Characterization of Cabernet Sauvignon Wines by Untargeted HS-SPME GC-QTOF-MS
Abstract
:1. Introduction
2. Results
2.1. Method Validation and Data Acquisition
2.2. Quality Control
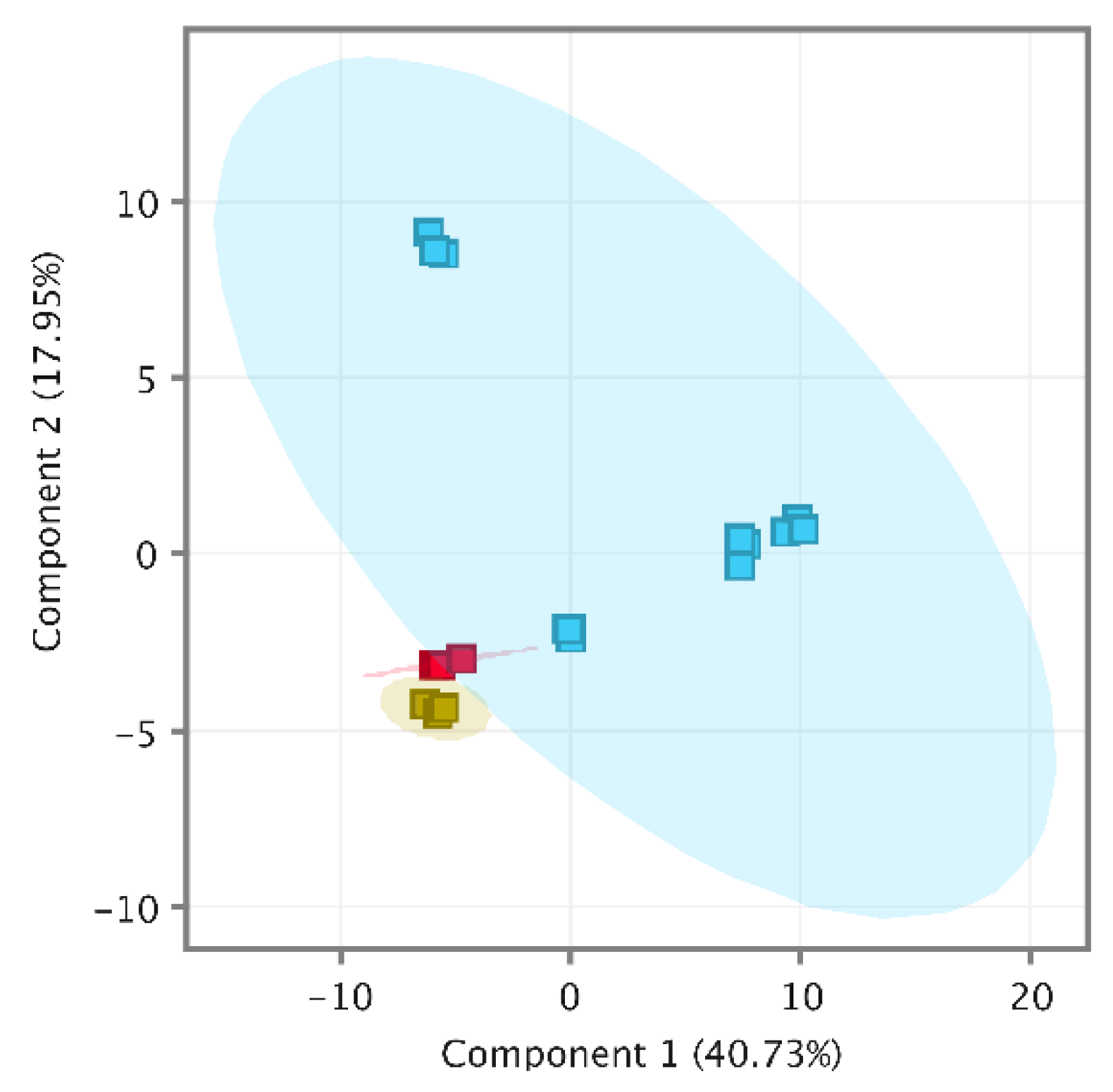
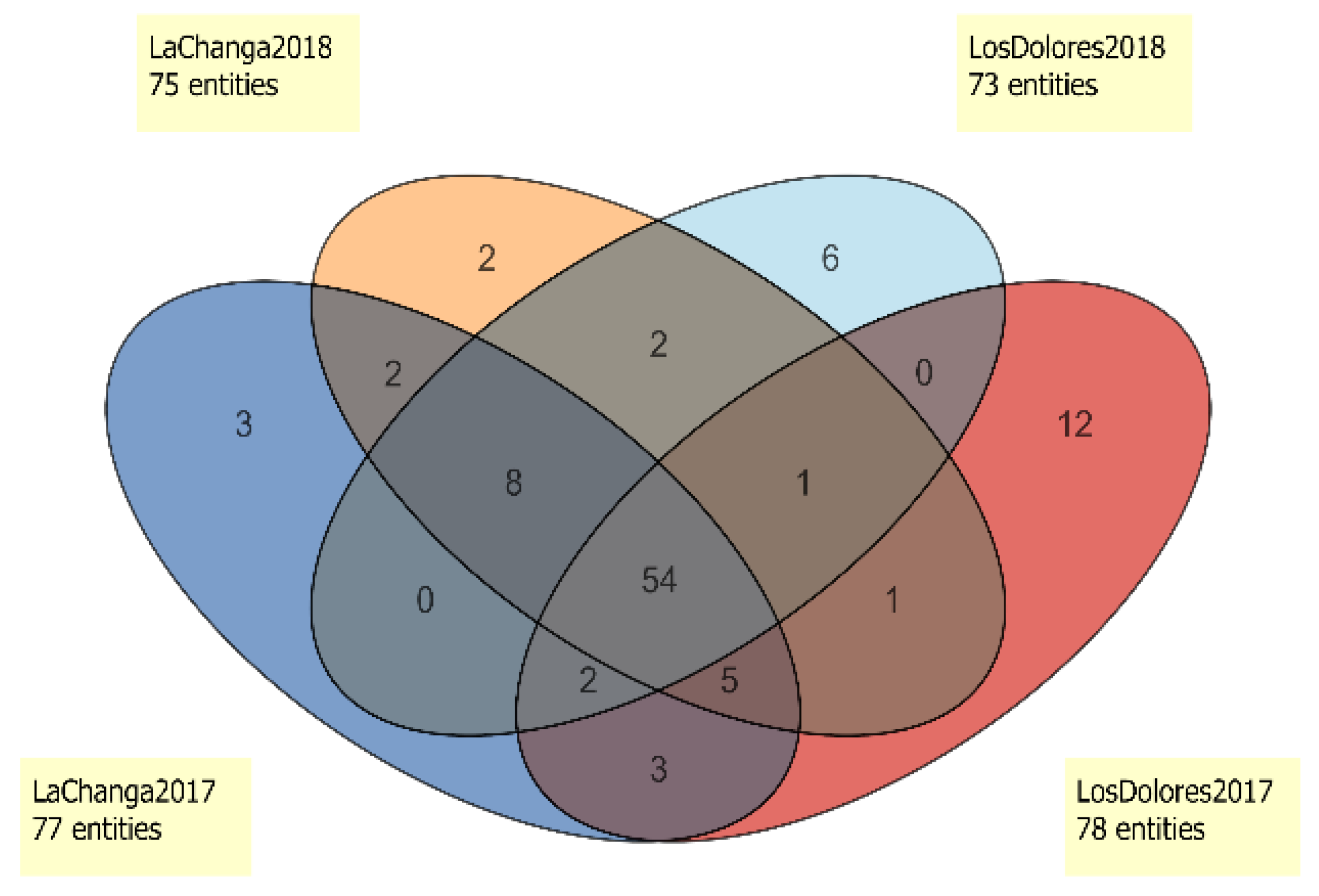
2.3. Wine Characterization
3. Conclusions
4. Materials and Methods
4.1. Samples
4.2. Data Acquisition
4.3. Method Validation
4.4. Quality Control
4.5. Data Processing/Mining and Identification
4.5.1. Data Processing/Mining
4.5.2. Retention Index
4.5.3. Internal Library
4.5.4. Recursive Analysis
4.6. Data Interpretation/Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lacalle-Bergeron, L.; Izquierdo-Sandoval, D.; Sancho, J.V.; López, F.J.; Hernández, F.; Portolés, T. Chromatography hyphenated to high resolution mass spectrometry in untargeted metabolomics for investigation of food (bio)markers. TrAC Trends Anal. Chem. 2021, 135, 116161. [Google Scholar] [CrossRef]
- Alañón, M.E.; Pérez-Coello, M.S.; Marina, M.L. Wine science in the metabolomics era. TrAC Trends Anal. Chem. 2015, 74, 1–20. [Google Scholar] [CrossRef]
- Klåvus, A.; Kokla, M.; Noerman, S.; Koistinen, V.M.; Tuomainen, M.; Zarei, I.; Meuronen, T.; Häkkinen, M.R.; Rummukainen, S.; Babu, A.F.; et al. “Notame”: Workflow for non-targeted LC-MS metabolic profiling. Metabolites 2020, 10, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunn, W.B.; Ellis, D.I. Metabolomics: Current analytical platforms and methodologies. TrAC Trends Anal. Chem. 2005, 24, 285–294. [Google Scholar] [CrossRef]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef] [Green Version]
- Spicer, R.A.; Salek, R.; Steinbeck, C. Comment: A decade after the metabolomics standards initiative it’s time for a revision. Sci. Data 2017, 4, 2–4. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Baran, R. Untargeted metabolomics suffers from incomplete raw data processing. Metabolomics 2017, 13, 107. [Google Scholar] [CrossRef]
- Beger, R.D.; Dunn, W.B.; Bandukwala, A.; Bethan, B.; Broadhurst, D.; Clish, C.B.; Dasari, S.; Derr, L.; Evans, A.; Fischer, S.; et al. Towards quality assurance and quality control in untargeted metabolomics studies. Metabolomics 2019, 15, 4. [Google Scholar] [CrossRef]
- Dudzik, D.; Barbas-bernardos, C.; García, A.; Barbas, C. Quality assurance procedures for mass spectrometry untargeted metabolomics a review. J. Pharm. Biomed. Anal. 2018, 147, 149–173. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.I.; Edison, A.; Guillou, C.; Viant, M.R.; Bearden, D.W.; Beger, R.D. Quality assurance and quality control processes: Summary of a metabolomics community questionnaire. Metabolomics 2017, 13, 50. [Google Scholar] [CrossRef]
- Martin, J.C.; Maillot, M.; Mazerolles, G.; Verdu, A.; Lyan, B.; Migné, C.; Defoort, C.; Canlet, C.; Junot, C.; Guillou, C.; et al. Can we trust untargeted metabolomics? Results of the metabo-ring initiative, a large-scale, multi-instrument inter-laboratory study. Metabolomics 2015, 11, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Capece, A.; Romaniello, R.; Siesto, G.; Pietrafesa, R.; Massari, C.; Poeta, C.; Romano, P. Selection of indigenous saccharomyces cerevisiae strains for nero d’avola wine and evaluation of selected starter implantation in pilot fermentation. Int. J. Food Microbiol. 2010, 144, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.L.; Boss, P.K.; Heymann, H.; Solomon, P.S.; Trengove, R.D. Development of a sensitive non-targeted method for characterizing the wine volatile profile using headspace solid-phase microextraction comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry. J. Chromatogr. A 2011, 1218, 504–517. [Google Scholar] [CrossRef]
- Arapitsas, P.; Scholz, M.; Vrhovsek, U.; Di Blasi, S.; Biondi, A.; Masuero, D.; Perenzoni, D.; Rigo, A.; Mattivi, F. A metabolomic approach to the study of wine micro-oxygenation. PLoS ONE 2012, 7, e377783. [Google Scholar] [CrossRef]
- Arapitsas, P.; Speri, G.; Angeli, A.; Perenzoni, D.; Mattivi, F. The influence of storage on the ‘chemical age’ of red wines. Metabolomics 2014, 10, 816–832. [Google Scholar] [CrossRef]
- López-Rituerto, E.; Savorani, F.; Avenoza, A.; Busto, J.H.; Peregrina, J.M.; Engelsen, S.B. Investigations of la rioja terroir for wine production using 1H NMR metabolomics. J. Agric. Food Chem. 2012, 60, 3452–3461. [Google Scholar] [CrossRef]
- Castro, C.C.; Martins, R.C.; Teixeira, J.A.; Ferreira, A.C.S. Application of a high-throughput process analytical technology metabolomics pipeline to port wine forced ageing process. Food Chem. 2014, 143, 384–391. [Google Scholar] [CrossRef] [Green Version]
- Silva Ferreira, A.C.; Monforte, A.R.; Silva Teixeira, C.; Martins, R.; Fairbairn, S.; Bauer, F.F. Monitoring alcoholic fermentation: An untargeted approach. J. Agric. Food Chem. 2014, 62, 6784–6793. [Google Scholar] [CrossRef]
- Alves, Z.; Melo, A.; Figueiredo, A.R.; Coimbra, M.A.; Gomes, C.; Rocha, S.M. Exploring the saccharomyces cerevisiae volatile metabolome: Indigenous versus commercial strains. PLoS ONE 2015, 10, e0143641. [Google Scholar] [CrossRef] [Green Version]
- Boss, P.K.; Kalua, C.M.; Nicholson, E.L.; Maffei, S.M.; Böttcher, C.; Davies, C. Fermentation of grapes throughout development identifies stages critical to the development of wine volatile composition. Aust. J. Grape Wine Res. 2018, 24, 24–37. [Google Scholar] [CrossRef]
- Brown, M.; Dunn, W.B.; Ellis, D.I.; Goodacre, R.; Handl, J.; Knowles, J.D.; O’Hagan, S.; Spasić, I.; Kell, D.B. A metabolome pipeline: From concept to data to knowledge. Metabolomics 2005, 1, 39–51. [Google Scholar] [CrossRef]
- Jones, C.M.; Dunn, W.B.; Raftery, D.; Hartung, T.; Wilson, I.D.; Lewis, M.R.; Tayyari, F.; Baljit, K.; Souza, A.; Ntai, I.; et al. Metabolomics Quality Assurance and Quality Control Consortium (MQACC): Reference and Test Material Working Group; Metabolomics Quality Assurance and Quality Control Consortium (mQACC): Bethesda, MD, USA, 2018. [Google Scholar]
- Bletsou, A.A.; Jeon, J.; Hollender, J.; Archontaki, E.; Thomaidis, N.S. Targeted and non-targeted liquid chromatography-mass spectrometric workflows for identification of transformation products of emerging pollutants in the aquatic environment. TrAC Trends Anal. Chem. 2015, 66, 32–44. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Bueno, M.J.; Gómez Ramos, M.J.; Bauer, A.; Fernández-Alba, A.R. An Overview of non-targeted screening strategies based on high resolution accurate mass spectrometry for the identification of migrants coming from plastic food packaging materials. TrAC Trends Anal. Chem. 2019, 110, 191–203. [Google Scholar] [CrossRef]
- Palermo, A.; Botre, F.; de la Torre, X.; Zamboni, N. Non-targeted LC-MS based metabolomics analysis of the urinary steroidal profile. Anal. Chim. Acta 2017, 964, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Redondo, J.M.; Puertas, B.; Pereira-Caro, G.; Ordóñez-Díaz, J.L.; Ruiz-Moreno, M.J.; Cantos-Villar, E.; Moreno-Rojas, J.M. A statistical workflow to evaluate the modulation of wine metabolome and its contribution to the sensory attributes. Fermentation 2021, 7, 72. [Google Scholar] [CrossRef]
- Covarrubias, J.; Thach, L. Wines of Baja Mexico: A qualitative study examining viticulture, enology, and marketing practices. Wine Econ. Policy 2015, 4, 110–115. [Google Scholar] [CrossRef] [Green Version]
- Larios Córdova, H. Iniciativa Con Proyecto de Decreto Que Expide La Ley General de Fomento a La Industria Vitivinícola. 2016. Available online: https://infosen.senado.gob.mx/sgsp/gaceta/63/2/2017-04-25-1/assets/documentos/Inic_PAN_Ley_Industria_Vitivinicila.pdf (accessed on 20 October 2017).
- OIV. Estadísticas de México de 1995 a 2016. Available online: https://www.oiv.int/es/statistiques/recherche (accessed on 8 September 2019).
- DOF. El Pleno del Senado Aprobó la Ley General de Fomento a la Industria Vitivinícola. Available online: http://comunicacion.senado.gob.mx/index.php/informacion/boletines/39130-el-pleno-del-senado-aprobo-la-ley-general-de-fomento-a-la-industria-vitivinicola.html (accessed on 5 November 2017).
- CMV. Marca Colectiva. Available online: https://vinomexicano.org.mx/marca-colectiva/ (accessed on 10 January 2020).
- FDA. Bioanalytical Method Validation, Guidance for Industry. Available online: https://www.fda.gov/media/70858/download (accessed on 4 January 2022).
- Dashko, S.; Zhou, N.; Tinta, T.; Sivilotti, P. Use of non-conventional yeast improves the wine aroma profile of Ribolla Gialla. J. Ind. Microbiol. Biotechnol. 2015, 42, 997–1010. [Google Scholar] [CrossRef]
- Parker, M.; Capone, D.L.; Francis, I.L.; Herderich, M.J. Aroma precursors in grapes and wine: Flavor release during wine production and consumption. J. Agric. Food Chem. 2017, 66, 2281–2286. [Google Scholar] [CrossRef]
- Ramirez-Gaona, M.; Marcu, A.; Pon, A.; Guo, A.C.; Sajed, T.; Wishart, N.A.; Karu, N.; Feunang, Y.D.; Arndt, D.; Wishart, D.S. YMDB 2.0: A significantly expanded version of the yeast metabolome database. Nucleic Acids Res. 2017, 45, D440–D445. [Google Scholar] [CrossRef]
- Martins, N.; Garcia, R.; Mendes, D.; Costa Freitas, A.M.; da Silva, M.G.; Cabrita, M.J. An ancient winemaking technology: Exploring the volatile composition of amphora wines. LWT Food Sci. Technol. 2018, 96, 288–295. [Google Scholar] [CrossRef]
- Lu, Y.; Sun, F.; Wang, W.; Liu, Y.; Wang, J.; Sun, J.; Mu, J.; Gao, Z. Effects of spontaneous fermentation on the microorganisms diversity and volatile compounds during ‘Marselan’ from grape to wine. LWT Food Sci. Technol. 2020, 134, 110193. [Google Scholar] [CrossRef]
- Lu, Y.; Guan, X.; Li, R.; Wang, J.; Liu, Y.; Ma, Y.; Lv, J.; Wang, S.; Mu, J. Comparative study of microbial communities and volatile profiles during the inoculated and spontaneous fermentation of persimmon wine. Process Biochem. 2021, 100, 49–58. [Google Scholar] [CrossRef]
- Varsha, K.K.; Devendra, L.; Shilpa, G.; Priya, S.; Pandey, A.; Nampoothiri, K.M. 2,4-Di-Tert-Butyl phenol as the antifungal, antioxidant bioactive purified from a newly isolated Lactococcus sp. Int. J. Food Microbiol. 2015, 211, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Wang, C.; Zhu, X.; Ning, C.; Gao, L.; Zhang, J.; Wang, Y.; Huang, R. A multi-step screening approach of suitable non-saccharomyces yeast for the fermentation of hawthorn wine. LWT Food Sci. Technol. 2020, 127, 109432. [Google Scholar] [CrossRef]
- Mendes, B.; Gonalves, J.; Câmara, J.S. Effectiveness of high-throughput miniaturized sorbent- and solid phase microextraction techniques combined with gas chromatography-mass spectrometry analysis for a rapid screening of volatile and semi-volatile composition of wines—A comparative study. Talanta 2012, 88, 79–94. [Google Scholar] [CrossRef]
- Câmara, J.S.; Arminda Alves, M.; Marques, J.C. Development of headspace solid-phase microextraction-gas chromatography-mass spectrometry methodology for analysis of terpenoids in Madeira wines. Anal. Chim. Acta 2006, 555, 191–200. [Google Scholar] [CrossRef]
- Hjelmeland, A.K.; King, E.S.; Ebeler, S.E.; Heymann, H. Characterizing the chemical and sensory profiles of United States cabernet sauvignon wines and blends. Am. J. Enol. Vitic. 2013, 64, 169–179. [Google Scholar] [CrossRef]
- Wylie, P.; Hjelmeland, A.; Runnebaum, R.; Ebeler, S. Analysis of pinot noir wines by HS-SPME GC/Q-TOF: Correlating geographical origin with volatile aroma profiles. Planta Med. 2016, 82, OA49. [Google Scholar] [CrossRef] [Green Version]
- Baumann, S.; Conjelko, T.; Aronova, S.; Lafond, S.; David, F.; Ebeler, S.E. Accurate mass retention time locked flavor database by GC-TOF. In Proceedings of the American Society for Mass Spectrometry Annual Conference, Minneapolis, MN, USA, 9–13 June 2013; p. MP-685. [Google Scholar]
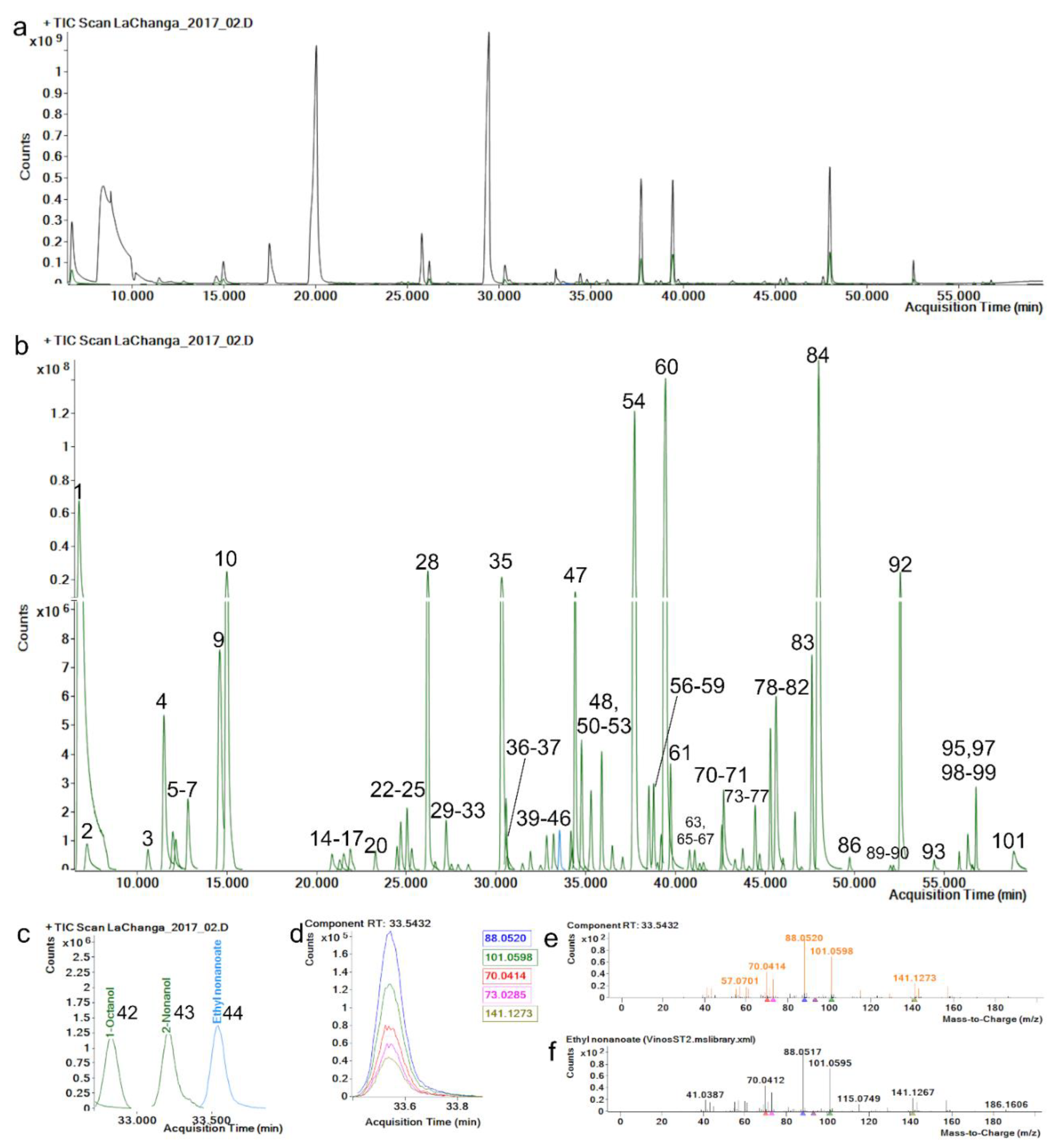

| # | RT | RI | Compound | Id | La Changa | Los Dolores | CAS Number | Formula | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2017 Ab (RelAb%) | 2018 Ab (RelAb%) | FC | 2017 Ab (RelAb%) | FC | 2018Ab (RelAb%) | FC | ||||||||||
| 1 | 6.7 | 900 | Ethyl Acetate ** | 2 | 9.2 × 108 (68) | 7.6 × 108 (67) | ↓ | 1.2 | 1.2 × 109 (52) | ↑ | 1.3 | 5.4 × 108 (27) | ↓ | 1.7 | 141-78-6 | C4H8O2 |
| 2 | 7.2 | 918 | Methyl Alcohol | 2 | 1.7 × 107 (1) | 1.5 × 107 (1) | ↓ | 1.2 | 1.4 × 107 (0.6) | ↓ | 1.2 | 1.7 × 107 (1) | ↓ | 1.0 | 67-56-1 | CH4O |
| 3 | 10.6 | 1028 | Isobutyl acetate * | 2 | 6.4 × 106 (0.5) | 6.4 × 106 (0.6) | ↓ | 1.0 | 8.2 × 106 (0.4) | ↑ | 1.3 | 6.4 × 106 (0.5) | ↑ | 1.0 | 110-19-0 | C6H12O2 |
| 4 | 11.5 | 1051 | Ethyl butyrate *** | 2 | 6.5 × 107 (5) | 5.6 × 107 (5) | ↓ | 1.2 | 4.6 × 107 (2) | ↓ | 1.4 | 6.9 × 107 (5) | ↑ | 1.1 | 105-54-4 | C6H12O2 |
| 5 | 12.0 | 1064 | 1-Propanol | 2 | 2.3 × 107 (2) | 1.4 × 107 (1) | ↓ | 1.6 | 2.7 × 107 (1) | ↑ | 1.2 | 2.7 × 107 (2) | ↑ | 1.2 | 71-23-8 | C3H8O |
| 6 | 12.2 | 1068 | Ethyl 2-methylbutyrate | 2 | 1.3 × 107 (1) | 2.9 × 106 (0.3) | ↓ | 4.5 | 1.2 × 107 (0.5) | ↓ | 1.1 | ND | - | - | 7452-79-1 | C7H14O2 |
| 7 | 12.8 | 1084 | Ethyl 3-methylbutyrate * | 2 | 2.5 × 107 (2) | 5.3 × 106 (0.5) | ↓ | 4.8 | 2.2 × 107 (1) | ↓ | 1.1 | 5.7 × 106 (0.4) | ↓ | 4.4 | 108-64-5 | C7H14O2 |
| 8 | 13.2 | 1093 | Unknown 13.2265 | 2 | ND | ND | - | - | 1.7 × 106 (0.1) | - | - | ND | - | - | - | C9H20O2 |
| 9 | 14.6 | 1127 | Isobutyl alcohol | 2 | 1.1 × 108 (8) | 1.2 × 108 (10) | ↑ | 1.1 | 9.5 × 106 (4) | ↓ | 1.2 | 1.1 × 108 (9) | ↑ | 1.0 | 78-83-1 | C4H10O |
| 10 | 15.0 | 1135 | Isoamyl acetate | 2 | 2.4 × 108 (18) | 2.3 × 108 (20) | ↓ | 1.0 | 2.8 × 106 (12) | ↑ | 1.2 | 2.7 × 108 (18) | ↑ | 1.1 | 123-92-2 | C7H14O2 |
| 11 | 15.3 | 1143 | p-Xylene | 3 | ND | ND | - | - | ND | - | - | 7.5 × 105 (0.1) | - | - | 106-42-3 | C8H10 |
| 2 | 16.6 | 1170 | Unknown 16.5912 | 4 | ND | ND | - | - | ND | - | - | 1.1 × 106 (0.1) | - | - | - | C10H16 |
| 13 | 17.3 | 1182 | 1-Butanol | 2 | ND | 4.6 × 106 (0.4) | - | - | ND | - | - | ND | - | - | 71-36-3 | C4H10O |
| 14 | 20.8 | 1261 | Styrene ** | 2 | 5.4 × 106 (0.4) | 5.4 × 106 (0.5) | ↑ | 1.0 | 2.9 × 106 (0.1) | ↓ | 1.9 | 7.2 × 106 (0.6) | ↑ | 1.3 | 100-42-5 | C8H8 |
| 15 | 21.2 | 1268 | p-Cymene | 1 | 3.4 × 106 (0.3) | 7.6 × 106 (0.1) | ↓ | 4.4 | ND | - | - | 1.3 × 106 (0.1) | ↓ | 2.6 | 99-87-6 | C10H14 |
| 16 | 21.4 | 1273 | Isoamyl butyrate | 2 | 7.1 × 106 (0.5) | 3.6 × 106 (0.3) | ↓ | 2.0 | 4.6 × 106 (0.2) | ↓ | 1.5 | 5.5 × 106 (0.4) | ↓ | 1.3 | 106-27-4 | C9H18O2 |
| 17 | 21.8 | 1284 | Hexyl acetate ** | 2 | 7.5 × 106 (0.6) | 8.6 × 106 (0.8) | ↑ | 1.1 | 7.6 × 106 (0.3) | ↑ | 1.0 | 1.0 × 107 (0.8) | ↑ | 1.3 | 142-92-7 | C8H16O2 |
| 18 | 22.7 | 1301 | Furfuryl ethyl ether | 2 | ND | ND | - | - | 1.6 × 106 (0.1) | - | - | ND | - | - | 1000450-02-5 | C7H10O2 |
| 19 | 22.8 | 1306 | Ethyl 3-hexenoate | 3 | ND | ND | - | - | ND | - | - | 2.7 × 106 (0.2) | - | - | 2396-83-0 | C8H14O2 |
| 20 | 23.3 | 1319 | Acetoin | 2 | 4.7 × 106 (0.4) | ND | - | - | ND | - | - | ND | - | - | 513-86-0 | C4H8O2 |
| 21 | 23.9 | 1328 | Unknown 23.8754 | 2 | ND | 2.3 × 106 (0.2) | - | - | 2.3 × 106 (0.1) | - | - | ND | - | - | - | C10H18O |
| 22 | 24.5 | 1343 | 4-Methyl-1-pentanol | 2 | 6.1 × 106 (0.5) | 3.3 × 106 (0.3) | ↓ | 1.9 | ND | - | - | 3.3 × 106 (0.3) | ↓ | 1.9 | 626-89-1 | C6H14O |
| 23 | 24.6 | 1345 | Ethyl heptanoate | 2 | 9.6 × 106 (0.7) | 6.5 × 106 (0.6) | ↓ | 1.5 | 8.5 × 106 (0.4) | ↓ | 1.1 | 6.7 × 106 (0.5) | ↓ | 1.4 | 106-30-9 | C9H18O2 |
| 24 | 25.1 | 1356 | 1-Hexanol | 2 | 1.5 × 107 (1) | 6.5 × 106 (0.6) | ↓ | 2.3 | 1.4 × 107 (0.6) | ↓ | 1.0 | ND | - | - | 111-27-3 | C7H14O2 |
| 25 | 25.3 | 1359 | Ethyl 2-hexanoate | 2 | 5.4 × 106 (0.4) | 3.9 × 106 (0.3) | ↓ | 1.4 | 6.1 × 106 (0.3) | ↑ | 1.1 | 7.8 × 106 (0.6) | ↑ | 1.4 | 1552-67-6 | C8H14O2 |
| 26 | 25.5 | 1366 | Isobutyl hexanoate | 2 | ND | ND | - | - | ND | - | - | 2.2 × 106 (0.2) | - | - | 105-79-3 | C10H20O2 |
| 27 | 25.8 | 1372 | Ethyl lactate † | 2 | ND | 1.2 × 108 (11) | - | - | ND | - | - | 1.0 × 108 (5) | ↓ | 1.2 | 97-64-3 | C5H10O3 |
| 28 | 26.2 | 1381 | Hexyl formate * | 2 | 1.7 × 108 (12) | 9.3 × 107 (8) | ↓ | 1.8 | 2.3 × 108 (10) | ↑ | 1.4 | 1.1 × 108 (8) | ↓ | 1.6 | 629-33-4 | C6H14O |
| 29 | 26.6 | 1391 | 2,6-Dimethyl-5-heptenal | 2 | 1.8 × 106 (0.1) | 1.2 × 106 (0.1) | ↓ | 1.5 | ND | - | - | 1.4 × 106 (0.1) | ↓ | 1.3 | 106-72-9 | C9H16O |
| 30 | 27.2 | 1402 | Methyl octanoate | 2 | 1.3 × 107 (0.9) | 1.3 × 107 (1) | ↑ | 1.0 | 7.5 × 106 (0.3) | ↓ | 1.7 | 1.9 × 107 (1) | ↑ | 1.5 | 111-11-5 | C9H18O2 |
| 31 | 27.5 | 1411 | trans-3-Hexen-1-ol ** | 2 | 1.4 × 106 (0.1) | 9.0 × 105 (0.1) | ↓ | 1.5 | 2.1 × 106 (0.1) | ↑ | 1.5 | 9.5 × 105 (0.1) | ↓ | 1.5 | 928-97-2 | C6H12O |
| 32 | 27.9 | 1419 | 3-Octanol | 2 | 1.3 × 106 (0.1) | 1.1 × 106 (0.1) | ↓ | 1.2 | 1.5 × 106 (0.1) | ↑ | 1.1 | 1.0 × 106 (0.1) | ↓ | 1.3 | 589-98-0 | C8H18O |
| 33 | 28.5 | 1433 | cis-2-Hexen-1-ol | 2 | 1.3 × 106 (0.1) | ND | - | - | 1.7 × 106 (0.1) | ↑ | 1.3 | ND | - | - | 928-94-9 | C6H12O |
| 34 | 29.3 | 1453 | Unknown 29.3449 | 4 | ND | ND | - | - | 2.3 × 109 (100) | - | - | ND | - | - | - | C10H20O2 |
| 35 | 30.3 | 1476 | Acetic acid ** | 2 | 2.3 × 108 (17) | 3.6 × 108 (32) | ↑ | 1.6 | 4.0 × 108 (17) | ↑ | 1.7 | 2.3 × 108 (14) | ↓ | 1.0 | 64-19-7 | C2H4O2 |
| 36 | 30.6 | 1482 | 1-Heptanol | 2 | 1.5 × 107 (1.2) | ND | - | - | 1.3 × 107 (0.6) | ↓ | 1.2 | 7.4 × 106 (0.6) | ↓ | 2.1 | 111-70-6 | C7H16O |
| 37 | 30.6 | 1482 | Unknown 30.6066 | 4 | 3.6 × 106 (0.3) | ND | - | - | ND | - | - | ND | - | - | - | C5H4O2 |
| 38 | 31.3 | 1499 | trans-Linalool oxide (furanoid) | 3 | ND | ND | - | - | 2.2 × 106 (0.1) | - | - | ND | - | - | 34995-77-2 | C10H18O2 |
| 39 | 31.4 | 1503 | Ethyl 7-octenoate | 3 | 1.4 × 106 (0.1) | 9.7 × 105 (0.1) | ↓ | 1.5 | ND | - | - | 1.1 × 106 (0.1) | ↓ | 1.3 | 35194-38-8 | C10H18O2 |
| 40 | 31.9 | 1513 | 2-Ethyl-1-hexanol | 2 | 4.0 × 106 (0.3) | 2.1 × 106 (0.2) | ↓ | 1.9 | 4.9 × 106 (0.2) | ↑ | 1.2 | 2.7 × 106 (0.2) | ↓ | 1.5 | 104-76-7 | C8H18O |
| 41 | 32.5 | 1527 | Geranyl ethyl ether 1 | 3 | 1.0 × 106 (0.1) | 7.3 × 105 (0.1) | ↓ | 1.4 | ND | - | - | 1.0 × 106 (0.1) | ↓ | 1.0 | 1000285-27-5 | C12H22O |
| 42 | 32.8 | 1536 | 1-Octanol | 2 | 8.1 × 106 (0.6) | 2.9 × 106 (0.3) | ↓ | 2.8 | 1.1 × 107 (0.5) | ↑ | 1.3 | 3.2 × 106 (0.3) | ↓ | 2.5 | 111-87-5 | C8H18O |
| 43 | 33.2 | 1545 | 2-Nonanol | 2 | 6.8 × 106 (0.5) | 6.2 × 106 (0.6) | ↓ | 1.1 | 7.8 × 106 (0.3) | ↑ | 1.1 | 5.7 × 106 (0.4) | ↓ | 1.2 | 628-99-9 | C9H20O |
| 44 | 33.5 | 1552 | Ethyl nonanoate | 2 | 8.4 × 106 (0.6) | 6.6 × 106 (0.6) | ↓ | 1.3 | ND | - | - | ND | - | - | 123-29-5 | C11H22O2 |
| 45 | 34.2 | 1569 | Ethyl 2-hydroxy-4-methylpentanoate | 2 | 1.1 × 107 (0.8) | 7.5 × 106 (0.7) | ↓ | 1.4 | 7.0 × 106 (0.3) | ↓ | 1.5 | 5.9 × 106 (0.5) | ↓ | 1.8 | 10348-47-7 | C8H16O3 |
| 46 | 34.2 | 1570 | β-Linalool | 2 | 4.8 × 106 (0.4) | 5.1 × 106 (0.5) | ↑ | 1.1 | 5.4 × 106 (0.2) | ↑ | 1.1 | 5.0 × 106 (0.3) | ↑ | 1.0 | 78-70-6 | C10H18O |
| 47 | 34.4 | 1575 | 2,3-Butanediol | 2 | 8.1 × 107 (6) | 9.7 × 107 (9) | ↑ | 1.2 | 1.2 × 108 (5) | ↑ | 1.5 | 1.1 × 108 (8) | ↑ | 1.3 | 513-85-9 | C4H10O2 |
| 48 | 34.8 | 1583 | Unknown 34.7591 | 4 | 2.8 × 107 (2) | 1.9 × 107 (2) | ↓ | 1.5 | 2.4 × 107 (1) | ↓ | 1.2 | 1.9 × 107 (1) | ↓ | 1.5 | - | C8H18O |
| 49 | 34.9 | 1588 | Unknown 34.9582 | 4 | 7.9 × 105 (0.1) | 2.6 × 105 (0.0) | ↓ | 3.0 | 7.0 × 105 (0.0) | ↓ | 1.1 | ND | - | - | - | - |
| 50 | 35.3 | 1596 | Unknown 35.2873 | 4 | 2.0 × 107 (2) | 8.8 × 106 (0.8) | ↓ | 2.3 | 2.4 × 107 (1) | ↑ | 1.2 | 9.5 × 106 (0.6) | ↓ | 2.1 | - | C8H16O3 |
| 51 | 35.9 | 1611 | Unknown 35.8931 | 4 | 2.6 × 107 (2) | 3.5 × 107 (3) | ↑ | 1.4 | 3.9 × 107 (2) | ↑ | 1.5 | 3.5 × 107 (2) | ↑ | 1.4 | - | C6H12O2 |
| 52 | 36.5 | 1627 | Propylene Glycol | 3 | 5.4 × 106 (0.4) | 4.4 × 106 (0.4) | ↓ | 1.2 | 6.5 × 106 (0.3) | ↑ | 1.2 | 5.5 × 106 (0.4) | ↑ | 1.0 | 57-55-6 | C3H8O2 |
| 53 | 37.0 | 1642 | Unknown 37.0675 | 4 | 2.7 × 106 (0.2) | 2.3 × 106 (0.2) | ↓ | 1.2 | 3.8 × 106 (0.1) | ↑ | 1.4 | ND | - | - | - | C12H24O |
| 54 | 37.7 | 1657 | Ethyl decanoate | 2 | 7.5 × 108 (55) | ND | - | - | 3.8 × 108 (17) | ↓ | 2.0 | 1.6 × 109 (100) | ↑ | 2.1 | 110-38-3 | C12H24O2 |
| 55 | 38.1 | 1668 | 4-methyl-benzaldehyde | 3 | ND | ND | - | - | 1.2 × 106 (0.1) | - | - | ND | - | - | 104-87-0 | C8H8O |
| 56 | 38.5 | 1678 | Isoamyl octanoate * | 2 | 1.7 × 107 (1) | 1.0 × 107 (0.9) | ↓ | 1.7 | 8.7 × 106 (0.4) | ↓ | 2.0 | 2.0 × 107 (2) | ↑ | 1.2 | 2035-99-6 | C13H26O2 |
| 57 | 38.8 | 1686 | 1-nonanol * | 2 | 1.8 × 107 (1) | 1.0 × 107 (0.9) | ↓ | 1.7 | 2.3 × 107 (1) | ↑ | 1.3 | 9.8 × 106 (0.8) | ↓ | 1.8 | 143-08-8 | C9H20O |
| 58 | 39.0 | 1691 | Unknown 38.9824 | 4 | 1.5 × 106 (0.1) | ND | - | - | 1.9 × 106 (0.8) | ↑ | 1.3 | ND | - | - | - | C15H32 |
| 59 | 39.2 | 1696 | Unknown 39.1966 | 4 | 9.2 × 106 (0.7) | 6.1 × 106 (0.5) | ↓ | 1.5 | 1.0 × 107 (0.4) | ↑ | 1.1 | 7.0 × 106 (0.5) | ↓ | 1.3 | - | C5H10O2 |
| 60 | 39.4 | 1702 | Diethyl succinate | 2 | 1.3 × 109 (94) | 2.9 × 108 (26) | ↓ | 4.3 | ND | - | - | 3.5 × 108 (21) | ↓ | 3.6 | 123-25-1 | C8H14O4 |
| 61 | 39.7 | 1710 | Ethyl 9-decenoate | 3 | 2.0 × 107 (2) | 1.8 × 107 (2) | ↓ | 1.1 | 2.1 × 107 (0.9) | ↑ | 1.0 | 3.2 × 107 (2) | ↑ | 1.6 | 67233-91-4 | C12H22O2 |
| 62 | 40.3 | 1725 | α-Terpineol † | 2 | ND | 1.7 × 106 (0.2) | - | - | 5.9 × 106 (0.3) | ↑ | 3.5 | 1.6 × 106 (0.1) | ↓ | 1.0 | 98-55-5 | C10H18O |
| 63 | 40.8 | 1738 | Unknown 40.7705 | 2 | 4.5 × 106 (0.3) | 1.8 × 106 (0.2) | ↓ | 2.5 | 5.6 × 106 (0.2) | ↑ | 1.2 | 2.6 × 106 (0.2) | ↓ | 1.7 | - | C12H16O2 |
| 64 | 41.0 | 1744 | 2-Undecanol | 3 | ND | ND | - | - | 1.6 × 106 (0.1) | - | - | ND | - | - | 1653-30-1 | C11H24O |
| 65 | 41.1 | 1746 | 3-(methylthio)-1-Propanol | 3 | 4.5 × 106 (0.3) | 2.7 × 106 (0.2) | ↓ | 1.7 | 5.0 × 106 (0.2) | ↑ | 1.1 | 3.4 × 106 (0.3) | ↓ | 1.3 | 505-10-2 | C4H10OS |
| 66 | 41.3 | 1754 | Unknown 41.3476 | 4 | 1.4 × 106 (0.1) | ND | - | - | 2.1 × 106 (0.1) | ↑ | 1.5 | ND | - | - | - | C11H12O3 |
| 67 | 41.5 | 1758 | Unknown 41.5185 * | 4 | 1.7 × 106 (0.1) | 5.0 × 105 (0.0) | ↓ | 3.5 | 8.8 × 105 (0.0) | ↓ | 2.0 | 9.0 × 105 (0.1) | ↓ | 1.9 | - | C13H16 |
| 68 | 41.8 | 1767 | Octyl ether † | 3 | ND | 7.5 × 105 (0.1) | - | - | ND | - | - | 8.9 × 105 (0.1) | ↑ | 1.2 | 629-82-3 | C16H34O |
| 69 | 42.2 | 1776 | trans-4-(1,1-dimethylethyl)-cyclohexanol | 3 | ND | 1.2 × 106 (0.1) | - | - | ND | - | - | ND | - | - | 21862-63-5 | C10H20O |
| 70 | 42.6 | 1787 | Decyl alcohol | 2 | 9.4 × 106 (0.7) | 9.8 × 106 (0.9) | ↑ | 1.0 | 9.1 × 106 (0.4) | ↓ | 1.0 | 9.7 × 106 (0.8) | ↑ | 1.0 | 112-30-1 | C10H22O |
| 71 | 42.7 | 1790 | Unknown 42.6886 | 2 | 3.8 × 107 (3) | 3.3 × 107 (3) | ↓ | 1.1 | 1.9 × 107 (0.8) | ↓ | 2.0 | 1.8 × 107 (1) | ↓ | 2.1 | - | C8H9NO2 |
| 72 | 42.9 | 1794 | Methyl Salicylate | 2 | ND | ND | - | - | 1.3 × 106 (0.1) | - | - | ND | - | - | 119-36-8 | C8H8O3 |
| 73 | 43.3 | 1807 | Ethyl phenylacetate | 2 | 2.3 × 106 (0.2) | 7.0 × 105 (0.1) | ↓ | 3.3 | 3.3 × 106 (0.2) | ↑ | 1.4 | 8.2 × 105 (0.1) | ↓ | 2.8 | 101-97-3 | C10H12O2 |
| 74 | 43.7 | 1818 | Unknown 43.7419 | 4 | 4.5 × 106 (0.3) | 1.8 × 106 (0.2) | ↓ | 2.5 | 3.7 × 106 (0.2) | ↓ | 1.2 | 2.1 × 106 (0.1) | ↓ | 2.1 | - | C8H14O4 |
| 75 | 44.1 | 1828 | Unknown 44.0789 | 4 | 1.0 × 106 (0.1) | 6.2 × 105 (0.1) | ↓ | 1.7 | ND | - | - | 7.5 × 105 (0.1) | ↓ | 1.4 | - | C14H22 |
| 76 | 44.4 | 1839 | Phenethyl acetate * | 2 | 1.5 × 107 (1) | 1.3 × 107 (1) | ↓ | 1.2 | 2.7 × 107 (1) | ↑ | 1.8 | 1.5 × 107 (0.8) | ↑ | 1.0 | 103-45-7 | C10H12O2 |
| 77 | 44.7 | 1846 | β-Damascenone | 2 | 3.5 × 106 (0.3) | 5.8 × 106 (0.5) | ↑ | 1.6 | 4.0 × 106 (0.2) | ↑ | 1.1 | 7.1 × 106 (0.6) | ↑ | 2.0 | 23726-93-4 | C13H18O |
| 78 | 45.3 | 1863 | Ethyl dodecanoate | 2 | 2.7 × 107 (2) | 2.9 × 107 (3) | ↑ | 1.1 | 1.7 × 107 (0.8) | ↓ | 1.5 | 2.4 × 107 (2) | ↓ | 1.1 | 106-33-2 | C14H28O2 |
| 79 | 45.6 | 1872 | Hexanoic acid * | 2 | 5.6 × 107 (4) | 3.3 × 107 (3) | ↓ | 1.7 | 5.9 × 107 (3) | ↑ | 1.1 | 5.7 × 107 (3) | ↑ | 1.0 | 142-62-1 | C6H12O2 |
| 80 | 46.0 | 1883 | Isoamyl decanoate * | 2 | 2.4 × 106 (0.2) | 1.1 × 106 (0.1) | ↓ | 2.1 | 1.1 × 106 (0.1) | ↓ | 2.3 | 1.6 × 106 (0.1) | ↓ | 1.5 | 2306-91-4 | C15H30O2 |
| 81 | 46.7 | 1902 | Benzyl alcohol *** | 2 | 1.3 × 107 (0.9) | 7.3 × 106 (0.6) | ↓ | 1.8 | 2.2 × 107 (1) | ↑ | 1.7 | 6.4 × 106 (0.5) | ↓ | 2.0 | 100-51-6 | C7H8O |
| 82 | 47.0 | 1912 | Unknown 47.0219 | 2 | 6.7 × 105 (0.1) | 6.0 × 105 (0.1) | ↓ | 1.1 | 7.0 × 105 (0.0) | ↑ | 1.1 | ND | - | - | - | C16H30O4 |
| 83 | 47.6 | 1929 | Ethyl 3-methylbutyl succinate | 3 | 4.7 × 107 (3) | 1.8 × 107 (2) | ↓ | 2.7 | 4.4 × 107 (2) | ↓ | 1.1 | 2.8 × 107 (2) | ↓ | 1.7 | 28024-16-0 | C11H20O4 |
| 84 | 48.0 | 1940 | 2-Phenylethanol | 2 | 1.4 × 109 (100) | 1.1 × 109 (100) | ↓ | 1.2 | ND | - | - | 1.3 × 109 (42) | ↓ | 1.1 | 60-12-8 | C8H10O |
| 85 | 48.9 | 1967 | Unknown 48.8990 | 4 | ND | ND | - | - | 6.8 × 106 (0.3) | - | - | ND | - | - | - | C7H5ClF3N |
| 86 | 49.7 | 1991 | 1-Dodecanol | 2 | 2.3 × 106 (0.2) | 2.4 × 106 (0.2) | ↑ | 1.0 | 2.4 × 106 (0.1) | ↑ | 1.0 | 3.2 × 106 (0.2) | ↑ | 1.4 | 112-53-8 | C12H26O |
| 87 | 50.9 | 2032 | Diphenyl ether | 2 | ND | ND | - | - | ND | - | - | 2.5 × 105 (0.0) | - | - | 101-84-8 | C12H10O |
| 88 | 51.7 | 2062 | 4-Ethylguaiacol | 3 | ND | ND | - | - | 1.8 × 106 (0.1) | - | - | ND | - | - | 2785-89-9 | C9H12O2 |
| 89 | 52.0 | 2072 | Unknown 51.9854 | 2 | 7.6 × 105 (0.1) | 1.4 × 106 (0.1) | ↑ | 1.8 | 8.6 × 105 (0.0) | ↑ | 1.1 | 1.5 × 106 (0.1) | ↑ | 1.9 | - | C15H26O3 |
| 90 | 52.1 | 2078 | Ethyl tetradecanoate | 3 | 7.0 × 105 (0.1) | 1.2 × 106 (0.1) | ↑ | 1.7 | 3.8 × 105 (0.0) | ↓ | 1.8 | 6.4 × 105 (0.0) | ↓ | 1.1 | 124-06-1 | C16H32O2 |
| 91 | 52.3 | 2084 | Unknown 52.3099 | 4 | ND | ND | - | - | 4.6 × 105 (0.0) | - | - | ND | - | - | - | C10H20O2 |
| 92 | 52.5 | 2093 | Octanoic acid | 2 | 1.2 × 108 (9) | 9.3 × 107 (8) | ↓ | 1.3 | 1.3 × 108 (6) | ↑ | 1.1 | 1.8 × 108 (9) | ↑ | 1.5 | 124-07-2 | C8H16O2 |
| 93 | 54.4 | 2194 | Unknown 54.4273 | 4 | 2.3 × 106 (0.1) | 1.7 × 106 (0.1) | ↓ | 1.4 | ND | - | - | ND | - | - | - | C8H7NO2 |
| 94 | 54.5 | 5197 | 4-Ethylphenol | 2 | ND | ND | - | - | 3.5 × 107 (2) | - | - | ND | - | - | 123-07-9 | C8H10O |
| 95 | 55.8 | 2274 | Ethyl hexadecanoate | 2 | 2.3 × 106 (0.2) | 1.1 × 106 (0.1) | ↓ | 2.0 | 1.7 × 106 (0.1) | ↓ | 1.4 | 1.3 × 106 (0.1) | ↓ | 1.8 | 628-97-7 | C18H36O2 |
| 96 | 56.1 | 2288 | Unknown 56.0757 | 4 | ND | ND | - | - | ND | - | - | 1.7 × 105 (0.0) | - | - | - | C16H18 |
| 97 | 56.3 | 2301 | Decanoic acid | 2 | 7.7 × 106 (0.6) | 1.0 × 107 (0.9) | ↑ | 1.4 | 6.1 × 106 (0.3) | ↓ | 1.3 | 1.9 × 107 (1) | ↑ | 2.5 | 334-48-5 | C10H20O2 |
| 98 | 56.5 | 2313 | Unknown 56.5471 | 4 | 8.2 × 105 (0.1) | 5.6 × 105 (0.1) | ↓ | 1.5 | 7.3 × 105 (0.0) | ↓ | 1.1 | 7.3 × 105 (0.1) | ↓ | 1.1 | - | C13H14ClF2NO3 |
| 99 | 56.8 | 2324 | 2,4-Di-tert-butylphenol | 2 | 1.2 × 107 (0.9) | 2.4 × 106 (0.2) | ↓ | 4.8 | 1.3 × 107 (0.6) | ↑ | 1.1 | 2.7 × 106 (0.2) | ↓ | 4.3 | 96-76-4 | C14H22O |
| 100 | 57.4 | 2354 | Ethyl trans-2-butenoate | 2 | ND | ND | - | - | 2.0 × 107 (0.9) | - | - | ND | - | - | 56-81-5 | C3H8O3 |
| 101 | 58.9 | 2426 | Ethyl hydrogen succinate | 3 | 8.6 × 106 (0.6) | ND | - | - | ND | - | - | ND | - | - | 1070-34-4 | C6H10O4 |
| RI | Name | Abundance (Area) | Fold Change | DataBase ID | Aromatic Properties 1 | ||
|---|---|---|---|---|---|---|---|
| 2017 | 2018 | ||||||
| 901 | Ethyl acetate | 1,048,176,640 | 640,235,900 | ↓ | 2 | YMDB00569 | Anise, balsam, ethereal |
| 1084 | Ethyl 3-methylbutyrate | 23,746,934 | 5,515,695 | ↓ | 4 | YMDB16003 | Apple, Fruity, pineapple |
| 1261 | Styrene † | 3,931,121 | 6,247,420 | ↓ | 1 | YMDB16080 | Balsam, floral, plastic |
| 1381 | Hexyl formate | 197,498,208 | 99,272,832 | ↓ | 2 | HMDB0032874 | Present in fruits |
| 1402 | Methyl octanoate † | 9,709,568 | 15,677,219 | ↓ | 1 | YMDB01339 | Aldehydic, green, herbal |
| 1411 | trans-3-Hexen-1-ol | 1,722,258 | 927,801 | ↓ | 2 | YMDB01421 | Green, cortex, leafy |
| 1419 | 3-Octanol | 1,443,536 | 1,057,637 | ↓ | 1 | HMDB0030070 | Earthy, mushroom, dairy |
| 1513 | 2-Ethyl-1-hexanol | 4,465,255 | 2,385,446 | ↓ | 2 | YMDB01330 | Citrus, floral, fresh, |
| 1536 | 1-Octanol | 9,221,439 | 3,018,129 | ↓ | 3 | YMDB00808 | Aldehyde, burnt, chemical |
| 1583 | Unknown 34.7591 | 25,799,506 | 18,890,604 | ↓ | 1 | - | - |
| 1596 | Unknown 35.2873 | 22,104,118 | 9,159,423 | ↓ | 2 | - | - |
| 1686 | 1-nonanol | 20,248,112 | 10,009,123 | ↓ | 2 | YMDB15917 | Bitter, fatty, floral |
| 1696 | Unknown 39.1966 | 9,593,656 | 6,539,898 | ↓ | 1 | - | - |
| 1738 | Unknown 40.7705 | 4,980,681 | 2,160,692 | ↓ | 2 | - | - |
| 1746 | 3-Methylthio-1-propanol | 4,779,528 | 2,999,897 | ↓ | 2 | YMDB1427 | Widely distributed aroma constituent of foods and beverages. |
| 1807 | Ethyl phenylacetate | 2,776,861 | 755,631 | ↓ | 4 | HMDB0032618 | Apricot, banana, brandy |
| 1818 | Unknown 43.7419 | 4,071,294 | 1,918,293 | ↓ | 2 | - | - |
| 1846 | β-Damascenone † | 3,772,885 | 6,413,788 | ↓ | 1 | YMDB15908 | Apple, honey, rose |
| 1902 | Benzyl alcohol | 16,751,528 | 6,809,303 | ↓ | 2 | YMDB01426 | Balsamic, cherry, floral |
| 1929 | Ethyl 3-methylbutyl succinate | 45,479,080 | 22,022,998 | ↓ | 2 | CID119794 | Found in wine and beer |
| 2072 | Unknown 51.9854 † | 810,844 | 1,414,013 | ↑ | 2 | - | - |
| 2274 | Ethyl hexadecanoate | 1,946,713 | 1,206,513 | ↓ | 2 | YMDB01349 | Balsam, creamy, fruity |
| 2301 | Decanoic acid † | 6,862,970 | 14,093,453 | ↑ | 2 | YMDB00677 | Citrus, fatty, rancid |
| 2324 | 2,4-Di-tert-butylphenol | 12,312,931 | 2,577,305 | ↓ | 5 | YMDB15942 | Phenolic |
| Extraction HS-SPME Fiber 50/30 μm DVB/CAR/PDMS | Detector MS-QToF | ||
|---|---|---|---|
| Sample | Conditioning at 40 °C/5 min Extraction at 40 °C/30 min | Ion source | Electron ionization (EI) |
| Source Temperature | 230 °C | ||
| Fiber conditioning | Pre-extraction at 250 °C/10 min Post-desorption at 250 °C/5 min | Emission energy | 15.2 μA |
| Electron energy | 70 eV | ||
| Desorption | 240 °C/10 min | Data storage | Profile |
| Separation GC column DB-WAX 30 m/250 μm/0.25 μm | Solvent delay | 3 min | |
| Quadropole TT1 cutoff mass | 30 amu | ||
| Inlet mode | Splitless | ||
| Flow rate | 1.0 mL/min He (RT Locked 2-Undecanone at 36.158 min) | Mass range | 30 to 400 amu |
| Acquisition rate | 2.5 spectra/s | ||
| Oven | 40 °C for 5 min | Acquisition time | 400 ms/spectrum |
| 3 °C/min to 180 °C | Transients/spectrum | 5443 | |
| 30 °C/min to 220 °C for 10 min | System with backflush and gas saver mode on. | ||
| Total run 63 min | |||
| Post-run | 2 min at 220 °C | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chávez-Márquez, A.; Gardea, A.A.; González-Rios, H.; Vazquez-Moreno, L. Characterization of Cabernet Sauvignon Wines by Untargeted HS-SPME GC-QTOF-MS. Molecules 2022, 27, 1726. https://doi.org/10.3390/molecules27051726
Chávez-Márquez A, Gardea AA, González-Rios H, Vazquez-Moreno L. Characterization of Cabernet Sauvignon Wines by Untargeted HS-SPME GC-QTOF-MS. Molecules. 2022; 27(5):1726. https://doi.org/10.3390/molecules27051726
Chicago/Turabian StyleChávez-Márquez, Alejandra, Alfonso A. Gardea, Humberto González-Rios, and Luz Vazquez-Moreno. 2022. "Characterization of Cabernet Sauvignon Wines by Untargeted HS-SPME GC-QTOF-MS" Molecules 27, no. 5: 1726. https://doi.org/10.3390/molecules27051726
APA StyleChávez-Márquez, A., Gardea, A. A., González-Rios, H., & Vazquez-Moreno, L. (2022). Characterization of Cabernet Sauvignon Wines by Untargeted HS-SPME GC-QTOF-MS. Molecules, 27(5), 1726. https://doi.org/10.3390/molecules27051726






