Chiral Recognition of Amino Acid Esters in Organic Solvents Using a Glucose-Based Receptor
Abstract
:1. Introduction
2. Results and Discussion
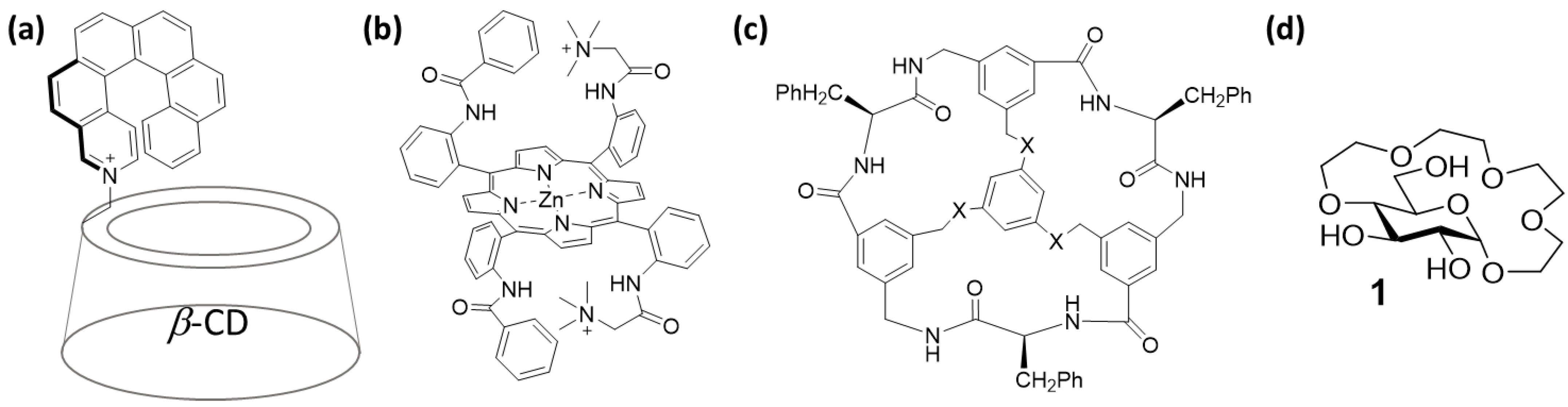
2.1. Experiment Design
2.2. Binding Studies
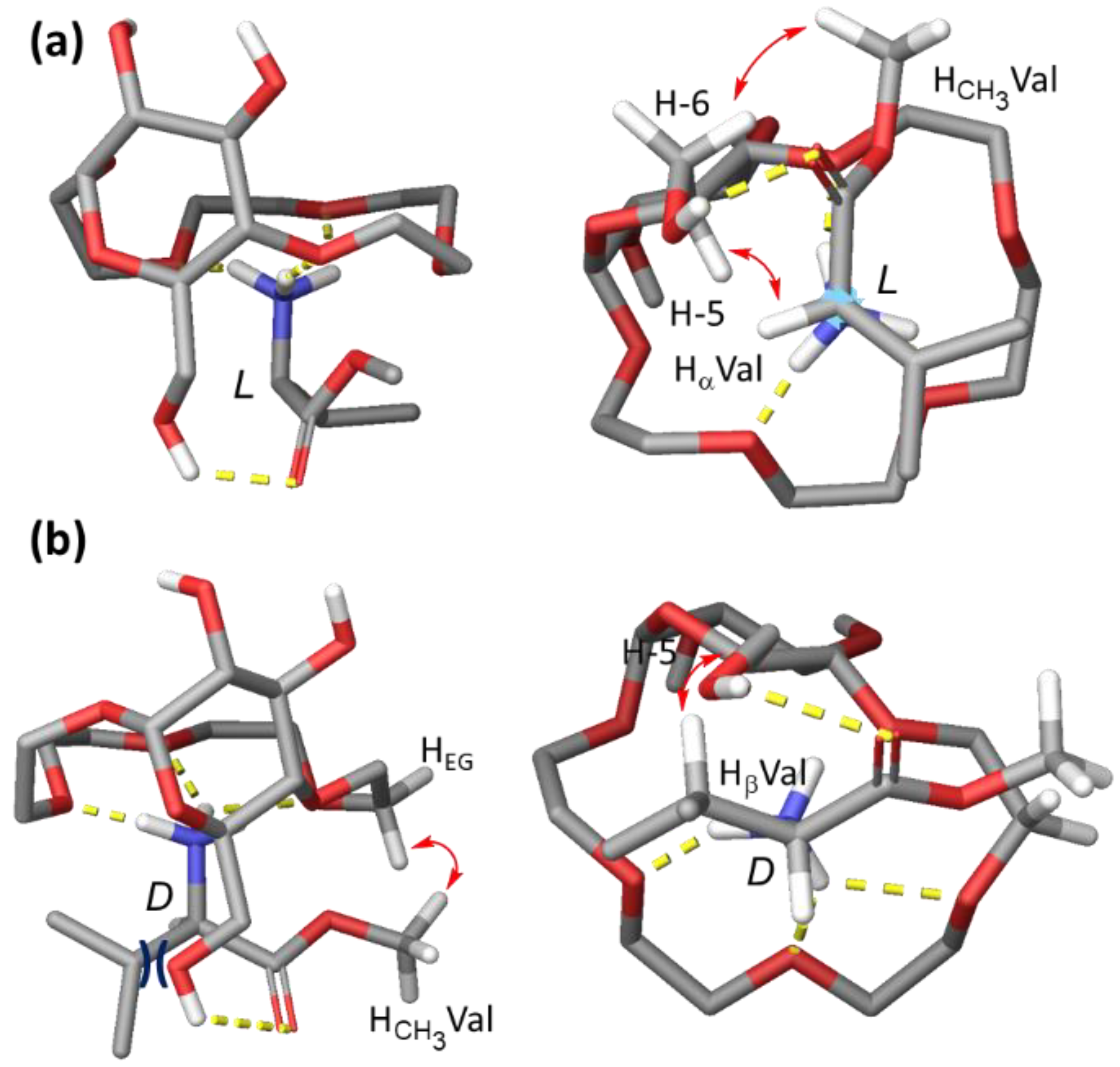
2.3. Conformational Studies and Binding Mode Elucidation in CDCl3
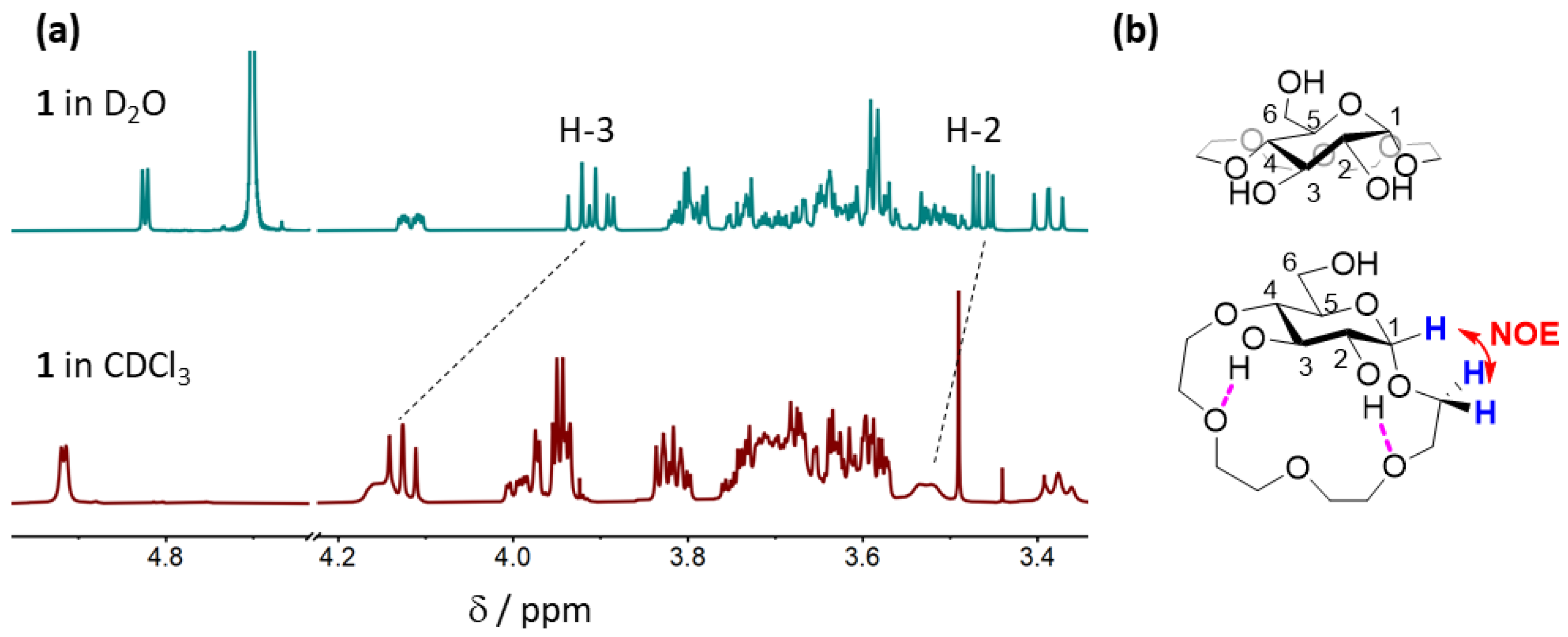
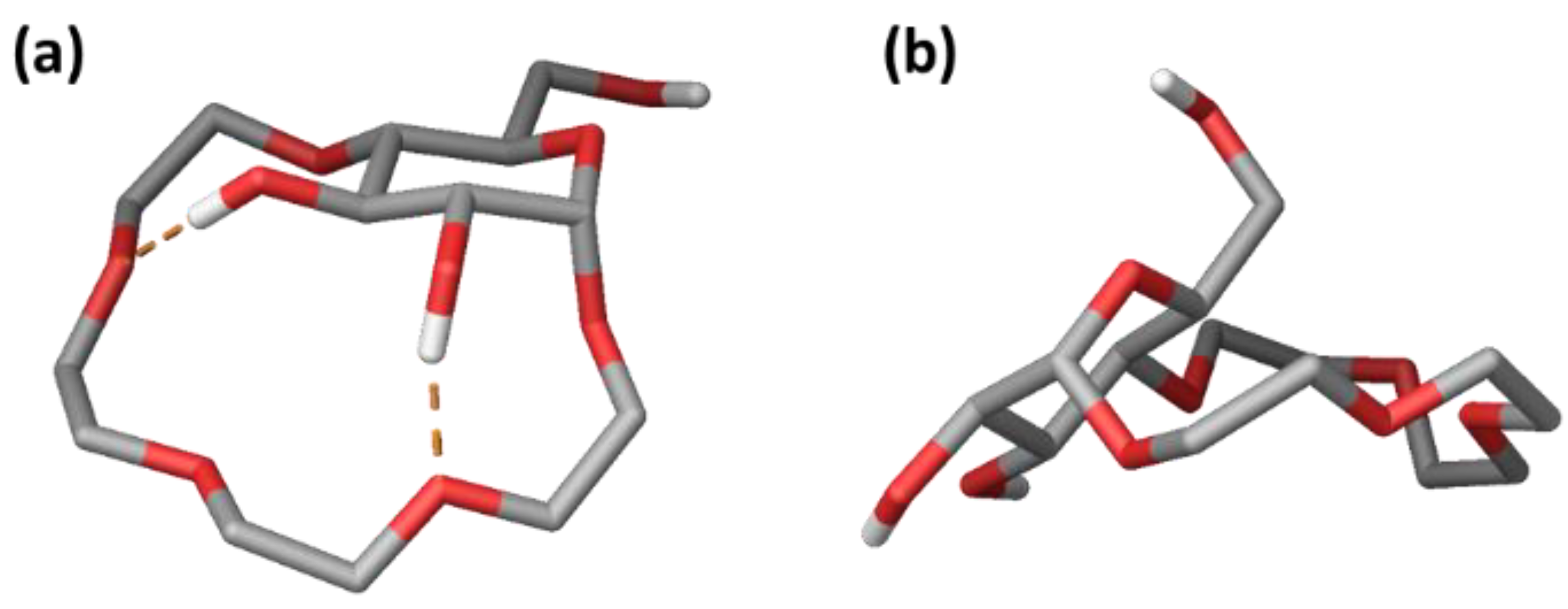
2.3.1. Conformational Analysis of Receptor 1
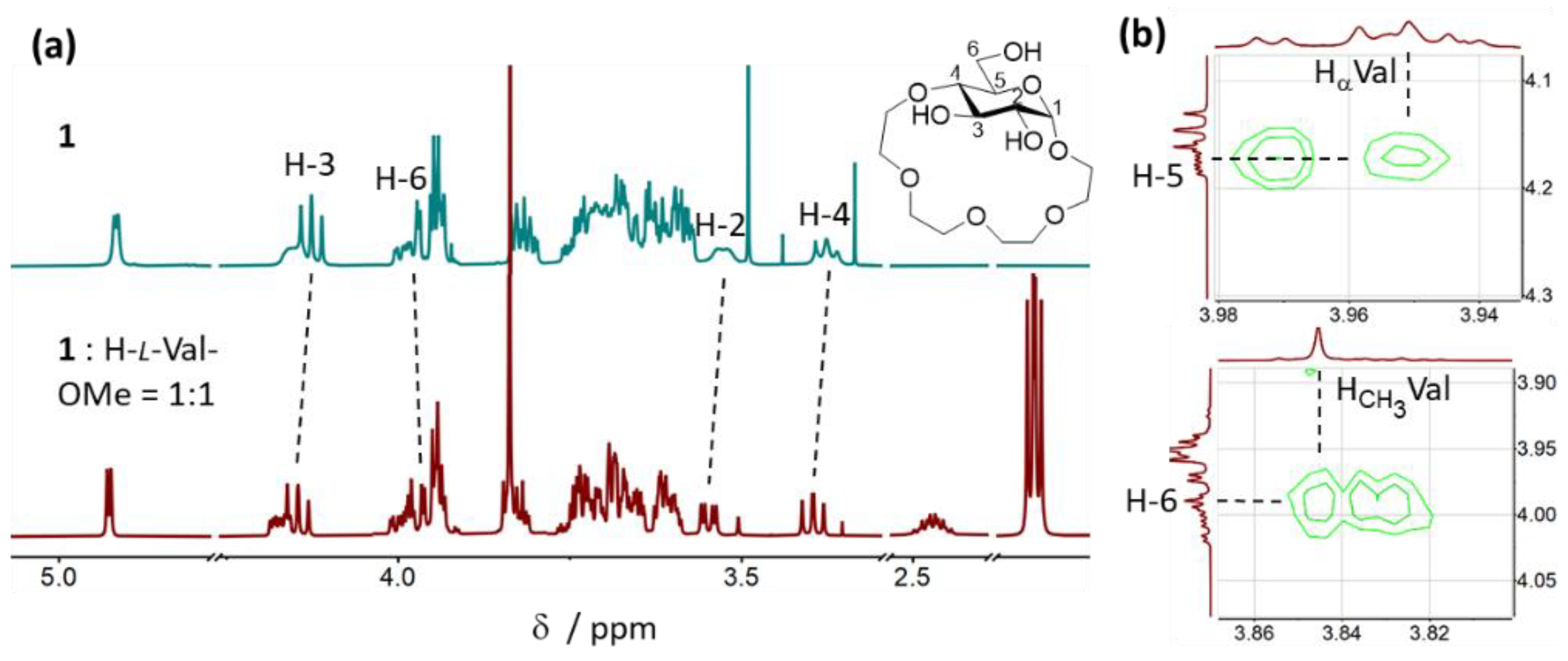
2.3.2. Binding Mode Analysis of 1 with H-Val-OMe × HCl
2.4. Discussion of the Proposed Binding Mode
2.4.1. Interpretation of Binding Data Obtained for Ala, Thr, Phe and Pro
2.4.2. Additional Titration Experiments
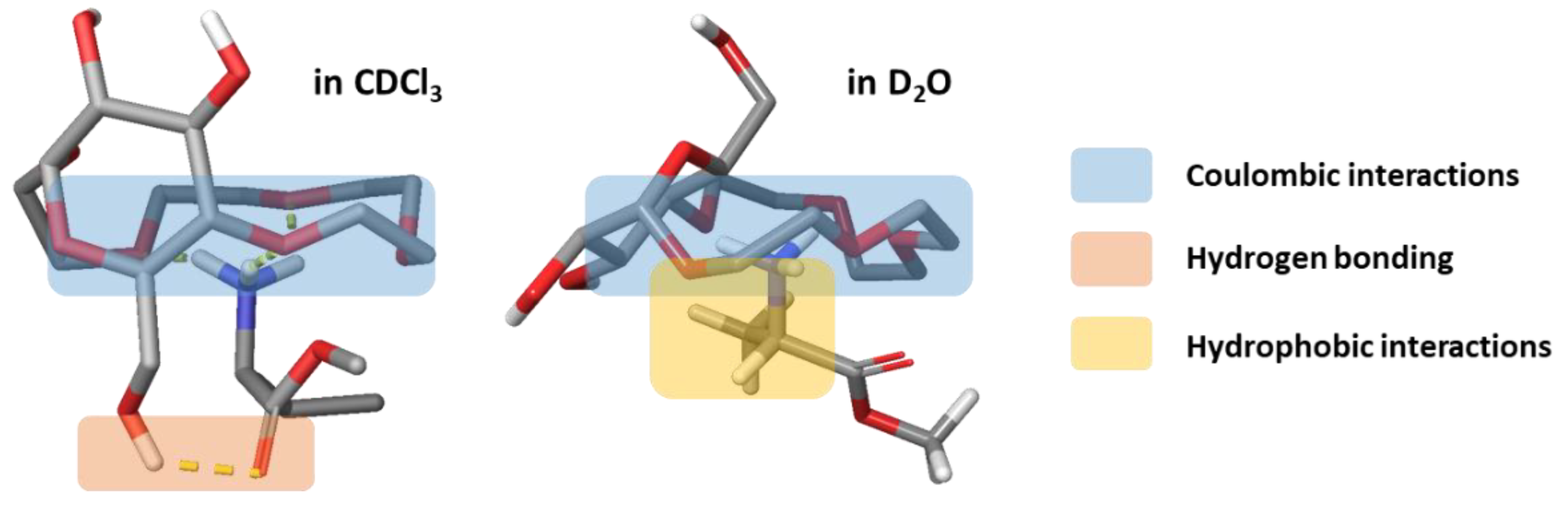
2.4.3. Comparison of Binding Modes in CDCl3 and D2O
3. Conclusions and Outlook
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hughes, A.B. Amino-Acids, Peptides and Proteins in Organic Chemistry; Wiley-VCH: Weinheim, Germany, 2010; Volume I–V. [Google Scholar]
- Mukherjee, S.; Yang, J.W.; Hoffmann, S.; List, B. Asymmetric enamine catalysis. Chem. Rev. 2007, 107, 5471–5569. [Google Scholar] [CrossRef] [PubMed]
- Lewandowski, B.; Wennemers, H. Asymmetric catalysis with short-chain peptides. Curr. Opin. Chem. Biol. 2014, 22, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Kuru, E.; Sachdeva, A.; Wendrell, M. Fluorescent amino acids as versatile building blocks for chemical biology. Nat. Rev. Chem. 2020, 4, 275–290. [Google Scholar] [CrossRef]
- Chakraborty, D.; Gazit, E. Amino acid based self-assembled nanostructures: Complex structures from remarkably simple building blocks. ChemNanoMat 2018, 4, 730–740. [Google Scholar] [CrossRef]
- Anderson, S.L.; Stylianou, K.C. Biologically derived metal organic frameworks. Coord. Chem. Rev. 2017, 349, 102–128. [Google Scholar] [CrossRef] [Green Version]
- Wu, G. Amino Acids: Biochemistry and Nutrition, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar]
- Easson, L.H.; Stedman, E. Studies on the relationship between chemical constitution and physiological action: Molecular dissymmetry and physiological activity. Biochem. J. 1933, 27, 1257–1266. [Google Scholar] [CrossRef]
- Metcalf, J.S.; Dunlop, R.A.; Powell, J.T.; Banack, S.A.; Cox, P.A. L-Serine: A naturally-occurring amino acid with therapeutic potential. Neurotox. Res. 2018, 33, 213–221. [Google Scholar] [CrossRef]
- Mothet, J.P.; Parent, A.T.; Wolosker, H.; Brady, R.O., Jr.; Linden, D.J.; Ferris, C.D.; Rogawski, M.A.; Snyder, S.H. D-Serine is an endogenous ligand for the glycine site of the N-methyl-D-aspartate receptor. Proc. Natl. Acad. Sci. USA 2000, 97, 4927–4931. [Google Scholar] [CrossRef] [Green Version]
- Helfman, P.M.; Bada, J.L. Aspartic acid racemization in tooth enamel from living humans. Proc. Natl. Acad. Sci. USA 1975, 72, 2891–2894. [Google Scholar] [CrossRef] [Green Version]
- Ohtani, S.; Yamamoto, T. Age estimation by amino acid racemization in human teeth. J. Forensic Sci. 2010, 55, 1630–1633. [Google Scholar] [CrossRef]
- Zhang, X.X.; Bradshaw, J.S.; Izatt, R.M. Enantiomeric recognition of amine compounds by chiral macrocyclic receptors. Chem. Rev. 1997, 97, 3313–3362. [Google Scholar] [CrossRef]
- Collins, A.N.; Sheldrake, G.N.; Crosby, J. Chirality in Industry; Wiley and Sons: Chichester, UK, 1997. [Google Scholar]
- Maity, D.; Schmuck, C. Synthetic receptors for amino acids and peptides. In Monographs in Supramolecular Chemistry; Smith, B., Ed.; The Royal Society of Chemistry: London, UK, 2015; pp. 326–368. [Google Scholar]
- Richard, G.I.; Marwani, H.M.; Jiang, S.; Fakadoye, S.O.; Lowry, M.; Strongin, R.M.; Warner, I.M. Chiral recognition of amino acids by use of a fluorescent resorcinarene. Appl. Spectrosc. 2008, 62, 476–480. [Google Scholar] [CrossRef] [Green Version]
- Bois, J.; Bonnamour, I.; Duchamp, C.; Parrot-Lopez, H.; Darbost, U.; Felix, C. Enantioselective recognition of amino acids by chiral peptido-calix[4]arenes and thiacalix[4]arenes. New J. Chem. 2009, 33, 2128–2135. [Google Scholar] [CrossRef]
- Impelizzeri, G.; Maccrrone, G.; Rizzarelli, E.; Vecchio, G.; Corradini, R.; Marchelli, R. 6-Deoxy-6-N-histamino-β-cyclodextrin copper(II) complex, a new enantioselective receptor for aromatic amino acids. Angew. Chem. Int. Ed. Engl. 1991, 30, 1348–1349. [Google Scholar] [CrossRef]
- Konishi, K.; Yahara, K.; Toshishige, H.; Aida, T.; Inoue, S. A novel anion-binding chiral receptor based on a metalloporphyrin with molecular asymmetry. Highly enantioselective recognition of amino acid derivatives. J. Am. Chem. Soc. 1994, 116, 1337–1344. [Google Scholar] [CrossRef]
- Huang, Q.; Jiang, L.; Liang, W.; Gui, J.; Xu, D.; Wu, W.; Nakai, Y.; Nishijima, M.; Fukuhara, G.; Mori, T.; et al. Inherently chiral azonia[6]helicene-modified β-cyclodextrin: Synthesis, characterization, and chirality sensing of underivatized amino acids in water. J. Org. Chem. 2016, 81, 3430–3434. [Google Scholar] [CrossRef]
- Imai, H.; Munakata, H.; Uemori, Y.; Sakura, N. Chiral recognition of amino acids and dipeptides by a water-soluble zinc porphyrin. Inorg. Chem. 2004, 43, 1211–1213. [Google Scholar] [CrossRef]
- Hong, J.-I.; Namgoong, S.K.; Bernardi, A.; Still, W.C. Highly selective binding of simple peptides by a C3 macrotricyclic receptor. J. Am. Chem. Soc. 1991, 113, 5111–5112. [Google Scholar] [CrossRef]
- Dominique, P.; Schnurr, M.; Lewandowski, B. Chiral recognition of amino-acid esters by a glucose-derived macrocyclic receptor. Chem. Commun. 2021, 57, 3476–3479. [Google Scholar] [CrossRef]
- Folmer-Andersen, J.F.; Lynch, V.M.; Anslyn, E.V. Colorimetric Enantiodiscrimination of α-Amino Acids in Protic Media. J. Am. Chem. Soc. 2005, 127, 7986–7987. [Google Scholar] [CrossRef]
- Pu, L. Enantioselective fluorescent recognition of free amino acids: Challenges and opportunities. Angew. Chem. Int. Ed. 2020, 59, 21814–21828. [Google Scholar] [CrossRef]
- Marcus, Y. Ion Solvation, 1st ed.; Wiley: Chichester, UK, 1985. [Google Scholar]

| Amino Acid | DMSO-d6 | CD3CN | CDCl3 | D2O | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kal 2 | Kad | Sel. | Kal | Kad | Sel. | Kal | Kad | Sel. | Kal | Kad | Sel. | |
| Ala | 98 ± 26 | 51 ± 14 | 1.8:1 | 299 ± 94 | 139 ± 55 | 2.2:1 | 707 ± 308 | 279 ± 103 | 2.5:1 | 133 ± 14 | 69 ± 9 | 1.9:1 |
| Thr | 104 ± 10 | 61 ± 11 | 1.4:1 | 306 ± 81 | 201 ± 44 | 1.5:1 | 680 ± 213 | 295 ± 80 | 2.2:1 | 166 ± 22 | 61 ± 9 | 2.7:1 |
| Val | 189 ± 72 | 52 ± 12 | 3.6:1 | 405 ± 98 | 117 ± 26 | 3.5:1 | 977 ± 266 | 201 ± 34 | 4.7:1 | 232 ± 43 | 46 ± 5 | 5.1:1 |
| Phe | 167 ± 43 | 54 ± 12 | 3.1:1 | 420 ± 161 3 | 141 ± 22 3 | 3.0:1 | 315 ± 59 3 | 212 ± 37 3 | 1.5:1 | 275 ± 30 | 45 ± 5 | 6.2:1 |
| Pro | 72 ± 15 | 32 ± 2 | 2.2:1 | 132 ± 32 | 72 ± 26 | 1.9:1 | 255 ± 74 | 162 ± 50 | 1.6:1 | 70 ± 5 | 44 ± 5 | 1.6:1 |
| Amino-Acid | Kal 2 | Kad | Sel. |
|---|---|---|---|
| Leu | 833 ± 354 | 258 ± 91 | 3.2:1 |
| tLeu | 699 ± 249 | 123 ± 29 | 5.6:1 |
| Asn | 322 ± 108 | 293 ± 64 | 1.1:1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mönkemöller, L.S.; Schnurr, M.; Lewandowski, B. Chiral Recognition of Amino Acid Esters in Organic Solvents Using a Glucose-Based Receptor. Molecules 2022, 27, 2177. https://doi.org/10.3390/molecules27072177
Mönkemöller LS, Schnurr M, Lewandowski B. Chiral Recognition of Amino Acid Esters in Organic Solvents Using a Glucose-Based Receptor. Molecules. 2022; 27(7):2177. https://doi.org/10.3390/molecules27072177
Chicago/Turabian StyleMönkemöller, Leah Susanne, Martin Schnurr, and Bartosz Lewandowski. 2022. "Chiral Recognition of Amino Acid Esters in Organic Solvents Using a Glucose-Based Receptor" Molecules 27, no. 7: 2177. https://doi.org/10.3390/molecules27072177







