abudahab, K.; Goater, R.; Giani, T.; Errico, G.; et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 2019, 4, 1919–1929. [Google Scholar] [CrossRef]
- Chen, C.M.; Guo, M.K.; Ke, S.C.; Lin, Y.P.; Li, C.R.; Nguyen, H.T.V.; Wu, L.T. Emergence and nosocomial spread of ST11 carbapenem-resistant Klebsiella pneumoniae co-producing OXA-48 and KPC-2 in a regional hospital in Taiwan. J. Med. Microbiol. 2018, 67, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Abdul Momin, M.; Liakopoulos, A.; Phee, L.; Wareham, D. Emergence and nosocomial spread of carbapenem-resistant OXA-232-producing Klebsiella pneumoniae in Brunei Darussalam. J. Glob. Antimicrob. Resist. 2017, 9, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Kuo, S.C.; Chang, S.C.; Wang, H.Y.; Lai, J.F.; Chen, P.C.; Shiau, Y.R.; Huang, I.W.; Lauderdale, T.L.; TSAR Hospitals. Emergence of extensively drug-resistant Acinetobacter baumannii complex over 10 years: Nationwide data from the Taiwan surveillance of antimicrobial resistance (TSAR) program. BMC Infect. Dis. 2012, 12, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dent, L.; Marshall, D.; Pratap, S.; Hulette, R. Multidrug-resistant Acinetobacter baumannii: A descriptive study in a city hospital. BMC Infect. Dis. 2010, 10, 196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kempf, M.; Rolain, J. Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: Clinical impact and therapeutic options. Int. J. Antimicrob. Agents 2012, 39, 105–114. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.K.; Peck, K.R.; Cheong, H.S.; Chung, D.R.; Song, J.H.; Ko, K.S. Extreme drug resistance in Acinetobacter baumannii infections in intensive care units, South Korea. Emerg. Infect. Dis. 2009, 15, 1325–1327. [Google Scholar] [CrossRef]
- Lautenbach, E.; Synnestvedt, M.; Weiner, M.G.; Bilker, W.B.; Vo, L.; Schein, J.; Kim, M. Imipenem resistance in Pseudomonas aeruginosa emergence, epidemiology, and impact on clinical and economic outcomes. Infect. Control Hosp. Epidemiol. 2010, 31, 47–53. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.; Lin, T.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Jalal, S.; Ciofu, O.; Høiby, N.; Gotoh, N.; Wretlind, B. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 2000, 44, 710–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryan, L.; Haraphongse, R.; Elzen, H. Gentamicin resistance in clinical isolates of Pseudomonas aeruginosa associated with diminished gentamicin accumulation and no detectable enzymatic modification. J. Antibiot. 1976, 29, 743–753. [Google Scholar] [CrossRef] [Green Version]
- Sindeldecker, D.; Stoodley, P. The many Antibiotic resistance and tolerance strategies of Pseudomonas aeruginosa. Biofilm 2021, 3, 100056. [Google Scholar] [CrossRef]
- Kumarasamy, K.; Toleman, M.; Walsh, T.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.; Irfan, S.; et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. [Google Scholar] [CrossRef]
- Mateos, M.; Hernández-García, M.; del Campo, R.; Martínez-García, L.; Gijón, D.; Morosini, M.; Ruiz-Garbajosa, P.; Cantón, R. Emergence and persistence over time of carbapenemase-producing Enterobacter isolates in a Spanish university hospital in Madrid, Spain (2005–2018). Microb. Drug Resist. 2021, 27, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Raymond, B.; Powell, N. We must follow these five rules to avert an antimicrobial resistance crisis. Pharm. J. 2019, 303, 7931. [Google Scholar]
- Dai, L.; Wu, T.Q.; Xiong, Y.S.; Ni, H.B.; Ding, Y.; Zhang, W.C.; Chu, S.P.; Ju, S.Q.; Yu, J. Ibuprofen-mediated potential inhibition of biofilm development and quorum sensing in Pseudomonas aeruginosa. Life Sci. 2019, 237, 116947. [Google Scholar] [CrossRef] [PubMed]
- Ahator, S.D.; Zhang, L. Small is mighty—Chemical communication systems in Pseudomonas aeruginosa. Annu. Rev. Microbiol. 2019, 73, 559–578. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.; Bassler, B. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009, 43, 197–222. [Google Scholar] [CrossRef] [Green Version]
- Tolker-Nielsen, T. Biofilm development. Microbiol. Spectr. 2015, 3, MB-0001-2014. [Google Scholar] [CrossRef] [Green Version]
- İnat, G.; Sırıken, B.; Başkan, C.; Erol, İ.; Yıldırım, T.; Çiftci, A. Quorum sensing systems and related virulence factors in Pseudomonas aeruginosa isolated from chicken meat and ground beef. Sci. Rep. 2021, 11, 15639. [Google Scholar] [CrossRef] [PubMed]
- Elnegery, A.; Mowafy, W.; Zahra, T.; Abou El-Khier, N. Study of quorum-sensing LasR and RhlR genes and their dependent virulence factors in Pseudomonas aeruginosa isolates from infected burn wounds. Access Microbiol. 2021, 3, 000211. [Google Scholar] [CrossRef]
- Zhao, X.; Yu, Z.; Ding, T. Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms 2020, 8, 425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, D. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakobsen, T.; Bjarnsholt, T.; Jensen, P.; Givskov, M.; Høiby, N. Targeting quorum sensing in Pseudomonas aeruginosa biofilms: Current and emerging inhibitors. Future Microbiol. 2013, 8, 901–921. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wang, M.; Smalley, N.; Kostylev, M.; Schaefer, A.; Greenberg, E.; Dandekar, A.; Xu, F. Modulation of Pseudomonas aeruginosa quorum sensing by glutathione. J. Bacteriol. 2019, 201, 7027–7032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, V.; Bushnell, D.; Passador, L.; Brooks, A.; Iglewski, B. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: Effects of growth phase and environment. J. Bacteriol. 2003, 185, 2080–2095. [Google Scholar] [CrossRef] [Green Version]
- Schuster, M.; Peter Greenberg, E. A network of networks: Quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 2006, 296, 73–81. [Google Scholar] [CrossRef]
- Pesci, E.; Milbank, J.; Pearson, J.; McKnight, S.; Kende, A.; Greenberg, E.; Iglewski, B. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1999, 96, 11229–11234. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, T.; Givskov, M. Quorum-sensing inhibitors as antipathogenic drugs. Int. J. Med. Microbiol. 2006, 296, 149–161. [Google Scholar] [CrossRef]
- Nouwens, A.; Beatson, S.; Whitchurch, C.; Walsh, B.; Schweizer, H.; Mattick, J.; Cordwell, S. Proteome analysis of extracellular proteins regulated by the las and rhl quorum sensing systems in Pseudomonas aeruginosa PAO1. Microbiology 2003, 149, 1311–1322. [Google Scholar] [CrossRef] [Green Version]
- Cigana, C.; Castandet, J.; Sprynski, N.; Melessike, M.; Beyria, L.; Ranucci, S.; Alcalá-Franco, B.; Rossi, A.; Bragonzi, A.; Zalacain, M.; et al. Pseudomonas aeruginosa elastase contributes to the establishment of chronic lung colonization and modulates the immune response in a murine model. Front. Microbiol. 2021, 11, 62081. [Google Scholar] [CrossRef]
- Saint-Criq, V.; Villeret, B.; Bastaert, F.; Kheir, S.; Hatton, A.; Cazes, A.; Xing, Z.; Sermet-Gaudelus, I.; Garcia-Verdugo, I.; Edelman, A.; et al. Pseudomonas aeruginosa LasB protease impairs innate immunity in mice and humans by targeting a lung epithelial cystic fibrosis transmembrane regulator–IL-6–antimicrobial–repair pathway. Thorax 2018, 73, 49–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potempa, J.; Pike, R.N. Corruption of innate immunity by bacterial proteases. J. Innate Immun. 2009, 1, 70–87. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; LaRock, D.; Skowronski, E.; Kimmey, J.; Olson, J.; Jiang, Z.; O’Donoghue, A.; Nizet, V.; LaRock, C. The Pseudomonas aeruginosa protease LasB directly activates IL-1Β. EBioMedicine 2020, 60, 102984. [Google Scholar] [CrossRef]
- Mandl, I.; Keller, S.; Cohen, B. Microbial elastases. A comparative Study. Proc. Soc. Exp. Biol. Med. 1962, 109, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Mull, J.; Callahan, W. Estimation of elastolytic activity of strains of Pseudomonas aeruginosa. J. Bacteriol. 1963, 85, 1178–1179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pamp, S.; Tolker-Nielsen, T. Multiple Roles of Biosurfactants in Structural Biofilm Development by Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 2531–2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davey, M.; Caiazza, N.; O’Toole, G. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2003, 185, 1027–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Chen, J.; Yan, Y.; Chen, S.; Xu, X.; Zhang, H.; Wang, H. Quorum sensing inhibitors from marine bacteria Oceanobacillus sp. XC22919. Nat. Prod. Res. 2019, 33, 1819–1823. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.; Alexandre, K.; Etienne, M. Tolerance and persistence of Pseudomonas aeruginosa in biofilms exposed to antibiotics: Molecular mechanisms, antibiotic strategies and therapeutic perspectives. Front. Microbiol. 2020, 11, 2057. [Google Scholar] [CrossRef]
- Shaw, E.; Wuest, W.M. Virulence attenuating combination therapy: A potential multi-target synergy approach to treat Pseudomonas aeruginosa infections in cystic fibrosis patients. RSC Med. Chem. 2020, 11, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Furiga, A.; Lajoie, B.; El Hage, S.; Baziard, G.; Roques, C. Impairment of Pseudomonas aeruginosa biofilm resistance to antibiotics by combining the drugs with a new quorum-sensing inhibitor. Antimicrob. Agents Chemother. 2015, 60, 1676–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ham, S.Y.; Kim, H.S.; Jo, M.J.; Lee, J.H.; Byun, Y.; Ko, G.J.; Park, H.D. Combined treatment of 6-gingerol analog and tobramycin for inhibiting Pseudomonas aeruginosa infections. Microbiol. Spectr. 2021, 9, e0019221. [Google Scholar] [CrossRef] [PubMed]
- Jing, Q.; Hu, X.; Ma, Y.; Mu, J.; Liu, W.; Xu, F.; Li, Z.; Bai, J.; Hua, H.; Li, D. Marine-derived natural lead compound disulfide-linked dimer psammaplin A: Biological activity and structural modification. Mar. Drugs 2019, 17, 384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabudravu, J.N.; Eijsink, V.G.H.; Gooday, G.W.; Jaspars, M.; Komander, D.; Legg, M.; Synstad, B.; van Aalten, D.M.F. Psammaplin A, a chitinase inhibitor isolated from the Fijian marine sponge Aplysinella rhax. Bioorg. Med. Chem. 2002, 10, 1123–1128. [Google Scholar] [CrossRef]
- Kim, D.; Lee, I.; Jung, J.; Yang, S. Psammaplin A, a natural bromotyrosine derivative from a sponge, possesses the antibacterial activity against methicillin-resistant Staphylococcus aureus and the DNA gyrase-inhibitory activity. Arch. Pharm. Res. 1999, 22, 25–29. [Google Scholar] [CrossRef]
- Lee, B.C.; Lee, A.; Jung, J.H.; Choi, S.H.; Kim, T.S. In vitro and in vivo anti-Vibrio vulnificus activity of psammaplin A, a natural marine compound. Mol. Med. Rep. 2016, 14, 2691–2696. [Google Scholar] [CrossRef]
- Oluwabusola, E.; Tabudravu, J.; Al Maqbali, K.; Annang, F.; Pérez-Moreno, G.; Reyes, F.; Jaspars, M. Antiparasitic activity of bromotyrosine alkaloids and new analogues isolated from the Fijian marine sponge Aplysinella rhax. Chem. Biodivers. 2020, 17, e200335. [Google Scholar] [CrossRef]
- Rodriguez, A.; Akee, R.; Scheuer, P. Two bromotyrosine-cysteine derived metabolites from a sponge. Tetrahedron Lett. 1987, 28, 4989–4992. [Google Scholar] [CrossRef]
- Jiménez, C.; Crews, P. Novel marine sponge derived amino acids 13. Additional psammaplin derivatives from Psammaplysilla purpurea. Tetrahedron 1991, 47, 2097–2102. [Google Scholar] [CrossRef]
- Park, Y.; Liu, Y.; Hong, J.; Lee, C.O.; Cho, H.; Kim, D.K.; Im, K.S.; Jung, J.H. New bromotyrosine derivatives from an association of two sponges, Jaspis wondoensis and Poecillastra wondoensis. J. Nat. Prod. 2003, 66, 1495–1498. [Google Scholar] [CrossRef]
- Kupchan, S.; Stevens, K.; Rohlfing, E.; Sickles, B.; Sneden, A.; Miller, R.; Bryan, R. Tumor inhibitors. 126. New cytotoxic neolignans from Aniba megaphylla Mez. J. Org. Chem. 1978, 43, 586–590. [Google Scholar] [CrossRef]
- Piña, I.; Gautschi, J.; Wang, G.; Sanders, M.; Schmitz, F.; France, D.; Cornell-Kennon, S.; Sambucetti, L.; Remiszewski, S.; Perez, L.; et al. Psammaplins from the sponge Pseudoceratina purpurea: Inhibition of both histone deacetylase and DNA methyltransferase. J. Org. Chem. 2003, 68, 3866–3873. [Google Scholar] [CrossRef]
- Hentzer, M.; Riedel, K.; Rasmussen, T.B.; Heydorn, A.; Andersen, J.B.; Parsek, M.R.; Rice, S.A.; Eberl, L.; Molin, S.; Høiby, N.; et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 2002, 148, 87–102. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Rybtke, M.T.; Jakobsen, T.H.; Hentzer, M.; Bjarnsholt, T.; Givskov, M.; Tolker-Nielsen, T. Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob. Agents Chemother. 2009, 53, 2432–2443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesci, E.C.; Pearson, J.P.; Seed, P.C.; Iglewski, B.H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1997, 179, 3127–3132. [Google Scholar] [CrossRef] [Green Version]
- Soberón-Chávez, G.; Aguirre-Ramírez, M.; Sánchez, R. The Pseudomonas aeruginosa RhlA enzyme is involved in rhamnolipid and polyhydroxyalkanoate production. J. Ind. Microbiol. Biotechnol. 2005, 32, 675–677. [Google Scholar] [CrossRef] [PubMed]
- Baud, M.G.; Leiser, T.; Haus, P.; Samlal, S.; Wong, A.C.; Wood, R.J.; Petrucci, V.; Gunaratnam, M.; Hughes, S.M.; Buluwela, L.; et al. Defining the mechanism of action and enzymatic selectivity of psammaplin A against its epigenetic targets. J. Med. Chem. 2012, 55, 1731–1750. [Google Scholar] [CrossRef] [PubMed]
- Tintillier, F.; Moriou, C.; Petek, S.; Fauchon, M.; Hellio, C.; Saulnier, D.; Ekins, M.; Hooper, J.; Al-Mourabit, A.; Debitus, C. Quorum sensing inhibitory and antifouling activities of new bromotyrosine metabolites from the Polynesian sponge Pseudoceratina n. sp. Mar. Drugs 2020, 18, 272. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, T.H.; van Gennip, M.; Phipps, R.K.; Shanmugham, M.S.; Christensen, L.D.; Alhede, M.; Skindersoe, M.E.; Rasmussen, T.B.; Friedrich, K.; Uthe, F.; et al. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob. Agents Chemother. 2012, 56, 2314–2325. [Google Scholar] [CrossRef] [Green Version]
- Fong, J.; Yuan, M.; Jakobsen, T.H.; Mortensen, K.T.; Delos Santos, M.M.; Chua, S.L.; Yang, L.; Tan, C.H.; Nielsen, T.E.; Givskov, M. Disulfide bond-containing ajoene analogues as novel quorum sensing inhibitors of Pseudomonas aeruginosa. J. Med. Chem. 2017, 60, 215–227. [Google Scholar] [CrossRef]
- Tan, S.Y.; Chua, S.L.; Chen, Y.; Rice, S.A.; Kjelleberg, S.; Nielsen, T.E.; Yang, L.; Givskov, M. Identification of five structurally unrelated quorum-sensing inhibitors of Pseudomonas aeruginosa from a natural-derivative database. Antimicrob. Agents Chemother. 2013, 57, 5629–5641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phyo, M.Y.; Goh, T.M.B.; Goh, J.X.; Tan, L.T. Trikoramides B–D, bioactive cyanobactins from the marine cyanobacterium Symploca hydnoides. Mar. Drugs 2021, 19, 548. [Google Scholar] [CrossRef] [PubMed]
- Soukarieh, F.; Williams, P.; Stocks, M.J.; Cámara, M. Pseudomonas aeruginosa quorum sensing systems as drug discovery targets: Current position and future perspectives. J. Med. Chem. 2018, 61, 10385–10402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qais, F.A.; Khan, M.S.; Ahmad, I.; Husain, F.M.; Khan, R.A.; Hassan, I.; Shahzad, S.A.; AlHarbi, W. Coumarin exhibits broad-spectrum antibiofilm and antiquorum sensing activity against Gram-negative bacteria: In vitro and in silico investigation. ACS Omega 2021, 6, 18823–18835. [Google Scholar] [CrossRef]
- Nam, S.; Ham, S.Y.; Kwon, H.; Kim, H.S.; Moon, S.; Lee, J.H.; Lim, T.; Son, S.H.; Park, H.D.; Byun, Y. Discovery and characterization of pure RhlR antagonists against Pseudomonas aeruginosa infections. J. Med. Chem. 2020, 63, 8388–8407. [Google Scholar] [CrossRef]
- Pearson, J.; Pesci, E.; Iglewski, B. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 1997, 179, 5756–5767. [Google Scholar] [CrossRef] [Green Version]
- Everett, M.J.; Davies, D.T. Pseudomonas aeruginosa elastase (LasB) as a therapeutic target. Drug Discov. Today 2021, 26, 2108–2123. [Google Scholar] [CrossRef]
- Fong, J.; Mortensen, K.T.; Nørskov, A.; Qvortrup, K.; Yang, L.; Tan, C.H.; Nielsen, T.E.; Givskov, M. Itaconimides as novel quorum sensing inhibitors of Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2019, 8, 443. [Google Scholar] [CrossRef]
- Zhao, C.; Zheng, H.; Zhou, L.; Ji, H.; Zhao, L.; Yu, W.; Gong, Q. Falcarindiol isolated from Notopterygium incisum inhibits the quorum sensing of Pseudomonas aeruginosa. Molecules 2021, 26, 5896. [Google Scholar] [CrossRef]
- Givskov, M.; de Nys, R.; Manefield, M.; Gram, L.; Maximilien, R.; Eberl, L.; Molin, S.; Steinberg, P.; Kjelleberg, S. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacterial. 1996, 178, 6618–6622. [Google Scholar] [CrossRef] [Green Version]
- Skariyachan, S.; Sridhar, V.S.; Packirisamy, S.; Kumargowda, S.T.; Challapilli, S.B. Recent perspectives on the molecular basis of biofilm formation by Pseudomonas aeruginosa and approaches for treatment and biofilm dispersal. Folia Microbiol. 2018, 63, 413–432. [Google Scholar] [CrossRef] [PubMed]
- De Kievit, T.R.; Gillis, R.; Marx, S.; Brown, C.; Iglewski, B.H. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: Their role and expression patterns. Appl. Environ. Microbiol. 2001, 67, 1865–1873. [Google Scholar] [CrossRef] [Green Version]
- O’Loughlin, C.T.; Miller, L.C.; Siryaporn, A.; Drescher, K.; Semmelhack, M.F.; Bassler, B.L. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. USA 2013, 110, 17981–17986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malešević, M.; Di Lorenzo, F.; Filipić, B.; Stanisavljević, N.; Novović, K.; Senerovic, L.; Polović, N.; Molinaro, A.; Kojić, M.; Jovčić, B. Pseudomonas aeruginosa quorum sensing inhibition by clinical isolate Delftia tsuruhatensis 11304: Involvement of N-octadecanoylhomoserine lactones. Sci. Rep. 2019, 9, 16465. [Google Scholar] [CrossRef] [PubMed]
- Almohaywi, B.; Yu, T.T.; Iskander, G.; Chan, D.S.H.; Ho, K.K.K.; Rice, S.; Black, D.S.; Griffith, R.; Kumar, N. Dihydropyrrolones as bacterial quorum sensing inhibitors. Bioorg. Med. Chem. Lett. 2019, 29, 1054–1059. [Google Scholar] [CrossRef] [PubMed]
- De Kievit, T.R. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2009, 11, 279–288. [Google Scholar] [CrossRef]
- Yan, S.; Wu, G. Can biofilm be reversed through quorum sensing in Pseudomonas aeruginosa? Front. Microbiol. 2019, 10, 1582. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.H.; Shin, J.; Kwon, H.J. Psammaplin A is a natural prodrug that inhibits class I histone deacetylase. Exp. Mol. Med. 2007, 39, 47–55. [Google Scholar] [CrossRef]
- Bao, Y.; Xu, Q.; Wang, L.; Wei, Y.; Hu, B.; Wang, J.; Liu, D.; Zhao, L.; Jing, Y. Studying histone deacetylase inhibition and apoptosis induction of psammaplin A monomers with modified thiol group. ACS Med. Chem. Lett. 2021, 12, 39–47. [Google Scholar] [CrossRef]
- Saurav, K.; Costantino, V.; Venturi, V.; Steindler, L. Quorum sensing inhibitors from the sea discovered using bacterial N-acyl-homoserine lactone-based biosensors. Mar. Drugs 2017, 15, 53. [Google Scholar] [CrossRef] [Green Version]
- Skindersoe, M.; Ettinger-Epstein, P.; Rasmussen, T.; Bjarnsholt, T.; Nys, R.; Givskov, M. Quorum sensing antagonism from marine organisms. Mar. Biotechnol. 2008, 10, 56–63. [Google Scholar] [CrossRef]
- Mai, T.; Tintillier, F.; Lucasson, A.; Moriou, C.; Bonno, E.; Petek, S.; Magré, K.; Al Mourabit, A.; Saulnier, D.; Debitus, C. Quorum sensing inhibitors from Leucetta chagosensis Dendy, 1863. Lett. Appl. Microbiol. 2015, 61, 311–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costantino, V.; Della Sala, G.; Saurav, K.; Teta, R.; Bar-Shalom, R.; Mangoni, A.; Steindler, L. Plakofuranolactone as a quorum quenching agent from the Indonesian sponge Plakortis cf. lita. Mar. Drugs 2017, 15, 59. [Google Scholar] [CrossRef] [Green Version]
- Alam, P.; Alqahtani, A.S.; Mabood Husain, F.; Tabish Rehman, M.; Alajmi, M.F.; Noman, O.M.; El Gamal, A.A.; Al-Massarani, S.M.; Shavez Khan, M. Siphonocholin isolated from red sea sponge Siphonochalina siphonella attenuates quorum sensing controlled virulence and biofilm formation. Saudi Pharm. J. 2020, 28, 1383–1391. [Google Scholar] [CrossRef]
- Dobretsov, S.; Teplitski, M.; Bayer, M.; Gunasekera, S.; Proksch, P.; Paul, V.J. Inhibition of marine biofouling by bacterial quorum sensing inhibitors. Biofouling 2011, 27, 893–905. [Google Scholar] [CrossRef]
- Saurav, K.; Borbone, N.; Burgsdorf, I.; Teta, R.; Caso, A.; Bar-Shalom, R.; Esposito, G.; Britstein, M.; Steindler, L.; Costantino, V. Identification of quorum sensing activators and inhibitors in the marine sponge Sarcotragus spinosulus. Mar. Drugs 2020, 18, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovalerchik, D.; Singh, R.P.; Schlesinger, P.; Mahajni, A.; Shefer, S.; Fridman, M.; Ilan, M.; Carmeli, S. Bromopyrrole alkaloids of the sponge Agelas oroides collected near the Israeli Mediterranean coastline. J. Nat. Prod. 2020, 83, 374–384. [Google Scholar] [CrossRef]
- Clark, D.; Maaløe, O. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 1967, 23, 99–112. [Google Scholar] [CrossRef]
- Yang, L.; Barken, K.B.; Skindersoe, M.E.; Christensen, A.B.; Givskov, M.; Tolker-Nielsen, T. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 2007, 153, 1318–1328. [Google Scholar]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P.; et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar]






| Compound | IC50 (μM) | % Inhibition (100 μg/mL) | ||
|---|---|---|---|---|
| lasB-gfp | rhlA-gfp | lasB-gfp | rhlA-gfp | |
| 1 | 14.02 | 4.99 | 85.4% | 63.3% |
| 2 | 3.53 | 2.41 | 80.1% | 68.9% |
| Hemifistularin 3 | - | - | 31.4% | 49.1% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oluwabusola, E.T.; Katermeran, N.P.; Poh, W.H.; Goh, T.M.B.; Tan, L.T.; Diyaolu, O.; Tabudravu, J.; Ebel, R.; Rice, S.A.; Jaspars, M. Inhibition of the Quorum Sensing System, Elastase Production and Biofilm Formation in Pseudomonas aeruginosa by Psammaplin A and Bisaprasin. Molecules 2022, 27, 1721. https://doi.org/10.3390/molecules27051721
Oluwabusola ET, Katermeran NP, Poh WH, Goh TMB, Tan LT, Diyaolu O, Tabudravu J, Ebel R, Rice SA, Jaspars M. Inhibition of the Quorum Sensing System, Elastase Production and Biofilm Formation in Pseudomonas aeruginosa by Psammaplin A and Bisaprasin. Molecules. 2022; 27(5):1721. https://doi.org/10.3390/molecules27051721
Chicago/Turabian StyleOluwabusola, Emmanuel T., Nursheena Parveen Katermeran, Wee Han Poh, Teo Min Ben Goh, Lik Tong Tan, Oluwatofunmilayo Diyaolu, Jioji Tabudravu, Rainer Ebel, Scott A. Rice, and Marcel Jaspars. 2022. "Inhibition of the Quorum Sensing System, Elastase Production and Biofilm Formation in Pseudomonas aeruginosa by Psammaplin A and Bisaprasin" Molecules 27, no. 5: 1721. https://doi.org/10.3390/molecules27051721