Modification of Lipid-Based Nanoparticles: An Efficient Delivery System for Nucleic Acid-Based Immunotherapy
Abstract
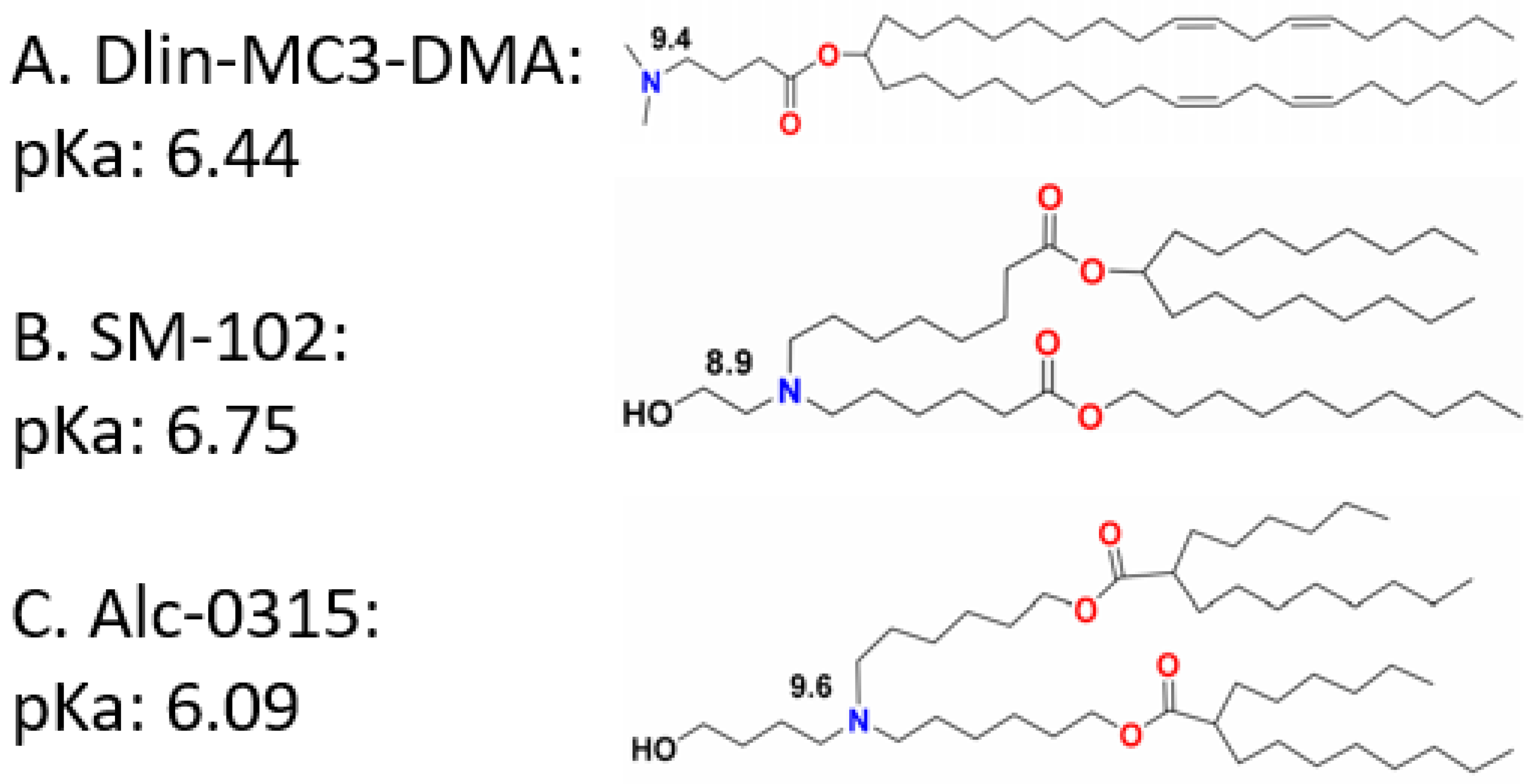
:1. Introduction
2. Modifications of Lipid-Based Nanoparticles
2.1. Types of Lipid-Based Nanoparticles
2.1.1. Liposomes
2.1.2. Lipid Nanoparticles
2.1.3. Lipid Nanoemulsions
2.1.4. Solid Lipid Nanoparticles
2.1.5. Nanostructured Lipid Carriers
2.1.6. Other Nanostructured Lipid Carriers
2.2. Development and Modification Strategies
2.2.1. Component Modification
2.2.2. Surface Modification
3. Applications of Lipid-Based Nanoparticles
3.1. LBNP in Treatment of Inherited Disease
3.2. LBNP in the Treatment of Infectious Diseases
3.3. LBNP in the Treatment of COVID-19
3.4. LBNP in the Treatment of Cancer Immunotherapy
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gonçalves, G.A.R.; Paiva, R.d.M.A. Gene therapy: Advances, challenges and perspectives. Einstein 2017, 15, 369–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramamoorth, M. Non Viral Vectors in Gene Therapy—An Overview. J. Clin. Diagn. Res. 2015, 9, GE01–GE06. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, C.; Wang, C.; Jankovic, K.E.; Dong, Y. Lipids and Lipid Derivatives for RNA Delivery. Chem. Rev. 2021, 121, 12181–12277. [Google Scholar] [CrossRef]
- Evers, M.M.; Toonen, L.J.A.; van Roon-Mom, W.M.C. Antisense oligonucleotides in therapy for neurodegenerative disorders. Adv. Drug Deliv. Rev. 2015, 87, 90–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinaldi, C.; Wood, M.J.A. Antisense oligonucleotides: The next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 2017, 14, 9–21. [Google Scholar] [CrossRef]
- Muntoni, F.; Wood, M.J.A. Targeting RNA to treat neuromuscular disease. Nat. Rev. Drug Discov. 2011, 10, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Opalinska, J.B.; Gewirtz, A.M. Nucleic-acid therapeutics: Basic principles and recent applications. Nat. Rev. Drug Discov. 2002, 1, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Di Fusco, D.; Dinallo, V.; Marafini, I.; Figliuzzi, M.M.; Romano, B.; Monteleone, G. Antisense Oligonucleotide: Basic Concepts and Therapeutic Application in Inflammatory Bowel Disease. Front. Pharmacol. 2019, 10, 305. [Google Scholar] [CrossRef] [Green Version]
- Scoles, D.R.; Minikel, E.V.; Pulst, S.M. Antisense oligonucleotides. Neurol. Genet. 2019, 5, e323. [Google Scholar] [CrossRef] [Green Version]
- Sharma, V.K.; Watts, J.K. Oligonucleotide therapeutics: Chemistry, delivery and clinical progress. Future Med. Chem. 2015, 7, 2221–2242. [Google Scholar] [CrossRef]
- Heidersbach, A.; Gaspar-Maia, A.; McManus, M.T.; Ramalho-Santos, M. RNA interference in embryonic stem cells and the prospects for future therapies. Gene Ther. 2006, 13, 478–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Levanova, A.; Poranen, M.M. RNA Interference as a Prospective Tool for the Control of Human Viral Infections. Front. Microbiol. 2018, 9, 2151. [Google Scholar] [CrossRef]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Speiser, J.J.; Erşahin, Ç.; Osipo, C. The Functional Role of Notch Signaling in Triple-Negative Breast Cancer. Vitnam Horm. 2013, 93, 277–306. [Google Scholar] [CrossRef]
- Laganà, A.; Veneziano, D.; Russo, F.; Pulvirenti, A.; Giugno, R.; Croce, C.M.; Ferro, A. Computational Design of Artificial RNA Molecules for Gene Regulation. In RNA Bioinformatics; Springer: Berlin/Heidelberg, Germany, 2015; pp. 393–412. [Google Scholar] [CrossRef]
- Kristen, A.V.; Ajroud-Driss, S.; Conceição, I.; Gorevic, P.; Kyriakides, T.; Obici, L. Patisiran, an RNAi therapeutic for the treatment of hereditary transthyretin-mediated amyloidosis. Neurodegener. Dis. Manag. 2019, 9, 5–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Das, G.M.; Shi, Y.; Liu, C.; Liu, X.; Tang, D.G.; Wang, J. The microRNA miR-34a Inhibits Non-Small Cell Lung Cancer (NSCLC) Growth and the CD44hi Stem-Like NSCLC Cells. PLoS ONE 2014, 9, e90022. [Google Scholar] [CrossRef]
- Yoo, B.; Jordan, V.C.; Sheedy, P.; Billig, A.-M.; Ross, A.; Pantazopoulos, P.; Medarova, Z. RNAi-Mediated PD-L1 Inhibition for Pancreatic Cancer Immunotherapy. Sci. Rep. 2019, 9, 4712. [Google Scholar] [CrossRef]
- Wiggins, J.F.; Ruffino, L.; Kelnar, K.; Omotola, M.; Patrawala, L.; Brown, D.; Bader, A.G. Development of a Lung Cancer Therapeutic Based on the Tumor Suppressor MicroRNA-34. Cancer Res. 2010, 70, 5923–5930. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Cheng, L.; Quek, C.; Bellingham, S.A.; Hill, A.F. Novel miR-29b target regulation patterns are revealed in two different cell lines. Sci. Rep. 2019, 9, 17449. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-R.; Yan, B.; Guo, Q.; Fu, F.-J.; Wang, Z.; Yin, Z.; Wei, Y.-B. The role of miR-29b in cancer: Regulation, function, and signaling. OncoTargets Ther. 2015, 2015, 539–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, A.; Kormann, M.; Rosenecker, J.; Rudolph, C. Current prospects for mRNA gene delivery. Eur. J. Pharm. Biopharm. 2009, 71, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.D.; Reidenbach, D.; Salomon, N.; Sahin, U.; Türeci, Ö.; Vormehr, M.; Kranz, L.M. mRNA therapeutics in cancer immunotherapy. Mol. Cancer 2021, 20, 69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Maruggi, G.; Shan, H.; Li, J. Advances in mRNA Vaccines for Infectious Diseases. Front. Immunol. 2019, 10, 594. [Google Scholar] [CrossRef] [Green Version]
- Wadhwa, A.; Aljabbari, A.; Lokras, A.; Foged, C.; Thakur, A. Opportunities and Challenges in the Delivery of mRNA-Based Vaccines. Pharmaceutics 2020, 12, 102. [Google Scholar] [CrossRef] [Green Version]
- Miao, L.; Zhang, Y.; Huang, L. mRNA vaccine for cancer immunotherapy. Mol. Cancer 2021, 20, 41. [Google Scholar] [CrossRef]
- McNamara, M.A.; Nair, S.K.; Holl, E.K. RNA-Based Vaccines in Cancer Immunotherapy. J. Immunol. Res. 2015, 2015, 794528. [Google Scholar] [CrossRef] [Green Version]
- Abu Abed, O.S. Gene therapy avenues and COVID-19 vaccines. Genes Immun. 2021, 22, 120–124. [Google Scholar] [CrossRef]
- Karikó, K.; Ni, H.; Capodici, J.; Lamphier, M.; Weissman, D. mRNA Is an Endogenous Ligand for Toll-like Receptor 3. J. Biol. Chem. 2004, 279, 12542–12550. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Zhang, S.; Fu, R.; Zhang, L.; Huang, K.; Peng, H.; Dai, L.; Chen, Q. Therapeutic Prospects of mRNA-Based Gene Therapy for Glioblastoma. Front. Oncol. 2019, 9, 1208. [Google Scholar] [CrossRef]
- Liang, X.; Li, D.; Leng, S.; Zhu, X. RNA-based pharmacotherapy for tumors: From bench to clinic and back. Biomed. Pharmacother. 2020, 125, 109997. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Su, H.-H.; Yang, Y.; Hu, Y.; Zhang, L.; Blancafort, P.; Huang, L. Systemic Delivery of Modified mRNA Encoding Herpes Simplex Virus 1 Thymidine Kinase for Targeted Cancer Gene Therapy. Mol. Ther. 2013, 21, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Hadas, Y.; Sultana, N.; Youssef, E.; Sharkar, M.T.K.; Kaur, K.; Chepurko, E.; Zangi, L. Optimizing Modified mRNA In Vitro Synthesis Protocol for Heart Gene Therapy. Mol. Ther. Methods Clin. Dev. 2019, 14, 300–305. [Google Scholar] [CrossRef]
- Pardi, N.; LaBranche, C.C.; Ferrari, G.; Cain, D.W.; Tombácz, I.; Parks, R.J.; Muramatsu, H.; Mui, B.L.; Tam, Y.K.; Karikó, K.; et al. Characterization of HIV-1 Nucleoside-Modified mRNA Vaccines in Rabbits and Rhesus Macaques. Mol. Ther. Nucleic Acids 2019, 15, 36–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, S.; Amorim, R.; Niu, M.; Breton, Y.; Tremblay, M.J.; Mouland, A.J. Host mRNA decay proteins influence HIV-1 replication and viral gene expression in primary monocyte-derived macrophages. Retrovirology 2019, 16, 3. [Google Scholar] [CrossRef]
- Jiang, C.; Mei, M.; Li, B.; Zhu, X.; Zu, W.; Tian, Y.; Wang, Q.; Guo, Y.; Dong, Y.; Tan, X. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 2017, 27, 440–443. [Google Scholar] [CrossRef]
- Wu, Y.; Liang, D.; Wang, Y.; Bai, M.; Tang, W.; Bao, S.; Yan, Z.; Li, D.; Li, J. Correction of a Genetic Disease in Mouse via Use of CRISPR-Cas9. Cell Stem Cell 2013, 13, 659–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bin Moon, S.; Lee, J.M.; Kang, J.G.; Lee, N.-E.; Ha, D.-I.; Kim, D.Y.; Kim, S.H.; Yoo, K.; Kim, D.; Ko, J.-H.; et al. Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3′-overhang. Nat. Commun. 2018, 9, 3651. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Niu, Y.; Ji, W.; Dong, Y. Strategies for the CRISPR-Based Therapeutics. Trends Pharmacol. Sci. 2020, 41, 55–65. [Google Scholar] [CrossRef]
- Li, B.; Zhao, W.; Luo, X.; Zhang, X.; Li, C.; Zeng, C.; Dong, Y. Engineering CRISPR–Cpf1 crRNAs and mRNAs to maximize genome editing efficiency. Nat. Biomed. Eng. 2017, 1, 0066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Zeng, C.; Dong, Y. Design and assessment of engineered CRISPR–Cpf1 and its use for genome editing. Nat. Protoc. 2018, 13, 899–914. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Glass, Z.; Chen, J.; Haas, M.; Jin, X.; Zhao, X.; Rui, X.; Ye, Z.; Li, Y.; Zhang, F.; et al. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc. Natl. Acad. Sci. USA 2021, 118, e2020401118. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, E.; Doudna, J.A. Rewriting a genome. Nature 2013, 495, 50–51. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Ma, Y.; Zhu, J.; Chen, Y.; Sun, Y.; Yao, Y.; Yang, Z.; Xie, J. A Review on Electroporation-Based Intracellular Delivery. Molecules 2018, 23, 3044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Yang, Y.; Li, L.; Zhang, P.; Guo, H.; Liu, N.; Yang, X.; Xu, F. Engineering extracellular matrix to improve drug delivery for cancer therapy. Drug Discov. Today 2020, 25, 1727–1734. [Google Scholar] [CrossRef]
- Butcher, D.T.; Alliston, T.; Weaver, V.M. A tense situation: Forcing tumour progression. Nat. Rev. Cancer 2009, 9, 108–122. [Google Scholar] [CrossRef]
- Levental, K.R.; Yu, H.; Kass, L.; Lakins, J.N.; Egeblad, M.; Erler, J.T.; Fong, S.F.T.; Csiszar, K.; Giaccia, A.; Weninger, W.; et al. Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. Cell 2009, 139, 891–906. [Google Scholar] [CrossRef] [Green Version]
- Bosman, F.T.; Stamenkovic, I. Functional structure and composition of the extracellular matrix. J. Pathol. 2003, 200, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Semple, S.C.; Chonn, A.; Cullis, P.R. Interactions of liposomes and lipid-based carrier systems with blood proteins: Relation to clearance behaviour in vivo. Adv. Drug Deliv. Rev. 1998, 32, 3–17. [Google Scholar] [CrossRef]
- Sorrentino, S. Human extracellular ribonucleases: Multiplicity, molecular diversity and catalytic properties of the major RNase types. Cell. Mol. Life Sci. 1998, 54, 785–794. [Google Scholar] [CrossRef]
- Harrison, E.B.; Azam, S.H.; Pecot, C.V. Targeting Accessories to the Crime: Nanoparticle Nucleic Acid Delivery to the Tumor Microenvironment. Front. Pharmacol. 2018, 9, 307. [Google Scholar] [CrossRef]
- Lin, Y.; Xu, J.; Lan, H. Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. J. Hematol. Oncol. 2019, 12, 76. [Google Scholar] [CrossRef]
- Zhou, J.; Tang, Z.; Gao, S.; Li, C.; Feng, Y.; Zhou, X. Tumor-Associated Macrophages: Recent Insights and Therapies. Front. Oncol. 2020, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 3151. [Google Scholar] [CrossRef] [PubMed]
- Weissleder, R.; Nahrendorf, M.; Pittet, M.J. Imaging macrophages with nanoparticles. Nat. Mater. 2014, 13, 125–138. [Google Scholar] [CrossRef]
- La-Beck, N.M.; Zamboni, B.A.; Gabizon, A.; Schmeeda, H.; Amantea, M.; Gehrig, P.A.; Zamboni, W.C. Factors affecting the pharmacokinetics of pegylated liposomal doxorubicin in patients. Cancer Chemother. Pharmacol. 2011, 69, 43–50. [Google Scholar] [CrossRef]
- Halma, C.; Daha, M.R.; Van Es, L.A. In vivo clearance by the mononuclear phagocyte system in humans: An overview of methods and their interpretation. Clin. Exp. Immunol. 1992, 89, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Monks, J.; Rosner, D.; Jon Geske, F.; Lehman, L.; Hanson, L.; Neville, M.C.; Fadok, V.A. Epithelial cells as phagocytes: Apoptotic epithelial cells are engulfed by mammary alveolar epithelial cells and repress inflammatory mediator release. Cell Death Differ. 2005, 12, 107–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gustafson, H.H.; Holt-Casper, D.; Grainger, D.W.; Ghandehari, H. Nanoparticle uptake: The phagocyte problem. Nano Today 2015, 10, 487–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stylianopoulos, T.; Munn, L.L.; Jain, R.K. Reengineering the Physical Microenvironment of Tumors to Improve Drug Delivery and Efficacy: From Mathematical Modeling to Bench to Bedside. Trends Cancer 2018, 4, 292–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitehead, K.A.; Langer, R.; Anderson, D.G. Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 2009, 8, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.P.; Gleeson, P.A. Macropinocytosis: An endocytic pathway for internalising large gulps. Immunol. Cell Biol. 2011, 89, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Commisso, C.; Davidson, S.M.; Soydaner-Azeloglu, R.G.; Parker, S.J.; Kamphorst, J.J.; Hackett, S.; Grabocka, E.; Nofal, M.; Drebin, J.A.; Thompson, C.B.; et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 2013, 497, 633–637. [Google Scholar] [CrossRef] [Green Version]
- Veiga, E.; Cossart, P. The role of clathrin-dependent endocytosis in bacterial internalization. Trends Cell Biol. 2006, 16, 499–504. [Google Scholar] [CrossRef]
- Robert, S.; Kleine-Vehn, J.; Barbez, E.; Sauer, M.; Paciorek, T.; Baster, P.; Vanneste, S.; Zhang, J.; Simon, S.; Čovanová, M.; et al. ABP1 Mediates Auxin Inhibition of Clathrin-Dependent Endocytosis in Arabidopsis. Cell 2010, 143, 111–121. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Liu, K.; Scally, L.; Manaloto, E.; Gunes, S.; Ng, S.W.; Maher, M.; Tiwari, B.; Byrne, H.J.; Bourke, P.; et al. Cold Atmospheric Plasma Stimulates Clathrin-Dependent Endocytosis to Repair Oxidised Membrane and Enhance Uptake of Nanomaterial in Glioblastoma Multiforme Cells. Sci. Rep. 2020, 10, 6985. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.-y.; Tang, X.-d.; Chen, J.; Wu, H.-b.; Chen, W.-s.; Chen, L. Rifampicin induces clathrin-dependent endocytosis and ubiquitin–proteasome degradation of MRP2 via oxidative stress-activated PKC-ERK/JNK/p38 and PI3K signaling pathways in HepG2 cells. Acta Pharmacol. Sin. 2019, 41, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.K.; Kim, S.-W.; Park, J.-Y.; Kang, W.C.; Kang, Y.-J.; Khang, D. Suppression of human arthritis synovial fibroblasts inflammation using dexamethasone-carbon nanotubes via increasing caveolin-dependent endocytosis and recovering mitochondrial membrane potential. Int. J. Nanomed. 2017, 12, 5761–5779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puzik, K.; Tonnier, V.; Opper, I.; Eckert, A.; Zhou, L.; Kratzer, M.-C.; Noble, F.l.; Nienhaus, G.U.; Gradl, D. Lef1 regulates caveolin expression and caveolin dependent endocytosis, a process necessary for Wnt5a/Ror2 signaling during Xenopus gastrulation. Sci. Rep. 2019, 9, 15645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vocelle, D.; Chan, C.; Walton, S.P. Endocytosis Controls siRNA Efficiency: Implications for siRNA Delivery Vehicle Design and Cell-Specific Targeting. Nucleic Acid Ther. 2020, 30, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Thakur, C.; Nayak, P.; Mishra, V.; Sharma, M.; Saraogi, G.K. Treating blood cancer with nanotechnology: A paradigm shift. In Nano Drug Delivery Strategies for the Treatment of Cancers; Elsevier: Amsterdam, The Netherlands, 2021; pp. 225–243. [Google Scholar] [CrossRef]
- Yu, M.; Zheng, J. Clearance Pathways and Tumor Targeting of Imaging Nanoparticles. ACS Nano 2015, 9, 6655–6674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Szoka, F.C. Lipid-based Nanoparticles for Nucleic Acid Delivery. Pharm. Res. 2007, 24, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Rehman, Z.U.; Zuhorn, I.S.; Hoekstra, D. How cationic lipids transfer nucleic acids into cells and across cellular membranes: Recent advances. J. Control. Release 2013, 166, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behr, J.P.; Demeneix, B.; Loeffler, J.P.; Perez-Mutul, J. Efficient gene transfer into mammalian primary endocrine cells with lipopolyamine-coated DNA. Proc. Natl. Acad. Sci. USA 1989, 86, 6982–6986. [Google Scholar] [CrossRef] [Green Version]
- Kunitake, T.; Okahata, Y. A totally synthetic bilayer membrane. J. Am. Chem. Soc. 2002, 99, 3860–3861. [Google Scholar] [CrossRef]
- Zuhorn, I.S.; Hoekstra, D. On the Mechanism of Cationic Amphiphile-mediated Transfection. To Fuse or not to Fuse: Is that the Question? J. Membr. Biol. 2002, 189, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Feng, J.; Liu, Y.; Che, J.; Bai, G.; Dong, X.; Wu, F.; Jin, T. A Synthetic Carrier of Nucleic Acids Structured as a Neutral Phospholipid Envelope Tightly Assembled on Polyplex Surface. Adv. Healthc. Mater. 2020, 9, 1901705. [Google Scholar] [CrossRef] [PubMed]
- García-Pinel, B.; Porras-Alcalá, C.; Ortega-Rodríguez, A.; Sarabia, F.; Prados, J.; Melguizo, C.; López-Romero, J.M. Lipid-Based Nanoparticles: Application and Recent Advances in Cancer Treatment. Nanomaterials 2019, 9, 638. [Google Scholar] [CrossRef] [Green Version]
- Sutradhar, K.B.; Amin, M.L. Nanoemulsions: Increasing possibilities in drug delivery. Eur. J. Nanomed. 2013, 5, 97–110. [Google Scholar] [CrossRef]
- Moss, K.H.; Popova, P.; Hadrup, S.R.; Astakhova, K.; Taskova, M. Lipid Nanoparticles for Delivery of Therapeutic RNA Oligonucleotides. Mol. Pharm. 2019, 16, 2265–2277. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez Gomez, A.; Syed, S.; Marshall, K.; Hosseinidoust, Z. Liposomal Nanovesicles for Efficient Encapsulation of Staphylococcal Antibiotics. ACS Omega 2019, 4, 10866–10876. [Google Scholar] [CrossRef] [PubMed]
- Lechanteur, A.; Sanna, V.; Duchemin, A.; Evrard, B.; Mottet, D.; Piel, G. Cationic Liposomes Carrying siRNA: Impact of Lipid Composition on Physicochemical Properties, Cytotoxicity and Endosomal Escape. Nanomaterials 2018, 8, 270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, T.; Yamada, K.; Fujiwara, Y.; Sato, Y.; Harashima, H. Reducing the Cytotoxicity of Lipid Nanoparticles Associated with a Fusogenic Cationic Lipid in a Natural Killer Cell Line by Introducing a Polycation-Based siRNA Core. Mol. Pharm. 2018, 15, 2142–2150. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, M.J.; Alishetty, S.; Alameh, M.-G.; Said, H.; Wright, L.; Paige, M.; Soliman, O.; Weissman, D.; Cleveland, T.E.; Grishaev, A.; et al. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun. Biol. 2021, 4, 956. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, Y.; Zhao, Y.; Lung, D.C.; Ye, Z.; Song, W.; Liu, F.-F.; Cai, J.-P.; Wong, W.-M.; Yip, C.C.-Y.; et al. Intravenous Injection of Coronavirus Disease 2019 (COVID-19) mRNA Vaccine Can Induce Acute Myopericarditis in Mouse Model. Clin. Infect. Dis. 2021, 73, 2372–2373. [Google Scholar] [CrossRef] [PubMed]
- Puri, A.; Loomis, K.; Smith, B.; Lee, J.-H.; Yavlovich, A.; Heldman, E.; Blumenthal, R. Lipid-Based Nanoparticles as Pharmaceutical Drug Carriers: From Concepts to Clinic. Crit. Rev. Ther. Drug Carr. Syst. 2009, 26, 523–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daraee, H.; Etemadi, A.; Kouhi, M.; Alimirzalu, S.; Akbarzadeh, A. Application of liposomes in medicine and drug delivery. Artif. Cells Nanomed. Biotechnol. 2014, 44, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Yadav, D.; Sandeep, K.; Pandey, D.; Dutta, R.K. Liposomes for Drug Delivery. J. Biotechnol. Biomater. 2017, 7, 276. [Google Scholar] [CrossRef]
- Fritze, A.; Hens, F.; Kimpfler, A.; Schubert, R.; Peschka-Süss, R. Remote loading of doxorubicin into liposomes driven by a transmembrane phosphate gradient. Biochim. Biophys. Acta Biomembr. 2006, 1758, 1633–1640. [Google Scholar] [CrossRef] [Green Version]
- Bolotin, E.M.; Cohen, R.; Bar, L.K.; Emanuel, N.; Ninio, S.; Barenholz, Y.; Lasic, D.D. Ammonium Sulfate Gradients for Efficient and Stable Remote Loading of Amphipathic Weak Bases into Liposomes and Ligandoliposomes. J. Liposome Res. 2008, 4, 455–479. [Google Scholar] [CrossRef]
- Alyane, M.; Barratt, G.; Lahouel, M. Remote loading of doxorubicin into liposomes by transmembrane pH gradient to reduce toxicity toward H9c2 cells. Saudi Pharm. J. 2016, 24, 165–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Cui, F.-D.; Choi, M.-K.; Lin, H.; Chung, S.-J.; Shim, C.-K.; Kim, D.-D. Liposome Formulation of Paclitaxel with Enhanced Solubility and Stability. Drug Deliv. 2008, 14, 301–308. [Google Scholar] [CrossRef]
- Lim, S.-J.; Hong, S.-S.; Choi, J.Y.; Kim, J.O.; Lee, M.-K.; Kim, S.H. Development of paclitaxel-loaded liposomal nanocarrier stabilized by triglyceride incorporation. Int. J. Nanomed. 2016, 11, 4465–4477. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Huang, L. Chapter Two—Lipid Nanoparticles for Gene Delivery. Adv. Genet. 2014, 88, 13–36. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, J.A.; Cullis, P.R.; van der Meel, R. Lipid Nanoparticles Enabling Gene Therapies: From Concepts to Clinical Utility. Nucleic Acid Ther. 2018, 28, 146–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulkarni, J.A.; Myhre, J.L.; Chen, S.; Tam, Y.Y.C.; Danescu, A.; Richman, J.M.; Cullis, P.R. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1377–1387. [Google Scholar] [CrossRef] [PubMed]
- Yung, B.C.; Li, J.; Zhang, M.; Cheng, X.; Li, H.; Yung, E.M.; Kang, C.; Cosby, L.E.; Liu, Y.; Teng, L.; et al. Lipid Nanoparticles Composed of Quaternary Amine–Tertiary Amine Cationic Lipid Combination (QTsome) for Therapeutic Delivery of AntimiR-21 for Lung Cancer. Mol. Pharm. 2016, 13, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.-C.; Wang, C.-C. Cationic solid lipid nanoparticles with primary and quaternary amines for release of saquinavir and biocompatibility with endothelia. Colloids Surf. B Biointerfaces 2013, 101, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Hsu, S.-H.; Zhou, C.; Wang, X.; Terp, M.C.; Wu, Y.; Teng, L.; Mao, Y.; Wang, F.; Xue, W.; et al. Lipid nanoparticles for hepatic delivery of small interfering RNA. Biomaterials 2012, 33, 5924–5934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, B.; Wang, X.; Zhou, C.; Teng, L.; Ren, W.; Yang, Z.; Shih, C.-H.; Wang, T.; Lee, R.J.; Tang, S.; et al. Insight into Mechanisms of Cellular Uptake of Lipid Nanoparticles and Intracellular Release of Small RNAs. Pharm. Res. 2014, 31, 2685–2695. [Google Scholar] [CrossRef] [PubMed]
- Varkouhi, A.K.; Scholte, M.; Storm, G.; Haisma, H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release 2011, 151, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Sabnis, S.; Kumarasinghe, E.S.; Salerno, T.; Mihai, C.; Ketova, T.; Senn, J.J.; Lynn, A.; Bulychev, A.; McFadyen, I.; Chan, J.; et al. A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates. Mol. Ther. 2018, 26, 1509–1519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulkarni, J.A.; Darjuan, M.M.; Mercer, J.E.; Chen, S.; van der Meel, R.; Thewalt, J.L.; Tam, Y.Y.C.; Cullis, P.R. On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and siRNA. ACS Nano 2018, 12, 4787–4795. [Google Scholar] [CrossRef] [Green Version]
- Kedmi, R.; Ben-Arie, N.; Peer, D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials 2010, 31, 6867–6875. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, A.; Yang, Z.; Wang, X.; Chang, L.; Chen, Z.; James Lee, L. Application of DODMA and Derivatives in Cationic Nanocarriers for Gene Delivery. Curr. Org. Chem. 2016, 20, 1813–1819. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.-B.; Dammer, E.B.; Ren, R.-J.; Wang, G. The endosomal-lysosomal system: From acidification and cargo sorting to neurodegeneration. Transl. Neurodegener. 2015, 4, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, R.J.; Wang, S.; Low, P.S. Measurement of endosome pH following folate receptor-mediated endocytosis. Biochim. Biophys. Acta Mol. Cell Res. 1996, 1312, 237–242. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Wang, X.-L.; Lu, Z.-R. New Amphiphilic Carriers Forming pH-Sensitive Nanoparticles for Nucleic Acid Delivery. Langmuir 2010, 26, 13874–13882. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Eral, H.B.; Hatton, T.A.; Doyle, P.S. Nanoemulsions: Formation, properties and applications. Soft Matter 2016, 12, 2826–2841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaiswal, M.; Dudhe, R.; Sharma, P.K. Nanoemulsion: An advanced mode of drug delivery system. 3 Biotech 2014, 5, 123–127. [Google Scholar] [CrossRef] [Green Version]
- Desai, J.; Thakkar, H. Enhanced oral bioavailability and brain uptake of Darunavir using lipid nanoemulsion formulation. Colloids Surf. B Biointerfaces 2019, 175, 143–149. [Google Scholar] [CrossRef]
- Anuchapreeda, S.; Fukumori, Y.; Okonogi, S.; Ichikawa, H. Preparation of Lipid Nanoemulsions Incorporating Curcumin for Cancer Therapy. J. Nanotechnol. 2012, 2012, 270383. [Google Scholar] [CrossRef] [Green Version]
- Singh, Y.; Meher, J.G.; Raval, K.; Khan, F.A.; Chaurasia, M.; Jain, N.K.; Chourasia, M.K. Nanoemulsion: Concepts, development and applications in drug delivery. J. Control. Release 2017, 252, 28–49. [Google Scholar] [CrossRef]
- Chen, L.; Chen, B.; Deng, L.; Gao, B.; Zhang, Y.; Wu, C.; Yu, N.; Zhou, Q.; Yao, J.; Chen, J. An optimized two-vial formulation lipid nanoemulsion of paclitaxel for targeted delivery to tumor. Int. J. Pharm. 2017, 534, 308–315. [Google Scholar] [CrossRef]
- Zhang, X.; Gan, Y.; Gan, L.; Nie, S.; Pan, W. PEGylated nanostructured lipid carriers loaded with 10-hydroxycamptothecin: An efficient carrier with enhanced anti-tumour effects against lung cancer. J. Pharm. Pharmacol. 2008, 60, 1077–1087. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.-P.; He, S.-N.; Li, Y.-L.; Feng, D.-L.; Lu, X.-Y.; Du, Y.-Z.; Yu, H.-Y.; Hu, F.-Q.; Yuan, H. Preparation and characteristics of lipid nanoemulsion formulations loaded with doxorubicin. Int. J. Nanomed. 2013, 8, 3141–3150. [Google Scholar] [CrossRef] [Green Version]
- Xue, H.; Guo, P.; Wen, W.-C.; Wong, H. Lipid-Based Nanocarriers for RNA Delivery. Curr. Pharm. Des. 2015, 21, 3140–3147. [Google Scholar] [CrossRef]
- Del Pozo-Rodríguez, A.; Solinís, M.Á.; Rodríguez-Gascón, A. Applications of lipid nanoparticles in gene therapy. Eur. J. Pharm. Biopharm. 2016, 109, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, C.; Sánchez-Hernández, N.; García-Montoya, E.; Pérez-Lozano, P.; Suñé-Negre, J.M.; Ticó, J.R.; Suñé, C.; Miñarro, M. DNA delivery via cationic solid lipid nanoparticles (SLNs). Eur. J. Pharm. Sci. 2013, 49, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Pink, D.L.; Loruthai, O.; Ziolek, R.M.; Wasutrasawat, P.; Terry, A.E.; Lawrence, M.J.; Lorenz, C.D. On the Structure of Solid Lipid Nanoparticles. Small 2019, 15, 1903156. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.M.; Mata, J.P.; Bryant, G.; Campo, L.; Ife, A.; Karpe, A.V.; Jadhav, S.R.; Eldridge, D.S.; Palombo, E.A.; Harding, I.H. Structure Analysis of Solid Lipid Nanoparticles for Drug Delivery: A Combined USANS/SANS Study. Part. Part. Syst. Charact. 2018, 36, 1800359. [Google Scholar] [CrossRef] [Green Version]
- Scioli Montoto, S.; Muraca, G.; Ruiz, M.E. Solid Lipid Nanoparticles for Drug Delivery: Pharmacological and Biopharmaceutical Aspects. Front. Mol. Biosci. 2020, 7, 587997. [Google Scholar] [CrossRef]
- Naseri, N.; Valizadeh, H.; Zakeri-Milani, P. Solid Lipid Nanoparticles and Nanostructured Lipid Carriers: Structure, Preparation and Application. Adv. Pharm. Bull. 2015, 5, 305–313. [Google Scholar] [CrossRef] [Green Version]
- Hu, F.-Q.; Jiang, S.-P.; Du, Y.-Z.; Yuan, H.; Ye, Y.-Q.; Zeng, S. Preparation and characteristics of monostearin nanostructured lipid carriers. Int. J. Pharm. 2006, 314, 83–89. [Google Scholar] [CrossRef]
- Müller, R.H.; Radtke, M.; Wissing, S.A. Solid lipid nanoparticles (SLN) and nanostructured lipid carriers (NLC) in cosmetic and dermatological preparations. Adv. Drug Deliv. Rev. 2002, 54, S131–S155. [Google Scholar] [CrossRef]
- Beloqui, A.; Solinís, M.Á.; Rodríguez-Gascón, A.; Almeida, A.J.; Préat, V. Nanostructured lipid carriers: Promising drug delivery systems for future clinics. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 143–161. [Google Scholar] [CrossRef] [PubMed]
- Muller, R.; Petersen, R.; Hommoss, A.; Pardeike, J. Nanostructured lipid carriers (NLC) in cosmetic dermal products. Adv. Drug Deliv. Rev. 2007, 59, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ma, Y.; Zhang, J.; Yuan, Y.; Wang, J. Exosomes: A Novel Therapeutic Agent for Cartilage and Bone Tissue Regeneration. Dose Response 2019, 17, 1559325819892702. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Dong, S.; Li, X.; Kim, B.Y.S.; Yang, Z.; Jiang, W. Extracellular Vesicles: An Emerging Nanoplatform for Cancer Therapy. Front. Oncol. 2020, 10, 606906. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yang, Z.; Huntoon, K.; Jiang, W.; Kim, B.Y.S. Advanced Immunotherapy Approaches for Glioblastoma. Adv. Ther. 2021, 4, 2100046. [Google Scholar] [CrossRef]
- Yang, X.; Ma, Y.; Xie, H.; Dong, S.; Rao, G.; Yang, Z.; Zhang, J.; Wu, Q. Extracellular Vesicles in the Treatment of Parkinson’s Disease: A Review. Curr. Med. Chem. 2021, 28, 6375–6394. [Google Scholar] [CrossRef]
- Yang, Z.; Ma, Y.; Zhao, H.; Yuan, Y.; Kim, B.Y.S. Nanotechnology platforms for cancer immunotherapy. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1590. [Google Scholar] [CrossRef]
- He, X.; Ma, Y.; Xie, H.; Rao, G.; Yang, Z.; Zhang, J.; Feng, Z. Biomimetic Nanostructure Platform for Cancer Diagnosis Based on Tumor Biomarkers. Front. Bioeng. Biotechnol. 2021, 9, 687664. [Google Scholar] [CrossRef]
- Ma, C.; Jiang, F.; Ma, Y.; Wang, J.; Li, H.; Zhang, J. Isolation and Detection Technologies of Extracellular Vesicles and Application on Cancer Diagnostic. Dose Response 2019, 17, 1559325819891004. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, N.; Whiteside, T.L.; Reichert, T.E. Challenges in Exosome Isolation and Analysis in Health and Disease. Int. J. Mol. Sci. 2019, 20, 4684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Shi, J.; Xie, J.; Wang, Y.; Sun, J.; Liu, T.; Zhao, Y.; Zhao, X.; Wang, X.; Ma, Y.; et al. Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nat. Biomed. Eng. 2020, 4, 69–83. [Google Scholar] [CrossRef]
- Mikamori, M.; Yamada, D.; Eguchi, H.; Hasegawa, S.; Kishimoto, T.; Tomimaru, Y.; Asaoka, T.; Noda, T.; Wada, H.; Kawamoto, K.; et al. MicroRNA-155 Controls Exosome Synthesis and Promotes Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma. Sci. Rep. 2017, 7, 42339. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, X.; Zhang, T.; Wang, C.; Huang, Z.; Luo, X.; Deng, Y. A review on phospholipids and their main applications in drug delivery systems. Asian J. Pharm. Sci. 2015, 10, 81–98. [Google Scholar] [CrossRef]
- Mudgil, P.; Borchman, D.; Yappert, M.C.; Duran, D.; Cox, G.W.; Smith, R.J.; Bhola, R.; Dennis, G.R.; Whitehall, J.S. Lipid order, saturation and surface property relationships: A study of human meibum saturation. Exp. Eye Res. 2013, 116, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Jendrasiak, G.L.; Mendible, J.C. The effect of the phase transition on the hydration and electrical conductivity of phospholipids. Biochim. Biophys. Acta Lipids Lipid Metab. 1976, 424, 133–148. [Google Scholar] [CrossRef]
- Stanton, M.G.; Colletti, S.L. Medicinal Chemistry of siRNA Delivery. J. Med. Chem. 2010, 53, 7887–7901. [Google Scholar] [CrossRef] [PubMed]
- Semple, S.C.; Akinc, A.; Chen, J.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.Y.; Stebbing, D.; Crosley, E.J.; Yaworski, E.; et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.R.; Weston, N.; Coombes, A.G.A.; Fitzgerald, M.; Perrie, Y. Liposome formulation of poorly water soluble drugs: Optimisation of drug loading and ESEM analysis of stability. Int. J. Pharm. 2004, 285, 23–34. [Google Scholar] [CrossRef]
- Funakoshi, Y.; Iwao, Y.; Noguchi, S.; Itai, S. Effect of Alkyl Chain Length and Unsaturation of the Phospholipid on the Physicochemical Properties of Lipid Nanoparticles. Chem. Pharm. Bull. 2015, 63, 731–736. [Google Scholar] [CrossRef] [Green Version]
- Hajj, K.A.; Melamed, J.R.; Chaudhary, N.; Lamson, N.G.; Ball, R.L.; Yerneni, S.S.; Whitehead, K.A. A Potent Branched-Tail Lipid Nanoparticle Enables Multiplexed mRNA Delivery and Gene Editing In Vivo. Nano Lett. 2020, 20, 5167–5175. [Google Scholar] [CrossRef] [PubMed]
- Kraft, J.C.; Freeling, J.P.; Wang, Z.; Ho, R.J.Y. Emerging Research and Clinical Development Trends of Liposome and Lipid Nanoparticle Drug Delivery Systems. J. Pharm. Sci. 2014, 103, 29–52. [Google Scholar] [CrossRef] [Green Version]
- Wasungu, L.; Hoekstra, D. Cationic lipids, lipoplexes and intracellular delivery of genes. J. Control. Release 2006, 116, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.-L.; Chen, H.-L.; Liou, W.; Lin, H.-K.; Liu, W.-L. Mesomorphic Complexes of DNA with the Mixtures of a Cationic Surfactant and a Neutral Lipid. Langmuir 2005, 21, 9426–9431. [Google Scholar] [CrossRef] [PubMed]
- Hayes, M.E.; Drummond, D.C.; Kirpotin, D.B.; Zheng, W.W.; Noble, C.O.; Park, J.W.; Marks, J.D.; Benz, C.C.; Hong, K. Genospheres: Self-assembling nucleic acid-lipid nanoparticles suitable for targeted gene delivery. Gene Ther. 2005, 13, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Colombani, T.; Peuziat, P.; Dallet, L.; Haudebourg, T.; Mével, M.; Berchel, M.; Lambert, O.; Habrant, D.; Pitard, B. Self-assembling complexes between binary mixtures of lipids with different linkers and nucleic acids promote universal mRNA, DNA and siRNA delivery. J. Control. Release 2017, 249, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, M.; Nawaz, M.; Papadimitriou, A.; Angerfors, A.; Camponeschi, A.; Na, M.; Hölttä, M.; Skantze, P.; Johansson, S.; Sundqvist, M.; et al. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 2019, 10, 4333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felgner, P.L.; Gadek, T.R.; Holm, M.; Roman, R.; Chan, H.W.; Wenz, M.; Northrop, J.P.; Ringold, G.M.; Danielsen, M. Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 1987, 84, 7413–7417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Templeton, N.S.; Lasic, D.D.; Frederik, P.M.; Strey, H.H.; Roberts, D.D.; Pavlakis, G.N. Improved DNA: Liposome complexes for increased systemic delivery and gene expression. Nat. Biotechnol. 1997, 15, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Pirollo, K.F.; Zon, G.; Rait, A.; Zhou, Q.; Yu, W.; Hogrefe, R.; Chang, E.H. Tumor-Targeting Nanoimmunoliposome Complex for Short Interfering RNA Delivery. Hum. Gene Ther. 2006, 17, 117–124. [Google Scholar] [CrossRef] [Green Version]
- Ciani, L.; Ristori, S.; Salvati, A.; Calamai, L.; Martini, G. DOTAP/DOPE and DC-Chol/DOPE lipoplexes for gene delivery: Zeta potential measurements and electron spin resonance spectra. Biochim. Biophys. Acta Biomembr. 2004, 1664, 70–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Z.; Li, J.; He, F.; Wilson, A.; Pitt, B.; Li, S. Cationic lipids enhance siRNA-mediated interferon response in mice. Biochem. Biophys. Res. Commun. 2005, 330, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.-H.; Yu, B.; Wang, X.; Lu, Y.; Schmidt, C.R.; Lee, R.J.; Lee, L.J.; Jacob, S.T.; Ghoshal, K. Cationic lipid nanoparticles for therapeutic delivery of siRNA and miRNA to murine liver tumor. Nanomed. Nanotechnol. Biol. Med. 2013, 9, 1169–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flieger, J.; Flieger, M. Ionic Liquids Toxicity—Benefits and Threats. Int. J. Mol. Sci. 2020, 21, 6267. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, Z.; Huang, Z.; Tang, X.; Zhang, X. Antimicrobial cationic polymers: From structural design to functional control. Polym. J. 2017, 50, 33–44. [Google Scholar] [CrossRef]
- Wang, D.; Galla, H.-J.; Drücker, P. Membrane interactions of ionic liquids and imidazolium salts. Biophys. Rev. 2018, 10, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Richter, C.; Rühling, A.; Drücker, P.; Siegmund, D.; Metzler-Nolte, N.; Glorius, F.; Galla, H.-J. A Remarkably Simple Class of Imidazolium-Based Lipids and Their Biological Properties. Chem. Eur. J. 2015, 21, 15123–15126. [Google Scholar] [CrossRef]
- Ilies, M.A.; Seitz, W.A.; Ghiviriga, I.; Johnson, B.H.; Miller, A.; Thompson, E.B.; Balaban, A.T. Pyridinium Cationic Lipids in Gene Delivery: A Structure-Activity Correlation Study. J. Med. Chem. 2004, 47, 3744–3754. [Google Scholar] [CrossRef]
- Guénin, E.; Hervé, A.-C.; Floch, V.; Loisel, S.; Yaouanc, J.-J.; Clément, J.-C.; Férec, C.; des Abbayes, H. Cationic Phosphonolipids Containing Quaternary Phosphonium and Arsonium Groups for DNA Transfection with Good Efficiency and Low Cellular Toxicity. Angew. Chem. Int. Ed. 2000, 39, 629–631. [Google Scholar] [CrossRef]
- Vigneron, J.P.; Oudrhiri, N.; Fauquet, M.; Vergely, L.; Bradley, J.C.; Basseville, M.; Lehn, P.; Lehn, J.M. Guanidinium-cholesterol cationic lipids: Efficient vectors for the transfection of eukaryotic cells. Proc. Natl. Acad. Sci. USA 1996, 93, 9682–9686. [Google Scholar] [CrossRef] [Green Version]
- Meka, R.R.; Godeshala, S.; Marepally, S.; Thorat, K.; Reddy Rachamalla, H.K.; Dhayani, A.; Hiwale, A.; Banerjee, R.; Chaudhuri, A.; Vemula, P.K. Asymmetric cationic lipid based non-viral vectors for an efficient nucleic acid delivery. RSC Adv. 2016, 6, 77841–77848. [Google Scholar] [CrossRef]
- Khazanov, E.; Simberg, D.; Barenholz, Y. Lipoplexes prepared from cationic liposomes and mammalian DNA induce CpG-independent, direct cytotoxic effects in cell cultures and in mice. J. Gene Med. 2006, 8, 998–1007. [Google Scholar] [CrossRef] [PubMed]
- Dass, C.R. Cytotoxicity issues pertinent to lipoplex-mediated gene therapy in-vivo. J. Pharm. Pharmacol. 2002, 54, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.L.; Nguyen, J.; Tiffany, M.R.; Szoka, F.C. Synthesis, Characterization, and Evaluation of Ionizable Lysine-Based Lipids for siRNA Delivery. Bioconjug. Chem. 2012, 24, 36–43. [Google Scholar] [CrossRef] [Green Version]
- Gilleron, J.; Querbes, W.; Zeigerer, A.; Borodovsky, A.; Marsico, G.; Schubert, U.; Manygoats, K.; Seifert, S.; Andree, C.; Stöter, M.; et al. Image-based analysis of lipid nanoparticle–mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013, 31, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Heyes, J.; Palmer, L.; Bremner, K.; MacLachlan, I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release 2005, 107, 276–287. [Google Scholar] [CrossRef]
- Koynova, R.; Tenchov, B. Cationic Lipids: Molecular Structure/Transfection Activity Relationships and Interactions with Biomembranes. In Nucleic Acid Transfection; Springer: Berlin/Heidelberg, Germany, 2010; pp. 51–93. [Google Scholar] [CrossRef]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef]
- Hajj, K.A.; Ball, R.L.; Deluty, S.B.; Singh, S.R.; Strelkova, D.; Knapp, C.M.; Whitehead, K.A. Branched-Tail Lipid Nanoparticles Potently Deliver mRNA In Vivo due to Enhanced Ionization at Endosomal pH. Small 2019, 15, 1805097. [Google Scholar] [CrossRef]
- Miao, L.; Lin, J.; Huang, Y.; Li, L.; Delcassian, D.; Ge, Y.; Shi, Y.; Anderson, D.G. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 2020, 11, 2424. [Google Scholar] [CrossRef]
- Ramezanpour, M.; Schmidt, M.L.; Bodnariuc, I.; Kulkarni, J.A.; Leung, S.S.W.; Cullis, P.R.; Thewalt, J.L.; Tieleman, D.P. Ionizable amino lipid interactions with POPC: Implications for lipid nanoparticle function. Nanoscale 2019, 11, 14141–14146. [Google Scholar] [CrossRef] [Green Version]
- Jayaraman, M.; Ansell, S.M.; Mui, B.L.; Tam, Y.K.; Chen, J.; Du, X.; Butler, D.; Eltepu, L.; Matsuda, S.; Narayanannair, J.K.; et al. Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo. Angew. Chem. 2012, 124, 8657–8661. [Google Scholar] [CrossRef] [Green Version]
- Zukancic, D.; Suys, E.J.A.; Pilkington, E.H.; Algarni, A.; Al-Wassiti, H.; Truong, N.P. The Importance of Poly(ethylene glycol) and Lipid Structure in Targeted Gene Delivery to Lymph Nodes by Lipid Nanoparticles. Pharmaceutics 2020, 12, 1068. [Google Scholar] [CrossRef] [PubMed]
- Yanez Arteta, M.; Kjellman, T.; Bartesaghi, S.; Wallin, S.; Wu, X.; Kvist, A.J.; Dabkowska, A.; Székely, N.; Radulescu, A.; Bergenholtz, J.; et al. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. USA 2018, 115, E3351–E3360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viger-Gravel, J.; Schantz, A.; Pinon, A.C.; Rossini, A.J.; Schantz, S.; Emsley, L. Structure of Lipid Nanoparticles Containing siRNA or mRNA by Dynamic Nuclear Polarization-Enhanced NMR Spectroscopy. J. Phys. Chem. B 2018, 122, 2073–2081. [Google Scholar] [CrossRef]
- Brown, M.S.; Goldstein, J.L. Receptor-mediated endocytosis: Insights from the lipoprotein receptor system. Proc. Natl. Acad. Sci. USA 1979, 76, 3330–3337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.R.; Gil, S.; Andrieux, K.; Nicolas, V.; Appel, M.; Chacun, H.; Desmaële, D.; Taran, F.; Georgin, D.; Couvreur, P. Low-density lipoprotein receptor-mediated endocytosis of PEGylated nanoparticles in rat brain endothelial cells. Cell. Mol. Life Sci. 2007, 64, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Zhigaltsev, I.V.; Maurer, N.; Wong, K.F.; Cullis, P.R. Triggered release of doxorubicin following mixing of cationic and anionic liposomes. Biochim. Biophys. Acta Biomembr. 2002, 1565, 129–135. [Google Scholar] [CrossRef] [Green Version]
- Briuglia, M.-L.; Rotella, C.; McFarlane, A.; Lamprou, D.A. Influence of cholesterol on liposome stability and on in vitro drug release. Drug Deliv. Transl. Res. 2015, 5, 231–242. [Google Scholar] [CrossRef] [Green Version]
- Paunovska, K.; Gil, C.J.; Lokugamage, M.P.; Sago, C.D.; Sato, M.; Lando, G.N.; Gamboa Castro, M.; Bryksin, A.V.; Dahlman, J.E. Analyzing 2000 in Vivo Drug Delivery Data Points Reveals Cholesterol Structure Impacts Nanoparticle Delivery. ACS Nano 2018, 12, 8341–8349. [Google Scholar] [CrossRef]
- Kuai, R.; Li, D.; Chen, Y.E.; Moon, J.J.; Schwendeman, A. High-Density Lipoproteins: Nature’s Multifunctional Nanoparticles. ACS Nano 2016, 10, 3015–3041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jovanović, A.A.; Balanč, B.D.; Ota, A.; Ahlin Grabnar, P.; Djordjević, V.B.; Šavikin, K.P.; Bugarski, B.M.; Nedović, V.A.; Poklar Ulrih, N. Comparative Effects of Cholesterol and β-Sitosterol on the Liposome Membrane Characteristics. Eur. J. Lipid Sci. Technol. 2018, 120, 1800039. [Google Scholar] [CrossRef]
- Patel, S.; Ashwanikumar, N.; Robinson, E.; Xia, Y.; Mihai, C.; Griffith, J.P.; Hou, S.; Esposito, A.A.; Ketova, T.; Welsher, K.; et al. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 2020, 11, 983. [Google Scholar] [CrossRef] [Green Version]
- Herrera, M.; Kim, J.; Eygeris, Y.; Jozic, A.; Sahay, G. Illuminating endosomal escape of polymorphic lipid nanoparticles that boost mRNA delivery. Biomater. Sci. 2021, 9, 4289–4300. [Google Scholar] [CrossRef] [PubMed]
- Eygeris, Y.; Patel, S.; Jozic, A.; Sahay, G. Deconvoluting Lipid Nanoparticle Structure for Messenger RNA Delivery. Nano Lett. 2020, 20, 4543–4549. [Google Scholar] [CrossRef] [PubMed]
- Bernsdorff, C.; Winter, R. Differential Properties of the Sterols Cholesterol, Ergosterol, β-Sitosterol, trans-7-Dehydrocholesterol, Stigmasterol and Lanosterol on DPPC Bilayer Order. J. Phys. Chem. B 2003, 107, 10658–10664. [Google Scholar] [CrossRef]
- Gref, R.; Lück, M.; Quellec, P.; Marchand, M.; Dellacherie, E.; Harnisch, S.; Blunk, T.; Müller, R.H. ‘Stealth’ corona-core nanoparticles surface modified by polyethylene glycol (PEG): Influences of the corona (PEG chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption. Colloids Surf. B Biointerfaces 2000, 18, 301–313. [Google Scholar] [CrossRef]
- Morla-Folch, J.; Guerrini, L.; Pazos-Perez, N.; Arenal, R.; Alvarez-Puebla, R.A. Synthesis and Optical Properties of Homogeneous Nanoshurikens. ACS Photonics 2014, 1, 1237–1244. [Google Scholar] [CrossRef]
- Guerrini, L.; Alvarez-Puebla, R.; Pazos-Perez, N. Surface Modifications of Nanoparticles for Stability in Biological Fluids. Materials 2018, 11, 1154. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, J.A.; Witzigmann, D.; Leung, J.; Tam, Y.Y.C.; Cullis, P.R. On the role of helper lipids in lipid nanoparticle formulations of siRNA. Nanoscale 2019, 11, 21733–21739. [Google Scholar] [CrossRef]
- Suk, J.S.; Xu, Q.; Kim, N.; Hanes, J.; Ensign, L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016, 99, 28–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Vlerken, L.E.; Vyas, T.K.; Amiji, M.M. Poly(ethylene glycol)-modified Nanocarriers for Tumor-targeted and Intracellular Delivery. Pharm. Res. 2007, 24, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Suzuki, Y.; Hihara, T.; Kubara, K.; Kondo, K.; Hyodo, K.; Yamazaki, K.; Ishida, T.; Ishihara, H. PEG shedding-rate-dependent blood clearance of PEGylated lipid nanoparticles in mice: Faster PEG shedding attenuates anti-PEG IgM production. Int. J. Pharm. 2020, 588, 119792. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, H.; Akita, H.; Ishida, E.; Hashimoto, K.; Kobayashi, H.; Aoki, T.; Yasuda, J.; Obata, K.; Kikuchi, H.; Ishida, T.; et al. Tumor targeting of doxorubicin by anti-MT1-MMP antibody-modified PEG liposomes. Int. J. Pharm. 2007, 342, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Szebeni, J.; Baranyi, L.; Savay, S.; Milosevits, J.; Bunger, R.; Laverman, P.; Metselaar, J.M.; Storm, G.; Chanan-Khan, A.; Liebes, L.; et al. Role of Complement Activation in Hypersensitivity Reactions to Doxil and Hynic Peg Liposomes: Experimental and Clinical Studies. J. Liposome Res. 2002, 12, 165–172. [Google Scholar] [CrossRef]
- Huntington, J.; Pachauri, M.; Ali, H.; Giacca, M. RNA interference therapeutics for cardiac regeneration. Curr. Opin. Genet. Dev. 2021, 70, 48–53. [Google Scholar] [CrossRef]
- Smola, A.; Samadzadeh, S.; Müller, L.; Adams, O.; Homey, B.; Albrecht, P.; Meller, S. Omalizumab prevents anaphylactoid reactions to mRNA COVID-19 vaccine. J. Eur. Acad. Dermatol. Venereol. 2021, 35, e743–e745. [Google Scholar] [CrossRef]
- Hatziantoniou, S.; Maltezou, H.C.; Tsakris, A.; Poland, G.A.; Anastassopoulou, C. Anaphylactic reactions to mRNA COVID-19 vaccines: A call for further study. Vaccine 2021, 39, 2605–2607. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Sze, R.; Zhang, M. Folic acid-PEG conjugated superparamagnetic nanoparticles for targeted cellular uptake and detection by MRI. J. Biomed. Mater. Res. Part A 2006, 78A, 550–557. [Google Scholar] [CrossRef]
- Sega, E.I.; Low, P.S. Tumor detection using folate receptor-targeted imaging agents. Cancer Metastasis Rev. 2008, 27, 655–664. [Google Scholar] [CrossRef]
- Weitman, S.D.; Lark, R.H.; Coney, L.R.; Fort, D.W.; Frasca, V.; Zurawski, V.R., Jr.; Kamen, B.A. Distribution of the folate receptor GP38 in normal and malignant cell lines and tissues. Cancer Res. 1992, 52, 3396–3401. [Google Scholar]
- Sudimack, J.; Lee, R.J. Targeted drug delivery via the folate receptor. Adv. Drug Deliv. Rev. 2000, 41, 147–162. [Google Scholar] [CrossRef]
- Nag, M.; Gajbhiye, V.; Kesharwani, P.; Jain, N.K. Transferrin functionalized chitosan-PEG nanoparticles for targeted delivery of paclitaxel to cancer cells. Colloids Surf. B Biointerfaces 2016, 148, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Sun, Y.; Cai, C.; Fan, R.; Guo, R.; Xie, D. Targeted delivery of DOX by transferrin conjugated DSPE-PEG nanoparticles in leukemia therapy. Int. J. Polym. Mater. Polym. Biomater. 2019, 70, 27–36. [Google Scholar] [CrossRef]
- Hoang Thi, T.T.; Pilkington, E.H.; Nguyen, D.H.; Lee, J.S.; Park, K.D.; Truong, N.P. The Importance of Poly(ethylene glycol) Alternatives for Overcoming PEG Immunogenicity in Drug Delivery and Bioconjugation. Polymers 2020, 12, 298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kircheis, R.; Wightman, L.; Wagner, E. Design and gene delivery activity of modified polyethylenimines. Adv. Drug Deliv. Rev. 2001, 53, 341–358. [Google Scholar] [CrossRef]
- Song, X.; Huang, Q.; Jin, B.; Peng, R. Fabrication and characterization of CL-20/PEI/GO composites with enhanced thermal stability and desensitization via electrostatic self-assembly. Appl. Surf. Sci. 2021, 558, 149933. [Google Scholar] [CrossRef]
- Benjaminsen, R.V.; Mattebjerg, M.A.; Henriksen, J.R.; Moghimi, S.M.; Andresen, T.L. The Possible “Proton Sponge ” Effect of Polyethylenimine (PEI) Does Not Include Change in Lysosomal pH. Mol. Ther. 2013, 21, 149–157. [Google Scholar] [CrossRef] [Green Version]
- Nel, A.E.; Mädler, L.; Velegol, D.; Xia, T.; Hoek, E.M.V.; Somasundaran, P.; Klaessig, F.; Castranova, V.; Thompson, M. Understanding biophysicochemical interactions at the nano–bio interface. Nat. Mater. 2009, 8, 543–557. [Google Scholar] [CrossRef]
- Moghimi, S.M.; Symonds, P.; Murray, J.C.; Hunter, A.C.; Debska, G.; Szewczyk, A. A two-stage poly(ethylenimine)-mediated cytotoxicity: Implications for gene transfer/therapy. Mol. Ther. 2005, 11, 990–995. [Google Scholar] [CrossRef]
- Brunot, C.; Ponsonnet, L.; Lagneau, C.; Farge, P.; Picart, C.; Grosgogeat, B. Cytotoxicity of polyethyleneimine (PEI), precursor base layer of polyelectrolyte multilayer films. Biomaterials 2007, 28, 632–640. [Google Scholar] [CrossRef]
- Neu, M.; Germershaus, O.; Behe, M.; Kissel, T. Bioreversibly crosslinked polyplexes of PEI and high molecular weight PEG show extended circulation times in vivo. J. Control. Release 2007, 124, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Neu, M.; Germershaus, O.; Merkel, O.; Sitterberg, J.; Bakowsky, U.; Kissel, T. Influence of Polyethylene Glycol Chain Length on the Physicochemical and Biological Properties of Poly(ethylene imine)-graft-Poly(ethylene glycol) Block Copolymer/SiRNA Polyplexes. Bioconjug. Chem. 2006, 17, 1209–1218. [Google Scholar] [CrossRef] [PubMed]
- Merkel, O.M.; Beyerle, A.; Librizzi, D.; Pfestroff, A.; Behr, T.M.; Sproat, B.; Barth, P.J.; Kissel, T. Nonviral siRNA Delivery to the Lung: Investigation of PEG−PEI Polyplexes and Their In Vivo Performance. Mol. Pharm. 2009, 6, 1246–1260. [Google Scholar] [CrossRef] [PubMed]
- Gasset, M.; Hu, J.; Zhu, M.; Liu, K.; Fan, H.; Zhao, W.; Mao, Y.; Zhang, Y. A Biodegradable Polyethylenimine-Based Vector Modified by Trifunctional Peptide R18 for Enhancing Gene Transfection Efficiency In Vivo. PLoS ONE 2016, 11, e0166673. [Google Scholar] [CrossRef]
- Zheng, B.I.N.; Yang, S.; Tian, Q.; Xie, Y.I.N.; Zhang, S.; Lee, R.J. Delivery of Antisense Oligonucleotide LOR-2501 Using Transferrin-conjugated Polyethylenimine-based Lipid Nanoparticle. Anticancer Res. 2019, 39, 1785–1793. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, G.; He, B.; Li, L.; Li, C.; Lai, Y.; Xu, X.; Gu, Z. Cationic lipid-coated PEI/DNA polyplexes with improved efficiency and reduced cytotoxicity for gene delivery into mesenchymal stem cells. Int. J. Nanomed. 2012, 2012, 4637–4648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawant, R.R.; Sriraman, S.K.; Navarro, G.; Biswas, S.; Dalvi, R.A.; Torchilin, V.P. Polyethyleneimine-lipid conjugate-based pH-sensitive micellar carrier for gene delivery. Biomaterials 2012, 33, 3942–3951. [Google Scholar] [CrossRef] [Green Version]
- Wiseman, J.W.; Goddard, C.A.; McLelland, D.; Colledge, W.H. A comparison of linear and branched polyethylenimine (PEI) with DCChol/DOPE liposomes for gene delivery to epithelial cells in vitro and in vivo. Gene Ther. 2003, 10, 1654–1662. [Google Scholar] [CrossRef]
- Witzigmann, D.; Kulkarni, J.A.; Leung, J.; Chen, S.; Cullis, P.R.; van der Meel, R. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv. Drug Deliv. Rev. 2020, 159, 344–363. [Google Scholar] [CrossRef]
- Falzarano, M.S.; Passarelli, C.; Ferlini, A. Nanoparticle Delivery of Antisense Oligonucleotides and Their Application in the Exon Skipping Strategy for Duchenne Muscular Dystrophy. Nucleic Acid Ther. 2014, 24, 87–100. [Google Scholar] [CrossRef] [Green Version]
- Hair, P.; Cameron, F.; McKeage, K. Mipomersen Sodium: First Global Approval. Drugs 2013, 73, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.A. Cell and molecular biology of the assembly and secretion of apolipoprotein B-containing lipoproteins by the liver. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1999, 1440, 1–31. [Google Scholar] [CrossRef]
- Gelsinger, C.; Steinhagen-Thiessen, E.; Kassner, U. Therapeutic Potential of Mipomersen in the Management of Familial Hypercholesterolaemia. Drugs 2012, 72, 1445–1455. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.N.; Patel, N.J.; Bhowmik, T.; D’Souza, B.; Akalkotkar, A.; Etzlar, F.; Oettinger, C.W.; D’Souza, M. Enhanced bioavailability of orally administered antisense oligonucleotide to nuclear factor kappa B mRNA after microencapsulation with albumin. J. Drug Target. 2013, 21, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015, 87, 46–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, M.; Yokota, T. An Overview of Recent Advances and Clinical Applications of Exon Skipping and Splice Modulation for Muscular Dystrophy and Various Genetic Diseases. In Exon Skipping and Inclusion Therapies; Springer: Berlin/Heidelberg, Germany, 2018; pp. 31–55. [Google Scholar] [CrossRef]
- Dzierlega, K.; Yokota, T. Optimization of antisense-mediated exon skipping for Duchenne muscular dystrophy. Gene Ther. 2020, 27, 407–416. [Google Scholar] [CrossRef]
- Kole, R.; Leppert, B.J. Targeting mRNA splicing as a potential treatment for Duchenne muscular dystrophy. Discov. Med. 2012, 14, 59–69. [Google Scholar]
- Let’s talk about lipid nanoparticles. Nat. Rev. Mater. 2021, 6, 99. [CrossRef]
- Ruberg, F.L.; Berk, J.L. Transthyretin (TTR) Cardiac Amyloidosis. Circulation 2012, 126, 1286–1300. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Khan, S.; Rahman, S.; Singh, L.R. The Extracellular Protein, Transthyretin Is an Oxidative Stress Biomarker. Front. Physiol. 2019, 10, 5. [Google Scholar] [CrossRef]
- Waddington-Cruz, M.; Ackermann, E.J.; Polydefkis, M.; Heitner, S.B.; Dyck, P.J.; Barroso, F.A.; Wang, A.K.; Berk, J.L.; Dyck, P.J.B.; Monia, B.P.; et al. Hereditary transthyretin amyloidosis: Baseline characteristics of patients in the NEURO-TTR trial. Amyloid 2018, 25, 180–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J. Patisiran for the treatment of hereditary transthyretin-mediated amyloidosis. Expert Rev. Clin. Pharmacol. 2019, 12, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Balwani, M.; Sardh, E.; Ventura, P.; Peiró, P.A.; Rees, D.C.; Stölzel, U.; Bissell, D.M.; Bonkovsky, H.L.; Windyga, J.; Anderson, K.E.; et al. Phase 3 Trial of RNAi Therapeutic Givosiran for Acute Intermittent Porphyria. N. Engl. J. Med. 2020, 382, 2289–2301. [Google Scholar] [CrossRef]
- Ray, K.K.; Wright, R.S.; Kallend, D.; Koenig, W.; Leiter, L.A.; Raal, F.J.; Bisch, J.A.; Richardson, T.; Jaros, M.; Wijngaard, P.L.J.; et al. Two Phase 3 Trials of Inclisiran in Patients with Elevated LDL Cholesterol. N. Engl. J. Med. 2020, 382, 1507–1519. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J.; Keam, S.J. Lumasiran: First Approval. Drugs 2021, 81, 277–282. [Google Scholar] [CrossRef]
- Jaklevic, M.C. First Drug Approved for Rare Genetic Disorder Affecting Kidneys. JAMA 2021, 325, 214. [Google Scholar] [CrossRef] [PubMed]
- Springer, A.D.; Dowdy, S.F. GalNAc-siRNA Conjugates: Leading the Way for Delivery of RNAi Therapeutics. Nucleic Acid Ther. 2018, 28, 109–118. [Google Scholar] [CrossRef]
- Landen, C.N.; Chavez-Reyes, A.; Bucana, C.; Schmandt, R.; Deavers, M.T.; Lopez-Berestein, G.; Sood, A.K. TherapeuticEphA2Gene TargetingIn vivoUsing Neutral Liposomal Small Interfering RNA Delivery. Cancer Res. 2005, 65, 6910–6918. [Google Scholar] [CrossRef] [Green Version]
- Nabhan, J.F.; Wood, K.M.; Rao, V.P.; Morin, J.; Bhamidipaty, S.; LaBranche, T.P.; Gooch, R.L.; Bozal, F.; Bulawa, C.E.; Guild, B.C. Intrathecal delivery of frataxin mRNA encapsulated in lipid nanoparticles to dorsal root ganglia as a potential therapeutic for Friedreich’s ataxia. Sci. Rep. 2016, 6, 20019. [Google Scholar] [CrossRef] [Green Version]
- Truong, B.; Allegri, G.; Liu, X.-B.; Burke, K.E.; Zhu, X.; Cederbaum, S.D.; Häberle, J.; Martini, P.G.V.; Lipshutz, G.S. Lipid nanoparticle-targeted mRNA therapy as a treatment for the inherited metabolic liver disorder arginase deficiency. Proc. Natl. Acad. Sci. USA 2019, 116, 21150–21159. [Google Scholar] [CrossRef]
- Hauser, S.; Poenisch, M.; Schelling, Y.; Höflinger, P.; Schuster, S.; Teegler, A.; Betten, R.; Gustafsson, J.-Å.; Hübener-Schmid, J.; Schlake, T.; et al. mRNA as a Novel Treatment Strategy for Hereditary Spastic Paraplegia Type 5. Mol. Ther. Methods Clin. Dev. 2019, 15, 359–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stambas, J.; Magini, D.; Giovani, C.; Mangiavacchi, S.; Maccari, S.; Cecchi, R.; Ulmer, J.B.; De Gregorio, E.; Geall, A.J.; Brazzoli, M.; et al. Self-Amplifying mRNA Vaccines Expressing Multiple Conserved Influenza Antigens Confer Protection against Homologous and Heterosubtypic Viral Challenge. PLoS ONE 2016, 11, e0161193. [Google Scholar] [CrossRef]
- Hekele, A.; Bertholet, S.; Archer, J.; Gibson, D.G.; Palladino, G.; Brito, L.A.; Otten, G.R.; Brazzoli, M.; Buccato, S.; Bonci, A.; et al. Rapidly produced SAM® vaccine against H7N9 influenza is immunogenic in mice. Emerg. Microbes Infect. 2019, 2, 1–7. [Google Scholar] [CrossRef]
- Brazzoli, M.; Magini, D.; Bonci, A.; Buccato, S.; Giovani, C.; Kratzer, R.; Zurli, V.; Mangiavacchi, S.; Casini, D.; Brito, L.M.; et al. Induction of Broad-Based Immunity and Protective Efficacy by Self-amplifying mRNA Vaccines Encoding Influenza Virus Hemagglutinin. J. Virol. 2016, 90, 332–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindgren, G.; Ols, S.; Liang, F.; Thompson, E.A.; Lin, A.; Hellgren, F.; Bahl, K.; John, S.; Yuzhakov, O.; Hassett, K.J.; et al. Induction of Robust B Cell Responses after Influenza mRNA Vaccination Is Accompanied by Circulating Hemagglutinin-Specific ICOS+ PD-1+ CXCR3+ T Follicular Helper Cells. Front. Immunol. 2017, 8, 1539. [Google Scholar] [CrossRef] [Green Version]
- McCullough, K.C.; Bassi, I.; Milona, P.; Suter, R.; Thomann-Harwood, L.; Englezou, P.; Démoulins, T.; Ruggli, N. Self-replicating Replicon-RNA Delivery to Dendritic Cells by Chitosan-nanoparticles for Translation In Vitro and In Vivo. Mol. Ther. Nucleic Acids USA 2014, 3, e173. [Google Scholar] [CrossRef]
- Lutz, J.; Lazzaro, S.; Habbeddine, M.; Schmidt, K.E.; Baumhof, P.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; Hope, M.J.; Heidenreich, R.; et al. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. NPJ Vaccines 2017, 2, 29. [Google Scholar] [CrossRef] [Green Version]
- Aldrich, C.; Leroux–Roels, I.; Huang, K.B.; Bica, M.A.; Loeliger, E.; Schoenborn-Kellenberger, O.; Walz, L.; Leroux-Roels, G.; von Sonnenburg, F.; Oostvogels, L. Proof-of-concept of a low-dose unmodified mRNA-based rabies vaccine formulated with lipid nanoparticles in human volunteers: A phase 1 trial. Vaccine 2021, 39, 1310–1318. [Google Scholar] [CrossRef]
- Armbruster, N.; Jasny, E.; Petsch, B. Advances in RNA Vaccines for Preventive Indications: A Case Study of A Vaccine Against Rabies. Vaccines 2019, 7, 132. [Google Scholar] [CrossRef] [Green Version]
- Mehrjardi, M.Z. Is Zika Virus an Emerging TORCH Agent? An Invited Commentary. Virol. Res. Treat. 2017, 8, 1178122X17708993. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Pelc, R.S.; Muramatsu, H.; Andersen, H.; DeMaso, C.R.; Dowd, K.A.; Sutherland, L.L.; Scearce, R.M.; Parks, R.; et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 2017, 543, 248–251. [Google Scholar] [CrossRef]
- Richner, J.M.; Himansu, S.; Dowd, K.A.; Butler, S.L.; Salazar, V.; Fox, J.M.; Julander, J.G.; Tang, W.W.; Shresta, S.; Pierson, T.C.; et al. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell 2017, 168, 1114–1125.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erasmus, J.H.; Khandhar, A.P.; Guderian, J.; Granger, B.; Archer, J.; Archer, M.; Gage, E.; Fuerte-Stone, J.; Larson, E.; Lin, S.; et al. A Nanostructured Lipid Carrier for Delivery of a Replicating Viral RNA Provides Single, Low-Dose Protection against Zika. Mol. Ther. 2018, 26, 2507–2522. [Google Scholar] [CrossRef] [Green Version]
- Uhl, P.; Pantze, S.; Storck, P.; Parmentier, J.; Witzigmann, D.; Hofhaus, G.; Huwyler, J.; Mier, W.; Fricker, G. Oral delivery of vancomycin by tetraether lipid liposomes. Eur. J. Pharm. Sci. 2017, 108, 111–118. [Google Scholar] [CrossRef]
- Alhariri, M.; Majrashi, M.A.; Bahkali, A.H.; Almajed, F.S.; Azghani, A.O.; Khiyami, M.; Alyamani, E.J.; Aljohani, S.M.; Halwani, M.A. Efficacy of neutral and negatively charged liposome-loaded gentamicin on planktonic bacteria and biofilm communities. Int. J. Nanomed. 2017, 12, 6949–6961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carneiro, S.P.; Carvalho, K.V.; de Oliveira Aguiar Soares, R.D.; Carneiro, C.M.; de Andrade, M.H.G.; Duarte, R.S.; dos Santos, O.D.H. Functionalized rifampicin-loaded nanostructured lipid carriers enhance macrophages uptake and antimycobacterial activity. Colloids Surf. B Biointerfaces 2019, 175, 306–313. [Google Scholar] [CrossRef]
- Bogers, W.M.; Oostermeijer, H.; Mooij, P.; Koopman, G.; Verschoor, E.J.; Davis, D.; Ulmer, J.B.; Brito, L.A.; Cu, Y.; Banerjee, K.; et al. Potent Immune Responses in Rhesus Macaques Induced by Nonviral Delivery of a Self-amplifying RNA Vaccine Expressing HIV Type 1 Envelope with a Cationic Nanoemulsion. J. Infect. Dis. 2014, 211, 947–955. [Google Scholar] [CrossRef]
- Brito, L.A.; Kommareddy, S.; Maione, D.; Uematsu, Y.; Giovani, C.; Berlanda Scorza, F.; Otten, G.R.; Yu, D.; Mandl, C.W.; Mason, P.W.; et al. Self-Amplifying mRNA Vaccines. Adv. Genet. 2015, 89, 179–233. [Google Scholar] [CrossRef]
- Meyer, M.; Huang, E.; Yuzhakov, O.; Ramanathan, P.; Ciaramella, G.; Bukreyev, A. Modified mRNA-Based Vaccines Elicit Robust Immune Responses and Protect Guinea Pigs From Ebola Virus Disease. J. Infect. Dis. 2018, 217, 451–455. [Google Scholar] [CrossRef]
- Cully, M. New tricks to treat Ebola. Nat. Rev. Drug Discov. 2016, 15, 675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Sun, J.; Li, M.; Jin, X. Modified mRNA-LNP Vaccines Confer Protection against Experimental DENV-2 Infection in Mice. Mol. Ther. Methods Clin. Dev. 2020, 18, 702–712. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Lagniton, P.N.P.; Liu, Y.; Xu, R.-H. mRNA vaccines for COVID-19: What, why and how. Int. J. Biol. Sci. 2021, 17, 1446–1460. [Google Scholar] [CrossRef]
- Haynes, B.F. A New Vaccine to Battle Covid-19. N. Engl. J. Med. 2021, 384, 470–471. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, R.; Lentacker, I.; De Smedt, S.C.; Dewitte, H. The dawn of mRNA vaccines: The COVID-19 case. J. Control. Release 2021, 333, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Milane, L.; Amiji, M. Clinical approval of nanotechnology-based SARS-CoV-2 mRNA vaccines: Impact on translational nanomedicine. Drug Deliv. Transl. Res. 2021, 11, 1309–1315. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, W.; Nguyen, G.N.; Zhang, C.; Zeng, C.; Yan, J.; Du, S.; Hou, X.; Li, W.; Jiang, J.; et al. Functionalized lipid-like nanoparticles for in vivo mRNA delivery and base editing. Sci. Adv. 2020, 6, eabc2315. [Google Scholar] [CrossRef]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Love, K.T.; Chen, Y.; Eltoukhy, A.A.; Kastrup, C.; Sahay, G.; Jeon, A.; Dong, Y.; Whitehead, K.A.; Anderson, D.G. Rapid Discovery of Potent siRNA-Containing Lipid Nanoparticles Enabled by Controlled Microfluidic Formulation. J. Am. Chem. Soc. 2012, 134, 6948–6951. [Google Scholar] [CrossRef] [PubMed]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef]
- Jackson, N.A.C.; Kester, K.E.; Casimiro, D.; Gurunathan, S.; DeRosa, F. The promise of mRNA vaccines: A biotech and industrial perspective. NPJ Vaccines 2020, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Seth, R.B.; Sun, L.; Chen, Z.J. Antiviral innate immunity pathways. Cell Res. 2006, 16, 141–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crommelin, D.J.A.; Anchordoquy, T.J.; Volkin, D.B.; Jiskoot, W.; Mastrobattista, E. Addressing the Cold Reality of mRNA Vaccine Stability. J. Pharm. Sci. 2021, 110, 997–1001. [Google Scholar] [CrossRef]
- Srinivas, R.; Garu, A.; Moku, G.; Agawane, S.B.; Chaudhuri, A. A long-lasting dendritic cell DNA vaccination system using lysinylated amphiphiles with mannose-mimicking head-groups. Biomaterials 2012, 33, 6220–6229. [Google Scholar] [CrossRef]
- Sayour, E.J.; De Leon, G.; Pham, C.; Grippin, A.; Kemeny, H.; Chua, J.; Huang, J.; Sampson, J.H.; Sanchez-Perez, L.; Flores, C.; et al. Systemic activation of antigen-presenting cells via RNA-loaded nanoparticles. Oncoimmunology 2017, 6, e1256527. [Google Scholar] [CrossRef] [Green Version]
- Kapadia, C.H.; Perry, J.L.; Tian, S.; Luft, J.C.; DeSimone, J.M. Nanoparticulate immunotherapy for cancer. J. Control. Release 2015, 219, 167–180. [Google Scholar] [CrossRef]
- Kranz, L.M.; Diken, M.; Haas, H.; Kreiter, S.; Loquai, C.; Reuter, K.C.; Meng, M.; Fritz, D.; Vascotto, F.; Hefesha, H.; et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 2016, 534, 396–401. [Google Scholar] [CrossRef] [PubMed]



| Genetic Drugs | Target | Indication | Clinical Trials Identifier or Approved Year |
|---|---|---|---|
| siRNA | |||
| Patisiran (ALN-TTR02) | Transthyretin | Hereditary transthyretin-mediated amyloidosis | FDA-approved in 2018 |
| Givosiran (ALN-AS1) | 5-Aminolevulinic acid synthase | Acute hepatic porphyria | FDA-approved in 2019 |
| Lumasiran (ALN-GO1) | HAO1 | Primary hyperoxaluria type 1 | FDA-approved in 2020 |
| Inclisiran (ALN-PCSSC) | PCSK9 | Hypercholesterolemia | FDA-approved in 2021 |
| ALN-VSP02 | VEGF-A, KSP | Solid tumors | NCT01158079 |
| ARB-001467 | HBsAg | Hepatitis B, chronic | NCT02631096 |
| TKM-PLK1 | PLK1 | Adrenocortical carcinoma, hepatocellular carcinoma | NCT01437007 |
| siRNA-EphA2-DOPC | EphA2 | Advanced or recurrent solid tumors | NCT01591356 |
| Atu027 | Protein kinase N3 | Advanced solid cancer | NCT00938574 |
| ND-L02-s0201 | HSP47 | Hepatic fibrosis | NCT03241264 |
| DCR-MYC | Oncogene MYC | Solid tumors, hepatocellular carcinoma | NCT02314052 |
| PRO-040201 | Apo-B | Hypercholesterolemia | NCT00927459 |
| mRNA | |||
| mRNA-1273 | SARS-CoV-2 | COVID-19 vaccine | FDA-approved in 2022 |
| BNT162b2 | SARS-CoV-2 | COVID-19 vaccine | FDA-approved in 2021 |
| mRNA-2416 | OX40L | Metastatic solid tumor, lymphoma | NCT03323398 |
| mRNA-2752 | OX40L | Lymphoma, ovarian | NCT03739931 |
| mRNA-1647 | 6 CMV | Cytomegalovirus infection | NCT03382405 |
| Antisense oligonucleotides | |||
| Mipomersen | ApoB-100 | Homozygous familial hypercholesterolemia | FDA-approved in 2013 |
| Nusinersen | SMN2 gene | Spinal muscular atrophy | FDA-approved in 2016 |
| Golodirsen | Dystrophin | Duchenne muscular dystrophy | FDA-approved in 2019 |
| Volanesorsen | Apo-CIII | Familial chylomicronaemia syndrome | NCT02658175 |
| Viltolarsen | Exon 45 | Duchenne muscular dystrophy | FDA-approved in 2020 |
| Casimersen | Exon 45 | Duchenne muscular dystrophy | FDA-approved in 2021 |
| Drug Delivery System | Characteristics | Application |
|---|---|---|
| Liposome | Spherical vesicles with lipid bilayer | Delivery of small molecule drug, oligo nucleotide |
| Lipid nanoparticle | Multilamellar and faceted nano-sized vesicle with w/o/w phase | Delivery of nucleic acids for gene therapy, gene editing, and genomic engineering |
| Lipid nanoemulsion | Spherical vesicles with a lipid monolayer and a lipid core | Delivery of hydrophobic or unstable drugs |
| Solid lipid nanoparticle | Spherical vesicles with a core consisting of solid matrix | Delivery of Hydrophobic drugs, active lipid ingredients, and nucleic acids |
| Nanostructured lipid carrier | Spherical vesicles with a core consisting of both solid and liquid lipids | Delivery of virus, non-viral nucleic acids, small molecule drugs with low aqueous solubility |
| Exosome | Nano-sized lipid vesicles secreted from living cells, ranging from 30 to 200 nm, present in cell culture medium and other biological fluids | Carrier of various proteins and nucleic acids for intercellular communication |
| Products | Formulation | FDA Approval Year | Mechanism | Indication |
|---|---|---|---|---|
| Doxil | Liposomal doxorubicin | 1995 | Topoisomerase II inhibitor | Leukemias, multiple myeloma, Hodgkin’s lymphoma, various cancers |
| DaunoXome | Liposomal daunoribucin | 1996 | Topoisomerase II inhibitor | Various cancers, HIV-associated Kaposi’s sarcoma |
| AmBisome | liposomal amphotericin B | 1997 | Binding to ergosterol and cause ion leakage | Invasive fungal infection |
| Marqibo | Liposomal vincristine | 2012 | Tubulin inhibitor | Lymphoma, leukemia, melanoma, brain cancer |
| ONIVYDE | Liposomal irinotecan | 2015 | Topoisomerase I inhibitor | Colon cancer, small-cell lung cancer |
| Lipusu | Liposomal paclitaxel | 2016 | Microtubule inhibitor | Breast cancer, non-small-cell lung cancer |
| Vyxeos | Liposomal daunorubicin and cytarabine | 2017 | Topoisomerase II inhibitor, antimetabolic | Acute myeloid leukemia (AML) |
| ONPATTRO | Patisiran siRNA LNP | 2018 | siRNA target to transthyretin | Hereditary transthyretin-mediated amyloidosis |
| GIVLAARI | Givosiran siRNA LNP | 2019 | siRNA target to 5-aminolevulinic acid synthase | Acute hepatic porphyria |
| OXLUMO | Lumasiran siRNA LNP | 2020 | siRNA target to HAO1 | Primary hyperoxaluria type 1 |
| Leqvio | Inclisiran siRNA LNP | 2021 | siRNA target to PCSK9 | Hypercholesterolemia |
| mRNA-1273 | mRNA LNP | 2022 | Encoding SARS-CoV-2 S protein | COVID-19 vaccine |
| BNT162b2 | mRNA LNP | 2021 | Encoding SARS-CoV-2 S protein | COVID-19 vaccine |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Ma, Y.; Zhang, J.; Kuo, J.C.-T.; Zhang, Z.; Xie, H.; Zhu, J.; Liu, T. Modification of Lipid-Based Nanoparticles: An Efficient Delivery System for Nucleic Acid-Based Immunotherapy. Molecules 2022, 27, 1943. https://doi.org/10.3390/molecules27061943
Zhang C, Ma Y, Zhang J, Kuo JC-T, Zhang Z, Xie H, Zhu J, Liu T. Modification of Lipid-Based Nanoparticles: An Efficient Delivery System for Nucleic Acid-Based Immunotherapy. Molecules. 2022; 27(6):1943. https://doi.org/10.3390/molecules27061943
Chicago/Turabian StyleZhang, Chi, Yifan Ma, Jingjing Zhang, Jimmy Chun-Tien Kuo, Zhongkun Zhang, Haotian Xie, Jing Zhu, and Tongzheng Liu. 2022. "Modification of Lipid-Based Nanoparticles: An Efficient Delivery System for Nucleic Acid-Based Immunotherapy" Molecules 27, no. 6: 1943. https://doi.org/10.3390/molecules27061943
APA StyleZhang, C., Ma, Y., Zhang, J., Kuo, J. C.-T., Zhang, Z., Xie, H., Zhu, J., & Liu, T. (2022). Modification of Lipid-Based Nanoparticles: An Efficient Delivery System for Nucleic Acid-Based Immunotherapy. Molecules, 27(6), 1943. https://doi.org/10.3390/molecules27061943






