Methyl Jasmonate Activates the 2C Methyl-D-erithrytol 2,4-cyclodiphosphate Synthase Gene and Stimulates Tanshinone Accumulation in Salvia miltiorrhiza Solid Callus Cultures
Abstract
:1. Introduction
2. Results
2.1. Isolation of S. Miltiorrhiza MEC Promoter, 5′UTR and 5′fragment of CDS
2.2. In Silico Analysis of SmMEC Promoter
2.3. Microarray Co-Expression Studies
2.4. RT-PCR Analysis of SmMDS Promoter Activity after MeJa Administration
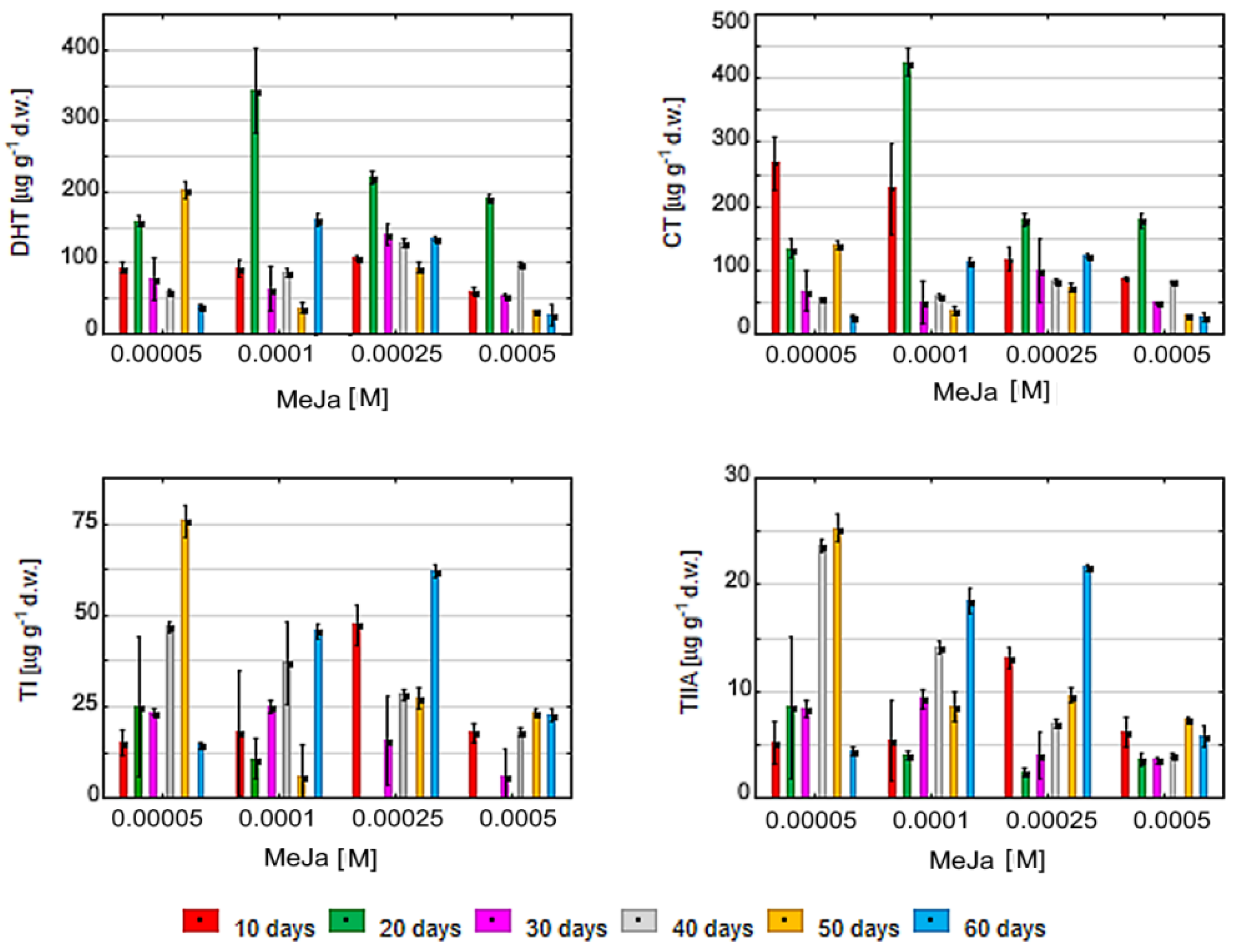
2.5. Treatment of S. miltiorrhiza Callus by MeJa
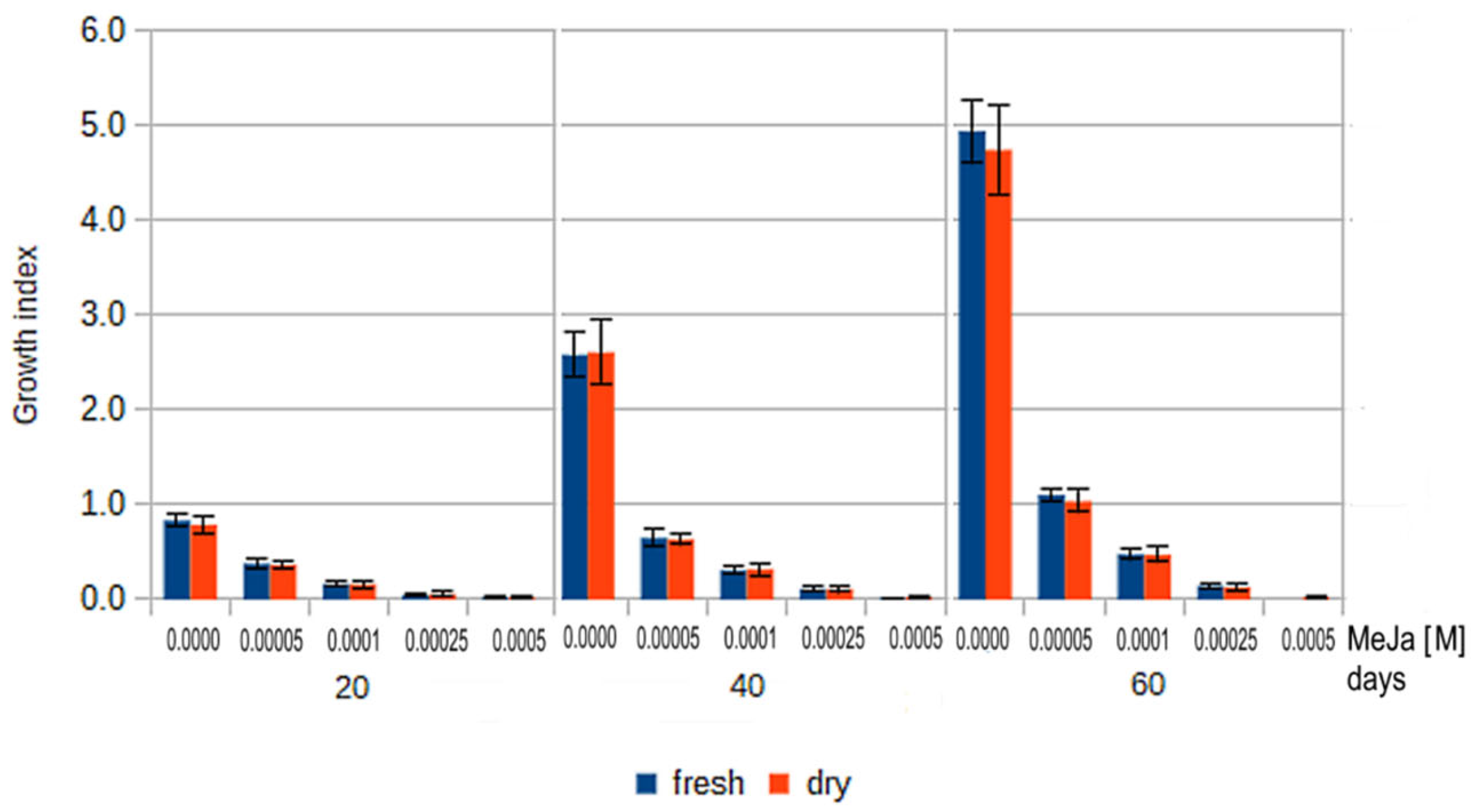
2.6. Growth Index Rates of Callus Treated by MeJa
2.7. Distribution of MeJa Responsive Cis-Active Elements within Proximal Promoters of A. thaliana and S. miltiorrhiza
3. Discussion
4. Material and Methods
4.1. Plant Cultivation
4.2. Isolation of Genomic DNA
4.3. Promoter Isolation and In Silico Characterization
4.4. Microarray Co-Expression Studies
4.5. Callus Induction and Methyl Jasmonate (MeJa) Treatment
4.6. Calculation of Callus Growth Index
4.7. Extraction of Callus
4.8. HPLC Analysis
4.9. Preparation of Calibration Standard Solutions
4.10. RNA Isolation and cDNA Synthesis
4.11. Real-Time PCR
4.12. Statistical Analysis
4.13. Promoter Analysis of MEP, MVA and Tanshinone Precursor Biosynthesis Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Kirby, J.; Keasling, J.D. Biosynthesis of Plant Isoprenoids: Perspectives for Microbial Engineering. Annu. Rev. Plant Biol. 2009, 60, 335–355. [Google Scholar] [CrossRef] [PubMed]
- Gryszczynska, A.; Opala, B.; Lowicki, Z.; Dreger, M.; Gorska-Paukszta, M.; Szulc, M.; Kaminska, E.; Litwin, E.; Struzik, P.; Dyr, W.; et al. Bioactive compounds determination in the callus and hydroalcoholic extracts from Salvia miltiorrhiza and Salvia przewalskii—Preliminary study on their anti-alcoholic activity effects. Phytochem. Lett. 2015, 11, 399–403. [Google Scholar] [CrossRef]
- Su, C.Y.; Ming, Q.L.; Rahman, K.; Han, T.; Qin, L.P. Salvia miltiorrhiza: Traditional medicinal uses, chemistry and pharmacology. Chin. J. Nat. Med. 2015, 13, 163–182. [Google Scholar] [CrossRef]
- Wang, L.; Ma, R.; Liu, C.; Liu, H.; Zhu, R.; Guo, S.; Tang, M.; Li, Y.; Niu, J.; Fu, M.; et al. Salvia miltiorrhiza: A potential red light to the development of cardiovascular diseases. Curr. Pharm. Des. 2017, 23, 1077–1097. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Zuo, Z.; Chow, M.S. Danshen: An overview of its chemistry, pharmacology, and clinical use. J. Clin. Pharmacol. 2005, 45, 1345–1389. [Google Scholar] [CrossRef]
- Sui, C. Salvia miltiorrhiza Resources, Cultivation and Breeding. In The Salvia miltiorrhiza Genome, 1st ed.; Lu, S., Ed.; Springer Nature: Cham, Switzerland, 2019; pp. 17–32. [Google Scholar] [CrossRef]
- Pu, X.; Dong, X.; Li, Q.; Chen, Z.; Liu, L. An update on the function and regulation of methylerythritol phosphate and mevalonate pathways and their evolutionary dynamics. J. Integr. Plant Biol. 2021, 63, 1211–1226. [Google Scholar] [CrossRef]
- Calisto, B.M.; Perez-Gil, J.; Bergua, M.; Querol-Audi, J.; Fita, I.; Imperial, S. Biosynthesis of isoprenoids in plants: Structure of the 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Arabidopsis thaliana. Comparison with bacterial enzymes. Protein Sci. 2007, 16, 2082–2088. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Du, X.; Liang, X.; Han, R.; Liang, Z.; Liu, Y.; Liu, F.; Zhao, J. Different roles of mevalonate and methylerythritol phosphate pathways in cell growth and tanshinone production in Salvia miltiorrhiza hairy roots. PLoS ONE 2012, 7, e46797. [Google Scholar] [CrossRef]
- Hao, G.; Shi, R.; Tao, R.; Fang, Q.; Jiang, X.; Ji, H.; Feng, L.; Huang, L. Cloning, molecular characterization and functional analysis of 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase (HDR) for diterpenoid tanshinone biosynthesis in Salvia miltiorrhiza Bge. f. alba. Plant Physiol. Biochem. 2013, 70, 21–32. [Google Scholar] [CrossRef]
- Liao, P.; Hemmerlin, A.; Bach, T.J.; Chye, M.L. The potential of the mevalonate pathway for enhanced isoprenoid production. Biotechnol. Adv. 2016, 34, 697–713. [Google Scholar] [CrossRef]
- Hemmerlin, A. Post-translational events and modifications regulating plant enzymes involved in isoprenoid precursor biosynthesis. Plant Sci. 2013, 203–204, 41–54. [Google Scholar] [CrossRef]
- Leivar, P.; Antolín-Llovera, M.; Ferrero, S.; Closa, M.; Arró, M.; Ferrer, A.; Boronat, A.; Campos, N. Multilevel control of Arabidopsis 3-hydroxy-3-methylglutaryl coenzyme A reductase by protein phosphatase 2A. Plant Cell 2011, 23, 1494–1511. [Google Scholar] [CrossRef] [Green Version]
- Wright, L.P.; Rohwer, J.M.; Ghirardo, A.; Hammerbacher, A.; Ortiz-Alcaide, M.; Raguschke, B.; Schnitzler, J.P.; Gershenzon, J.; Phillips, M.A. Deoxyxylulose 5-phosphate synthase controls flux through the methylerythritol 4-phosphate pathway in Arabidopsis. Plant. Physiol. 2014, 165, 1488–1504. [Google Scholar] [CrossRef]
- Rodríguez-Concepción, M.; Boronat, A. Breaking new ground in the regulation of the early steps of plant isopenoid biosynthesis. Curr. Opin. Plant Biol. 2015, 25, 17–22. [Google Scholar] [CrossRef]
- Lemos, M.; Xiao, Y.; Bjornson, M.; Wang, J.Z.; Hicks, D.; de Souza, A.; Wang, C.Q.; Yang, P.; Ma, S.; Dinesh-Kumar, S.; et al. The plastidial retrograde signal methyl erythritol cyclopyrophosphate is a regulator of salicylic and jasmonic acid crosstalk. J. Exp. Bot. 2016, 67, 1557–1566. [Google Scholar] [CrossRef] [Green Version]
- Walley, J.; Xiao, Y.; Wang, J.Z.; Baidoo, E.E.; Keasling, J.D.; Shen, Z.; Briggs, S.P.; Dehesh, K. Plastid-produced interorganellar stress signal MEcPP potentiates induction of the unfolded protein response in endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2015, 112, 6212–6217. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Savchenko, T.; Baidoo, E.E.K.; Chehab, W.E.; Hayden, D.M.; Tolstikov, V.; Corvin, J.A.; Kliebenstein, D.J.; Keasling, J.D.; Dehesh, K. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 2012, 149, 1525–1535. [Google Scholar] [CrossRef] [Green Version]
- Kai, G.; Hu, H.; Zhou, C.; Liao, P.; Xiao, J.; Luo, X.; You, L.; Zhang, L. Metabolic engineering tanshinone biosynthetic pathway in S. miltiorrhiza hairy root cultures. Metab. Eng. 2011, 13, 319–327. [Google Scholar] [CrossRef]
- Ma, Y.; Yuan, L.; Li, X.; Chen, S.; Lu, S. Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J. Exp. Bot. 2012, 63, 2809–2823. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Wei, H.; Movahedi, A.; Sun, W.; Ma, X.; Li, D.; Yin, T.; Zhuge, Q. Evaluation, characterization, expression profiling and functional analysis of DXS and DXR genes from Populus trichocarpa. Plant Physiol. Biochem. 2019, 142, 94–105. [Google Scholar] [CrossRef]
- Xu, C.; Li, H.; Gu, C.; Mu, H.; Yue, Y.; Wang, L. Cloning and expression analysis of MEP pathway enzyme-encoding genes in Osmanthus fragrans. Genes 2016, 7, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, H.; Kumar, S. A functional (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase exhibits diurnal regulation of expression in Stevia rebaudiana (Bertoni). Gene 2013, 527, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xiao, Y.; Di, P.; Yu, X.; Chen, W.; Zhang, L. Molecular cloning and characterization of a 2C methyl-D-erithrytol 2,4-cyclodiphosphate synthase gene from Cephalotaxus harringtonia. Mol. Biol. Rep. 2009, 36, 1749–1756. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.K.; Nargis, S.; Kim, S.M.; Kim, S.U. Distinct expression patterns of two Gingko biloba 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase/isopentenyl diphosphate synthase (HDR/IDS) promoters in Arabidopsis model. Plant Physiol. Biochem. 2013, 62, 47–53. [Google Scholar] [CrossRef]
- Wiberley, A.E.; Donohue, A.R.; Westphal, M.M.; Sharkey, T.D. Regulation of isoprene emission from poplar leaves throughout a day. Plant Cell Environ. 2009, 32, 939–947. [Google Scholar] [CrossRef]
- Ahn, C.S.; Pai, H.S. Physiological function of IspE, a plastid MEP pathway gene for isoprenoid biosynthesis in organelle biogenesis and cell morphogenesis in Nicotiana benthamiana. Plant. Mol. Biol. 2008, 66, 503–517. [Google Scholar] [CrossRef]
- Gong, Y.F.; Liao, Z.H.; Guo, B.H.; Sun, X.F.; Tang, K.X. Molecular cloning and expression profile analysis of Ginkgo biloba DXS gene encoding the 1-deoxy-D-xylulose 5-phosphate synthase, the first committed enzyme of the 2-C-methyl-D-erythritol 4-phosphate pathway. Planta Med. 2006, 72, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Stitz, M.; Hartl, M.; Baldwin, I.T.; Gaquerel, E. Jasmonoyl-L-isoleucine coordinates metabolic networks required for anthesis and floral attractant emission in wild tobacco (Nicotiana attenuata). Plant Cell 2014, 26, 3964–3983. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Li, H.; Li, S.; Yao, R.; Deng, H.; Xie, Q.; Xie, D. The Arabidopsis F-box protein CORONATINE INSENSITIVE1 is stabilized by SCFCOI1 and degraded via the 26S proteasome pathway. Plant Cell 2013, 25, 486–498. [Google Scholar] [CrossRef] [Green Version]
- Griffiths, G. Jasmonates: Biosynthesis, perception and signal transduction. Essays Biochem. 2020, 64, 501–512. [Google Scholar] [CrossRef]
- Zhou, M.; Memelink, J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 2016, 34, 441–449. [Google Scholar] [CrossRef]
- Hou, Z.; Li, Y.; Su, F.; Wang, Y.; Zhang, X.; Xu, L.; Yang, D.; Liang, Z. The exploration of the methyl jasmonate on the tanshinones biosynthesis in hair roots of Salvia miltiorrhiza Bunge and Salvia castanea f. tomentosa Stib. Ind. Crop. Prod. 2021, 167, 113563. [Google Scholar] [CrossRef]
- Zhao, J.L.; Zhou, L.G.; Wu, J.Y. Effects of biotic and abiotic elicitors on cell growth and tanshinone accumulation in Salvia miltiorrhiza cell cultures. Appl. Microbiol. Biotechnol. 2010, 87, 137–144. [Google Scholar] [CrossRef]
- Hao, X.; Shi, M.; Cui, L.; Xu, C.; Zhang, Y.; Kai, G. Effects of methyl jasmonate and salicylic acid on tanshinone production and biosynthetic gene expression in transgenic Salvia miltiorrhiza hairy roots. Biotechnol. Appl. Biochem. 2015, 62, 24–31. [Google Scholar] [CrossRef]
- Wang, C.H.; Zheng, L.P.; Tian, H.; Wang, J.W. Synergistic effect of ultraviolet-B and methyl jasmonate on tanshinone biosynthesis in Salvia miltiorrhiza hairy roots. J. Photochem. Photobiol. B Biol. 2016, 159, 93–100. [Google Scholar] [CrossRef]
- Wu, C.T.; Mulagabal, V.; Nalawade, S.M.; Chen, C.L.; Yang, T.F.; Tsay, H.S. Isolation and quantitative analysis of cryptotanshinone, an active quinoid diterpene formed in callus of Salvia miltiorrhiza Bunge. Biol. Pharm. Bull. 2003, 26, 845–848. [Google Scholar] [CrossRef] [Green Version]
- Szymczyk, P.; Szymańska, G.; Kochan, E.; Szemraj, J.; Grąbkowska, R. Elicitation of solid callus cultures of Salvia miltiorrhiza Bunge with salicylic acid and a synthetic auxin (1-naphthalenecetic acid). Plant Cell. Tiss. Organ Cult. 2021, 147, 491–502. [Google Scholar] [CrossRef]
- Chow, C.N.; Lee, T.Y.; Hung, Y.C.; Li, G.Z.; Tseng, K.C.; Liu, Y.H.; Kuo, P.L.; Zheng, H.Q.; Chang, W.C. PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 2019, 47, D1155–D1163. [Google Scholar] [CrossRef] [Green Version]
- Shahmuradov, I.A.; Gammerman, A.J.; Hancock, J.M.; Bramley, P.M.; Solovyev, V.V. PlantProm: A database of plant promoter sequences. Nucleic Acids Res. 2003, 31, 114–117. [Google Scholar] [CrossRef] [Green Version]
- Shahmuradov, I.A.; Solovyev, V.V.; Gammerman, A.J. Plant promoter prediction with confidence estimation. Nucleic Acids Res. 2005, 33, 1069–1076. [Google Scholar] [CrossRef] [Green Version]
- Toufighi, K.; Brady, S.M.; Austin, R.; Ly, Y.; Provart, N.J. The botany array resource: E-Northerns, expression angling and promoter analyses. Plant J. 2005, 43, 153–163. [Google Scholar] [CrossRef]
- Rhee, S.Y.; Beavis, W.; Berardini, T.Z.; Chen, G.; Dixon, D.; Doyle, A.; Garcia-Hernandez, M.; Huala, E.; Lander, G.; Montoya, M.; et al. The Arabidopsis information Resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003, 31, 224–228. [Google Scholar] [CrossRef] [Green Version]
- NCBI Resource Coordinators. Database resources of the national center of biotechnology information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar] [CrossRef] [Green Version]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Majewska, M.; Wysokińska, H.; Kuźma, Ł.; Szymczyk, P. Eukaryotic and prokaryotic promoter databases as valuable tools in exploring the regulation of gene transcription: A comprehensive overview. Gene 2018, 644, 38–48. [Google Scholar] [CrossRef]
- Kiran, K.; Ansari, S.A.; Srivastava, R.; Lodhi, N.; Chaturvedi, C.P.; Sawant, S.V.; Tuli, R. The TATA-box sequence in the basal promoter contributes to determining light-dependent gene expression in plants. Plant Physiol. 2006, 142, 364–376. [Google Scholar] [CrossRef] [Green Version]
- Molina, C.; Grotewold, E. Genome wide analysis of Arabidopsis core promoters. BMC Genom. 2005, 6, 25. [Google Scholar] [CrossRef] [Green Version]
- Peres, A.L.G.L.; Soares, J.S.; Tavares, R.G.; Righetto, G.; Zullo, M.A.T.; Mandava, N.B.; Menossi, M. Brassinosteroids, the sixth class of phytohormones: A molecular view from the discovery to hormonal interactions in plant development and stress adaptation. Int. J. Mol. Sci. 2019, 20, 331. [Google Scholar] [CrossRef] [Green Version]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [Green Version]
- Ji, A.J.; Luo, H.M.; Xu, Z.C.; Zhang, X.; Zhu, Y.J.; Liao, B.S.; Yao, H.; Song, J.Y.; Chen, S.L. Genome-wide identification of the AP2/ERF gene family involved in active constituent biosynthesis in Salvia miltiorrhiza. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Pengda, M.; Liang, X.; Wei, Z.; Liang, Z.; Liu, Y.; Liu, F. PEG and ABA trigger methyl jasmonate accumulation to induce the MEP pathway and increase tanshinone production in Salvia miltiorrhiza hairy roots. Physiol. Plant. 2012, 146, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Zhu, Y.; Song, J.; Xu, L.; Sun, C.; Zhang, X.; Xu, Y.; He, L.; Sun, W.; Xu, H.; et al. Transcriptional data mining of Salvia miltiorrhiza in response to methyl jasmonate to examine the mechanism of bioactive compound biosynthesis and regulation. Physiol. Plant. 2014, 152, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Huang, Q.; Wu, X.; Zhou, Z.; Ding, M.; Shi, M.; Huang, F.; Li, S.; Wang, Y.; Kai, G. Comprehensive transcriptome profiling of Salvia miltiorrhiza for discovery of genes associated with the biosynthesis of tanshinones and phenolic acids. Sci. Rep. 2017, 7, 10554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Ma, Y. Biosynthetic pathway of tanshinones in Salvia miltiorhiza. In The Salvia miltiorrhiza Genome, 1st ed.; Lu, S., Ed.; Springer Nature: Cham, Switzerland, 2019; pp. 129–140. [Google Scholar] [CrossRef]
- Bolle, C.; Sopory, S.; Lubberstedt, T.; Herrmann, R.G.; Oelmuller, R. Segments encoding 5′-untranslated leaders of genes for thylakoid proteins contain cis-elements essential for transcription. Plant J. 1994, 6, 513–523. [Google Scholar] [CrossRef]
- Saulierè, J.; Sureau, A.; Expert-Bezancon, A.; Marie, J. The polypyrimidine tract binding protein (PTB) represses splicing of exon 6B from the β-tropomyosin pre-mRNA by directly interfering with the binding of the U2AF65 subunit. Mol. Cell. Biol. 2006, 26, 8755–8769. [Google Scholar] [CrossRef] [Green Version]
- Wachter, A.; Rühl, C.; Stauffer, E. The role of polypyrimidine tract-binding proteins and other hnRNP proteins in plant splicing regulation. Front. Plant Sci. 2012, 3, 81. [Google Scholar] [CrossRef] [Green Version]
- Vinces, M.D.; Legendre, M.; Caldara, M.; Hagihara, M.; Verstrepen, K.J. Unstable tandem repeats in promoters confer transcriptional evolvability. Science 2009, 324, 1213–1216. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Lang, Z.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell. Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, B.; Feng, D.; Jin, H.; Wang, P.; Liu, J.; Xiong, F.; Wang, J.; Wang, H.B. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 2015, 27, 787–805. [Google Scholar] [CrossRef] [Green Version]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant. Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Zhang, X.; Luo, H.; Xu, Z.; Zhu, Y.; Ji, A.; Song, J.; Chen, S. Genome-wide characterization and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza. Sci. Rep. 2015, 5, 11244. [Google Scholar] [CrossRef] [Green Version]
- Pei, T.; Ma, P.; Ding, K.D.; Liu, S.; Jia, Y.; Ru, M.; Dong, J.; Liang, Z. SmJAZ8 acts as a core repressor regulating JA-induced biosynthesis of salvianolic acid and tanshinones in Salvia miltiorrhiza hairy roots. J. Exp. Bot. 2018, 69, 1663–1678. [Google Scholar] [CrossRef]
- Yuan, J.M.; Tao, L.L.; Xu, J.T. Immobilization of callus tissue cells of Salvia miltiorrhiza and the characteristics of their products. Chin. J. Biotechnol. 1990, 6, 199–205. [Google Scholar]
- Hao, X.; Pu, Z.; Cao, G.; You, D.; Zhou, Y.; Deng, C.; Shi, M.; Nile, S.H.; Wang, Y.; Zhou, W.; et al. Tanshinone and salvianolic acid biosynthesis are regulated by SmMYB98 in Salvia miltiorrhiza hairy roots. J. Adv. Res. 2020, 23, 1–12. [Google Scholar] [CrossRef]
- Hu, Y.H.; Zhang, R.; Hu, Z.B.; Wu, Y.P.; Shen, X.M.; Zhang, G.Y.; Zhou, X.J. Callus culture and bioactive ingredients of Salvia miltiorrhiza. Plant Physiol. Comm. 1992, 28, 424–425. [Google Scholar]
- Li, M.H.; Peng, Y.; Xiao, P.G. Distribution of tanshinones in the genus Salvia (family Lamiaceae) from China and its systematic significance. J. Syst. Evol. 2010, 48, 118–122. [Google Scholar] [CrossRef]
- Cheng, Q.; He, Y.; Li, G.; Liu, Y.; Gao, W.; Huang, L. Effects of combined elicitors on tanshinone metabolic profiling and SmCPS expression in Salvia miltiorrhiza hairy root cultures. Molecules 2013, 18, 7473–7485. [Google Scholar] [CrossRef]
- Buchwald, W.; Forycka, A.; Mścisz, A.; Mielcarek, S.; Mrozikiewicz, P.M.; Baraniak, M. Zmiany zawartości substancji biologicznie czynnych w korzeniach Salvia miltiorrhza Bunge. Herba Pol. 2006, 52, 64. [Google Scholar]
- Li, B.; Wang, B.; Li, H.; Peng, L.; Ru, M.; Liang, Z.; Yan, X.; Zhu, Y. Establishment of Salvia castanea Diels f. tomentosa Sib. Hairy root culture and the promotion of taanshinone accumulation and gene expression with Ag+, methyl jasmonate, and yeast extract elicitation. Protoplasma 2016, 253, 87–100. [Google Scholar] [CrossRef]
- Li, J.; Li, B.; Luo, L.; Cao, F.; Yang, B.; Gao, J.; Yan, Y.; Zhang, G.; Peng, L.; Hu, B. Increased phenolic acids and tanshinone production and transcriptomic responses of biosynthetic genes in hairy root cultures of Salvia przewalskii Maxim. Treated with methyl jasmonate and salicylic acid. Mol. Biol. Rep. 2020, 47, 8565–8578. [Google Scholar] [CrossRef]
- Contreras, A.; Leroy, B.; Mariage, P.A.; Wattiez, R. Proteomic analysis reveals novel insights into tanshinones biosynthesis in Salvia miltiorrhiza hairy roots. Sci. Rep. 2019, 9, 5768. [Google Scholar] [CrossRef]
- Wei, T.; Gao, Y.; Deng, K.; Zhang, L.; Yang, M.; Liu, X.; Qi, C.; Wang, C.; Song, W.; Zhang, Y.; et al. Enhancement of tanshinone production in Salvia miltiorrhiza hairy root cultures by metabolic engineering. Plant Methods 2019, 15, 53. [Google Scholar] [CrossRef]
- Khan, S.; Qureshi, M.I.; Kamaluddin, A.T.; Abdin, M.Z. Protocol for isolation of genomic DNA from fresh and dry roots of medicinal plants suitable for RAPD and restriction digestion. Afr. J. Biotechnol. 2007, 6, 175–178. [Google Scholar] [CrossRef]
- Kibbe, W.A. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res. 2007, 35, W43–W46. [Google Scholar] [CrossRef]
- Birnboim, H.C.; Doly, J. A Rapid Alkaline Extraction Procedure for Screening Recombinant Plasmid DNA. Nucleic Acids Res. 1979, 7, 1513–1523. [Google Scholar] [CrossRef] [Green Version]
- Szymczyk, P.; Skała, E.; Grąbkowska, R.; Jeleń, A.; Żebrowska, M.; Balcerczak, E. Isolation and characterization of a copalyl diphosphate synthase gene promoter from Salvia miltiorrhiza. Acta Soc. Bot. Pol. 2016, 85, 3513. [Google Scholar] [CrossRef]
- Szymczyk, P.; Grąbkowska, R.; Skała, E.; Żebrowska, M.; Balcerczak, E.; Jeleń, A. Isolation and characterization of a 3-hydroxy-3-methylglutaryl coenzyme A reductase 2 promoter from Salvia miltiorrhiza. J. Plant Biochem. Biotechnol. 2018, 27, 223–236. [Google Scholar] [CrossRef]
- Usadel, B.; Obayashi, T.; Mutwil, M.; Giorgi, F.M.; Bassel, G.W.; Tanimoto, M.; Chow, A.; Steinhauser, D.; Persson, S.; Provart, N.J. Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant Cell Environ. 2009, 32, 1633–1651. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays for tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Godoy-Hernández, G.; Vázquez-Flota, F.A. Growth Measurements. In Plant Cell Culture Protocols. Methods in Molecular Biology; Loyola-Vargas, V.M., Vázquez-Flota, F., Eds.; Humana Press: Totowa, NJ, USA, 2006; Volume 318, pp. 51–58. [Google Scholar] [CrossRef]
- Wan, X.; Wang, Y.; Row, K.H. Separation of tanshinone I, tanshinone IIA, and cryptotanshinone from Salvia miltiorrhiza Bunge by normal phase HPLC. J. Liq. Chromatogr. Relat. Technol. 2009, 32, 544–552. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, J.; Tan, R.; Yang, L.; Zheng, X. Enhancing diterpenoid concentration in Salvia miltiorrhiza hairy roots through pathway engineering with maize C1 transcription factor. J. Exp. Bot. 2015, 66, 7211–7226. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, G.; Zhong, Z.; Ji, L.; Zhang, Y.; Zhou, J.; Zheng, X.; Deng, K. Overexpression of AtEDT1 promotes root elongation and affects medicinal secondary metabolite biosynthesis in roots of transgenic Salvia miltiorrhiza. Protoplasma 2017, 254, 1617–1625. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.H.; Lin, Y.H.; Yang, M.; Sun, J.H.; Guo, H.; Guo, D.A. High-performance liquid chromatographic determination of tanshinones in the roots of Salvia miltiorrhiza and related traditional Chinese medical preparations. J. Pharm. Sci. 2006, 9, 1–9. [Google Scholar]
- Yang, Y.; Hou, S.; Cui, G.; Chen, S.; Wei, J.; Huang, L. Characterization of reference genes for quantitative real time PCR analysis in various tissues of Salvia miltiorrhiza. Mol. Biol. Rep. 2010, 35, 507–513. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excell-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-DeltaDelta C(T)) method. Methods 2001, 25, 402–428. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Zakrajsek, B.A. Effects of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods 2000, 46, 69–81. [Google Scholar] [CrossRef]
- Kristiansson, E.; Thorsen, M.; Tamás, M.J.; Nerman, O. Evolutionary forces acts on promoter length: Identification of enriched cis-regulatory elements. Mol. Biol. Evol. 2009, 26, 1299–1307. [Google Scholar] [CrossRef]
- Mironova, V.V.; Omelyanchuk, N.A.; Wiebe, D.S.; Levitsky, V.G. Computational analysis of auxin responsive elements in the Arabidopsis thaliana L. genome. BMC Genomics 2014, 15, S4. [Google Scholar] [CrossRef] [Green Version]
- Keilwagen, J.; Grau, J.; Paponov, I.A.; Posch, S.; Strickert, M.; Grosse, I. De-novo discovery of differentially abundant transcription factor binding sites including their positional preference. PLoS Comput. Biol. 2011, 7, e1001070. [Google Scholar] [CrossRef]
- Yu, C.P.; Lin, J.J.; Li, W.H. Positional distribution of transcription factor binding sites in Arabidopsis thaliana. Sci. Rep. 2016, 6, 25164. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis Helix/basic-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Luo, Q.; Li, Y.; Zhou, N.; Li, X.; Gan, J.; Dong, A. Structural insights into target DNA recognition by R2R3-MYB transcription factors. Nucleic Acids Res. 2020, 48, 460–471. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant. Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Fonseca, J.P.; Menossi, M.; Thibaud-Nissen, F.; Town, C.D. Functional analysis of a TGA factor-binding site located in the promoter region controlling salicylic-acid induced NIMIN-1 expression in Arabidopsis. Genet. Mol. Res. 2010, 9, 167–175. [Google Scholar] [CrossRef]
- Sun, T.; Busta, L.; Zhang, Q.; Ding, P.; Jetter, R.; Zhang, Y. TGACG-binding factor 1 (TGA1) and TGA4 regulate salicylic acid and pipecolic acid biosynthesis by modulating the expression SYSTEMIC ACQUIRED RESISTANCE DEFICIENCT 1 (SARD1) and CALMODULIN-BINDING PROTEIN 60g (CBP60g). New Phytol. 2018, 217, 344–354. [Google Scholar] [CrossRef] [Green Version]
- Cao, W.; Wang, Y.; Shi, M.; Hao, X.; Zhao, W.; Wang, Y.; Ren, J.; Kai, G. Transcription factor SmWRKY1 positively promotes the biosynthesis of tanshinones in Salvia miltiorrhiza. Front. Plant. Sci. 2018, 9, 554. [Google Scholar] [CrossRef] [Green Version]
- Bai, Z.; Li, W.; Jia, Y.; Yue, Z.; Jiao, J.; Huang, W.; Xia, P.; Liang, Z. The ethylene response factor SmERF6 co-regulates the transcription of SmCPS1 and SmKSL1 and is involved in tanshinone biosynthesis in Salvia miltiorrhiza hairy roots. Planta 2018, 248, 243–255. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szymczyk, P.; Szymańska, G.; Kuźma, Ł.; Jeleń, A.; Balcerczak, E. Methyl Jasmonate Activates the 2C Methyl-D-erithrytol 2,4-cyclodiphosphate Synthase Gene and Stimulates Tanshinone Accumulation in Salvia miltiorrhiza Solid Callus Cultures. Molecules 2022, 27, 1772. https://doi.org/10.3390/molecules27061772
Szymczyk P, Szymańska G, Kuźma Ł, Jeleń A, Balcerczak E. Methyl Jasmonate Activates the 2C Methyl-D-erithrytol 2,4-cyclodiphosphate Synthase Gene and Stimulates Tanshinone Accumulation in Salvia miltiorrhiza Solid Callus Cultures. Molecules. 2022; 27(6):1772. https://doi.org/10.3390/molecules27061772
Chicago/Turabian StyleSzymczyk, Piotr, Grażyna Szymańska, Łukasz Kuźma, Agnieszka Jeleń, and Ewa Balcerczak. 2022. "Methyl Jasmonate Activates the 2C Methyl-D-erithrytol 2,4-cyclodiphosphate Synthase Gene and Stimulates Tanshinone Accumulation in Salvia miltiorrhiza Solid Callus Cultures" Molecules 27, no. 6: 1772. https://doi.org/10.3390/molecules27061772







