Preclassification of Broadband and Sparse Infrared Data by Multiplicative Signal Correction Approach
Abstract
:1. Introduction
2. Results
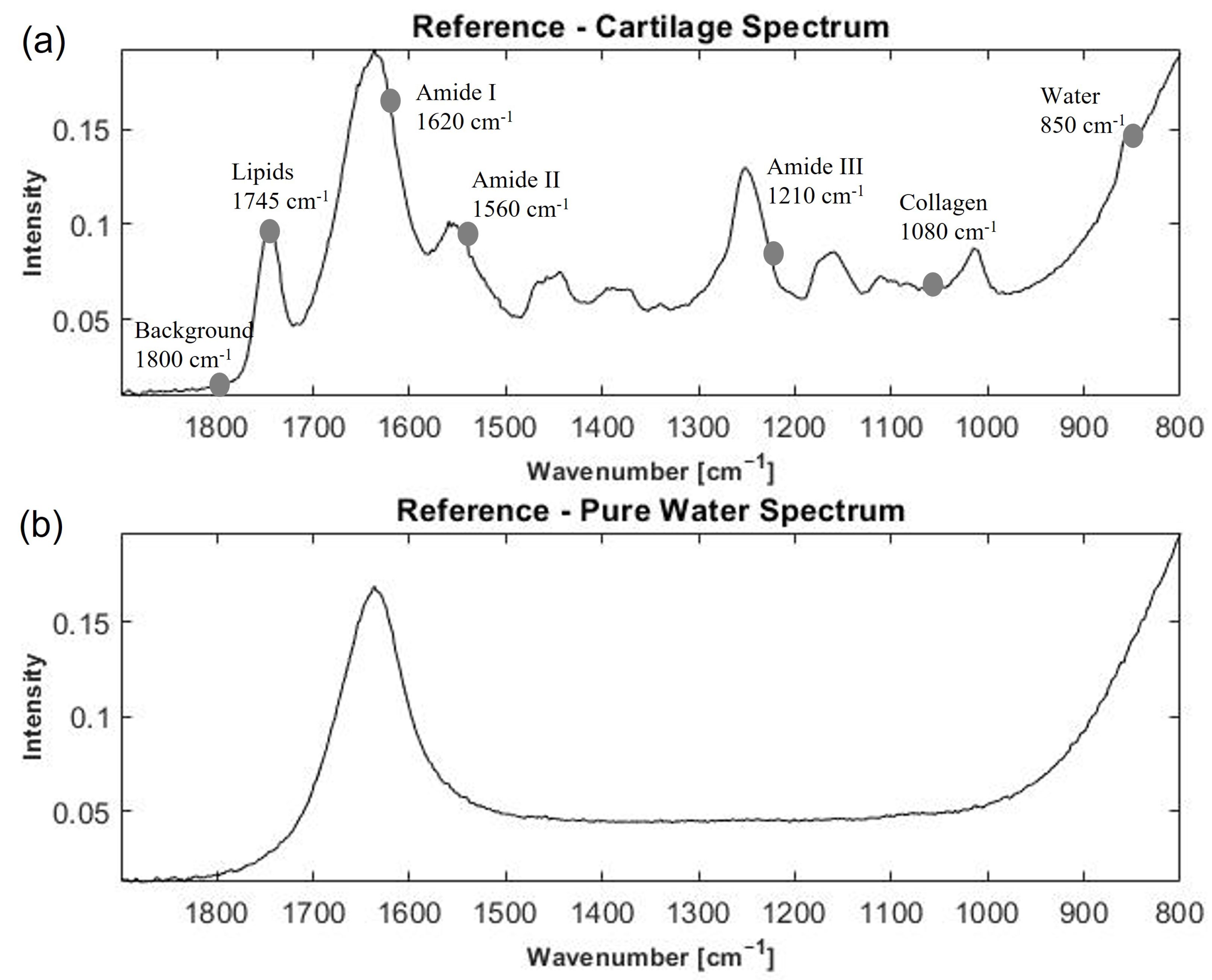
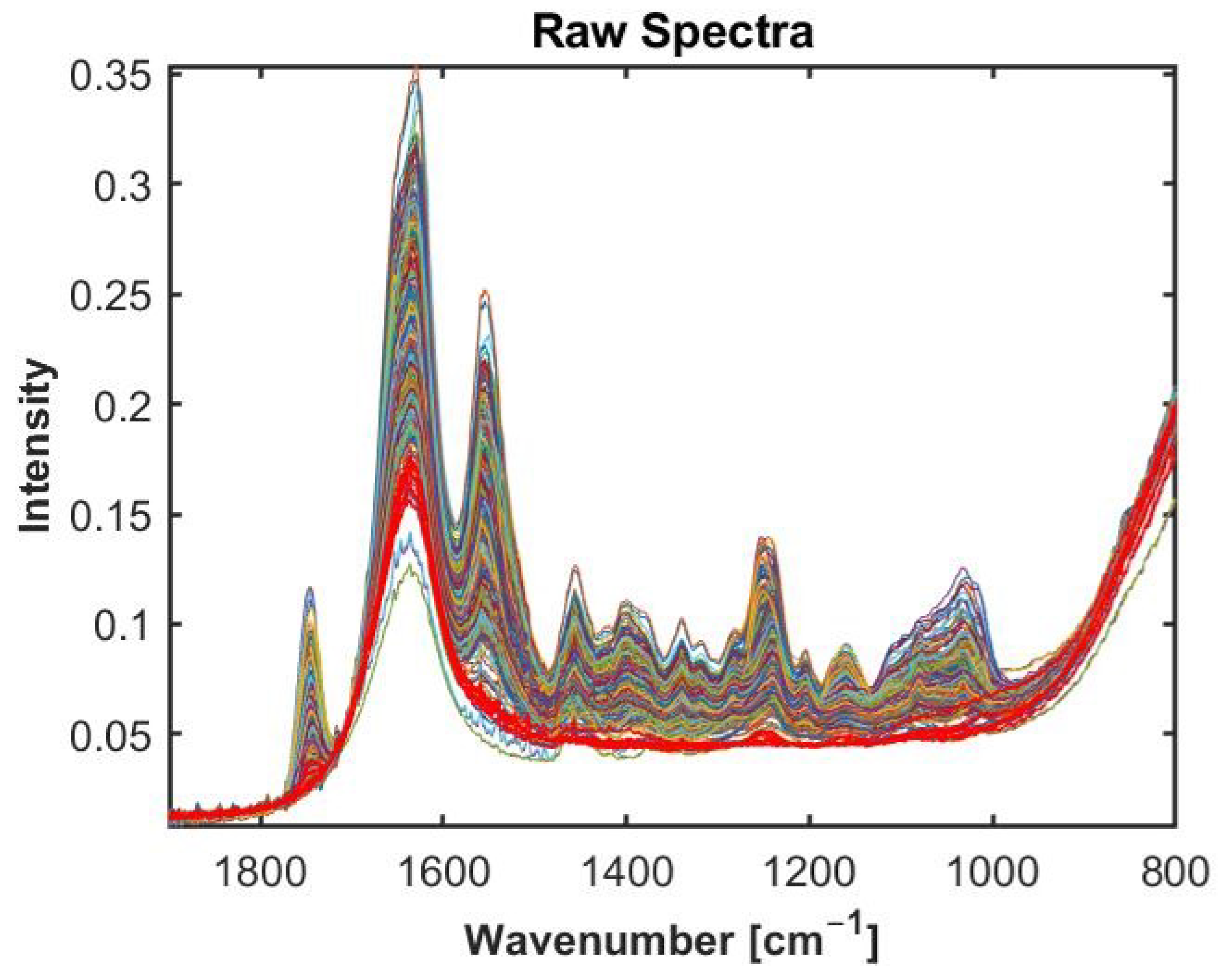
2.1. Broadband FTIR-ATR Spectra of Cartilage Samples
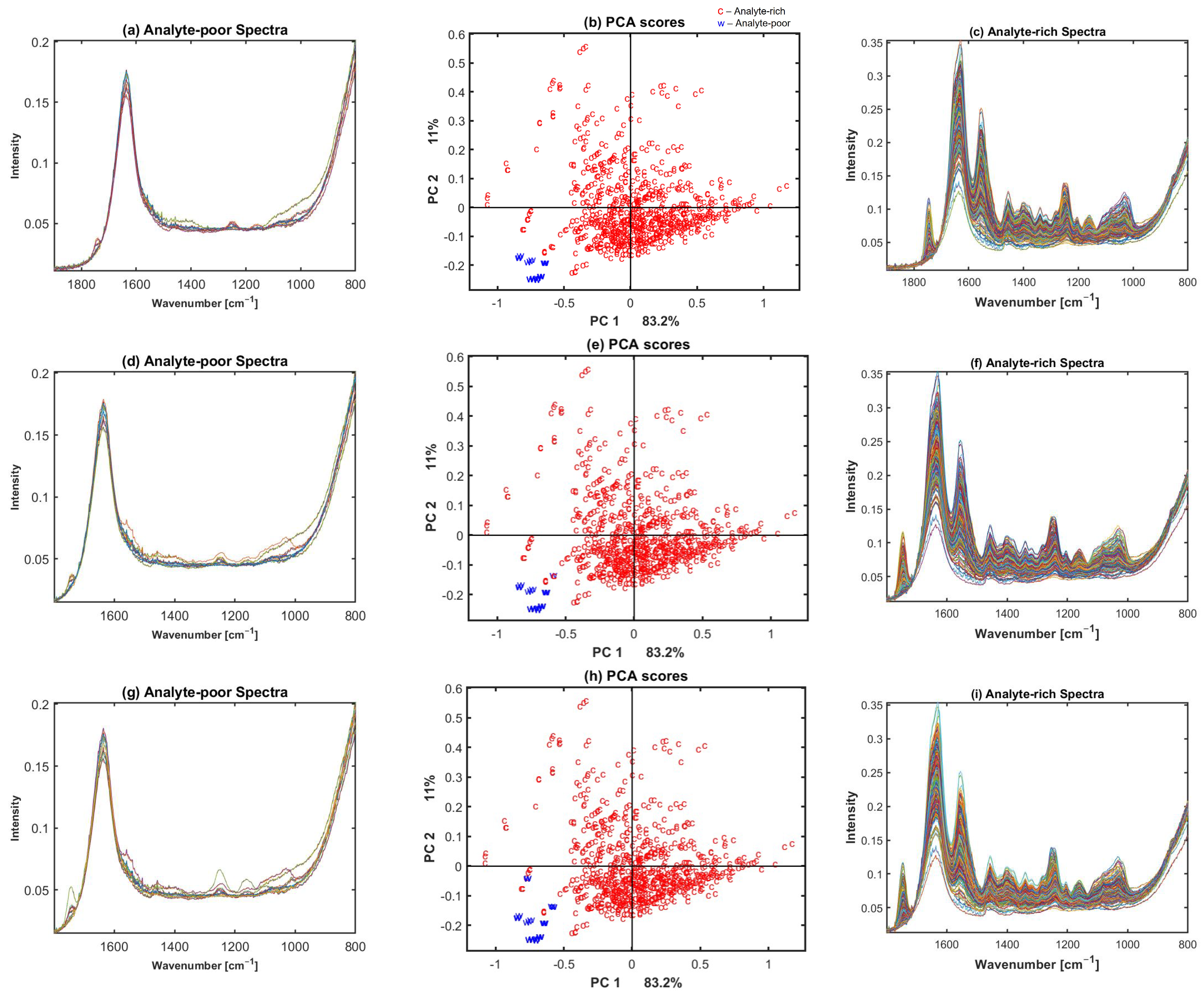
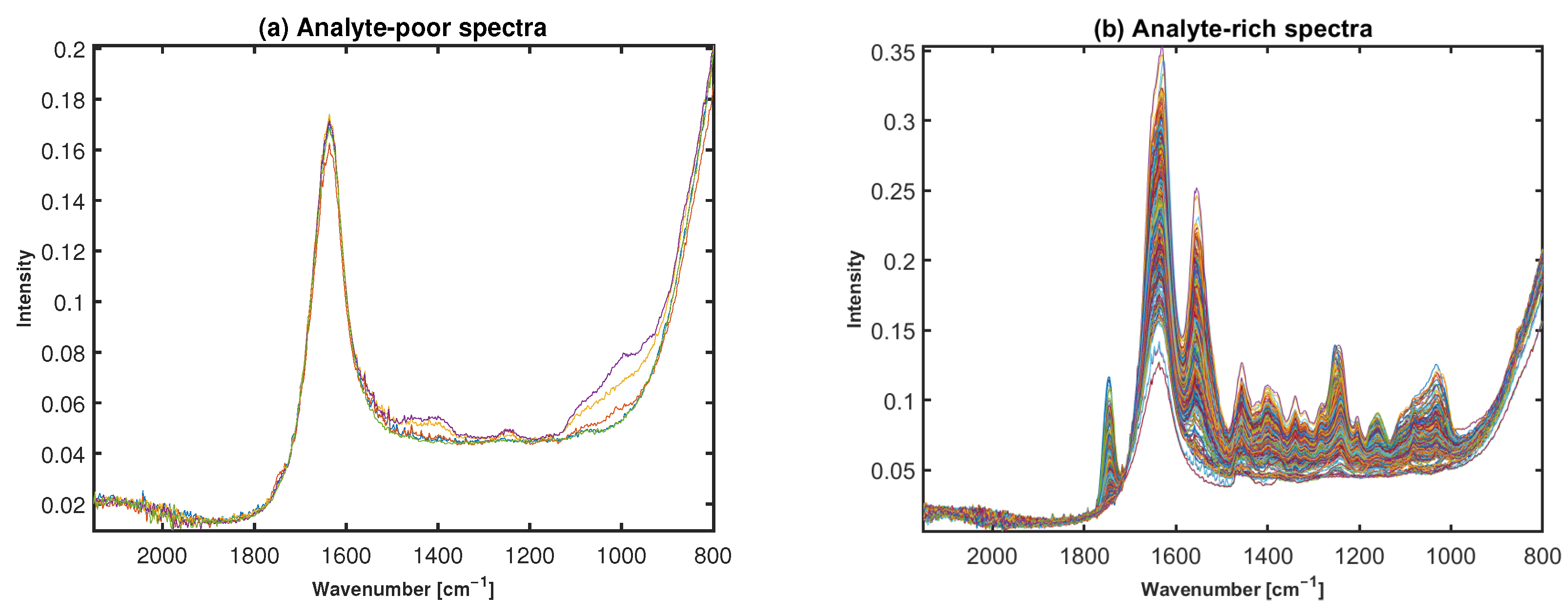
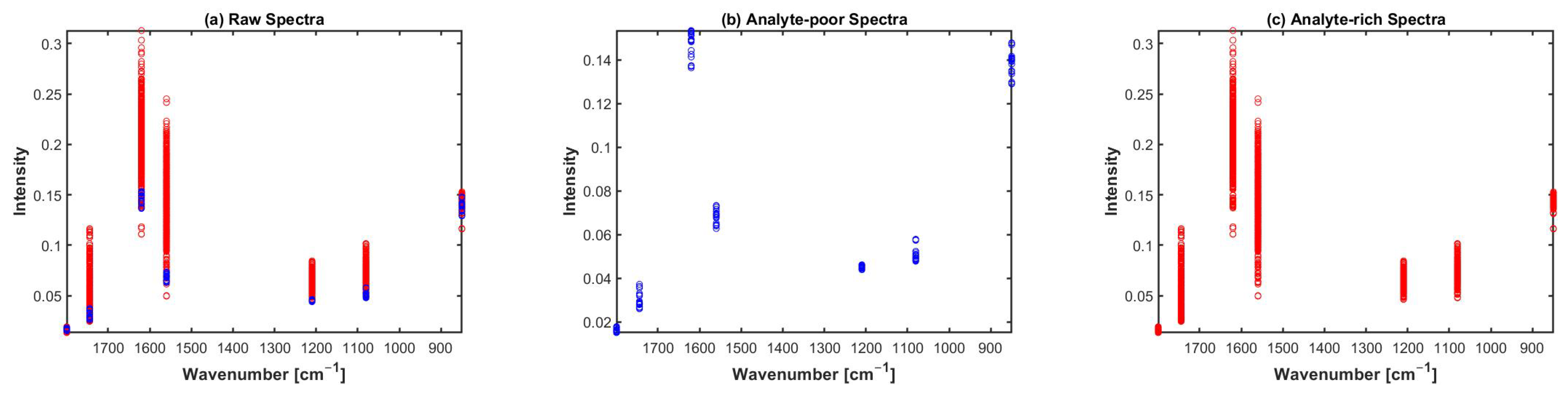
2.2. Preclassification Based on Broadband Spectra
2.3. Preclassification Based on Laser Wavelengths
3. Discussions
FTIR Spectra
4. Materials and Methods
4.1. Spectral Data
4.2. Annotation of Broadband Spectra for Water, Cartilage, and Low Signal Spectra
5. Theory
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Barth, A.; Haris, P.I. Biological and Biomedical Infrared Spectroscopy; IOS Press: Amsterdam, The Netherlands, 2009; Volume 2. [Google Scholar]
- Naumann, D. FT-infrared and FT-Raman spectroscopy in biomedical research. Appl. Spectrosc. Rev. 2001, 36, 239–298. [Google Scholar] [CrossRef]
- Figoli, C.; Garcea, M.; Bisioli, C.; Tafintseva, V.; Shapaval, V.; Gómez Peña, M.; Gibbons, L.; Althabe, F.; Miguel Yantorno, O.; Horton, M.; et al. A robust metabolomics approach for the evaluation of human embryos from in-vitro fertilization. Analyst 2021, 146, 6156–6169. [Google Scholar] [CrossRef]
- Sahu, R.K.; Mordechai, S. Spectroscopic techniques in medicine: The future of diagnostics. Appl. Spectrosc. Rev. 2016, 51, 484–499. [Google Scholar] [CrossRef]
- Zhao, Y.; Lu, Y.; Zhu, Y.; Wu, Y.; Zhai, M.; Wang, X.; Yin, J. Submillimetric FTIR detection of articular cartilage by home-made ATR-MIR-Hollow optical fiber probe. Infrared Phys. Technol. 2019, 98, 236–239. [Google Scholar] [CrossRef]
- Virtanen, V.K.O.; Nippolainen, E.; Shaikh, R.; Afara, I.; Toyras, J.; Solheim, J.; Tafintseva, V.; Zimmermann, B.; Kohler, A.; Saarakkala, S.; et al. Infrared fiber optic spectroscopy detects bovine articular cartilage degeneration. bioRxiv 2020, 13 (Suppl. 2), 285S–294S. [Google Scholar] [CrossRef]
- Cortizas, A.M.; López-Costas, O. Linking structural and compositional changes in archaeological human bone collagen: An FTIR-ATR approach. Sci. Rep. 2020, 10, 1–14. [Google Scholar]
- Rieppo, L.; Töyräs, J.; Saarakkala, S. Vibrational spectroscopy of articular cartilage. Appl. Spectrosc. Rev. 2017, 52, 249–266. [Google Scholar] [CrossRef]
- Oinas, J.; Rieppo, L.; Finnilä, M.; Valkealahti, M.; Lehenkari, P.; Saarakkala, S. Imaging of osteoarthritic human articular cartilage using fourier transform infrared microspectroscopy combined with multivariate and univariate analysis. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Camacho, N.P.; West, P.; Torzilli, P.A.; Mendelsohn, R. FTIR microscopic imaging of collagen and proteoglycan in bovine cartilage. Biopolym. Orig. Res. Biomol. 2001, 62, 1–8. [Google Scholar] [CrossRef]
- West, P.; Bostrom, M.P.G.; Torzilli, P.; Camacho, N.P. Fourier transform infrared spectral analysis of degenerative cartilage: An infrared fiber optic probe and imaging study. Appl. Spectrosc. 2004, 58, 376–381. [Google Scholar] [CrossRef]
- Rieppo, L.; Rieppo, J.; Jurvelin, J.S.; Saarakkala, S. Fourier transform infrared spectroscopic imaging and multivariate regression for prediction of proteoglycan content of articular cartilage. PLoS ONE 2012, 7, e32344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boskey, A.; Pleshko Camacho, N. FT-IR imaging of native and tissue-engineered bone and cartilage. Biomaterials 2007, 28, 2465–2478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, X.; Yang, X.; Bostrom, M.P.; Camacho, N.P. Fourier transform infrared imaging spectroscopy investigations in the pathogenesis and repair of cartilage. Biochim. Biophys. Acta-(BBA)-Biomembr. 2006, 1758, 934–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, N.; Rinnerthaler, S.; Paschalis, E.; Mendelsohn, R.; Boskey, A.; Fratzl, P. Complementary information on bone ultrastructure from scanning small angle X-ray scattering and Fourier-transform infrared microspectroscopy. Bone 1999, 25, 287–293. [Google Scholar] [CrossRef]
- Bi, X.; Yang, X.; Bostrom, M.P.; Bartusik, D.; Ramaswamy, S.; Fishbein, K.W.; Spencer, R.G.; Camacho, N.P. Fourier transform infrared imaging and MR microscopy studies detect compositional and structural changes in cartilage in a rabbit model of osteoarthritis. Anal. Bioanal. Chem. 2007, 387, 1601–1612. [Google Scholar] [CrossRef] [Green Version]
- Baykal, D.; Irrechukwu, O.; Lin, P.C.; Fritton, K.; Spencer, R.G.; Pleshko, N. Nondestructive assessment of engineered cartilage constructs using near-infrared spectroscopy. Appl. Spectrosc. 2010, 64, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- McGoverin, C.M.; Hanifi, A.; Palukuru, U.P.; Yousefi, F.; Glenn, P.B.; Shockley, M.; Spencer, R.G.; Pleshko, N. Nondestructive assessment of engineered cartilage composition by near infrared spectroscopy. Ann. Biomed. Eng. 2016, 44, 680–692. [Google Scholar] [CrossRef] [Green Version]
- Sarin, J.K.; Te Moller, N.; Mohammadi, A.; Prakash, M.; Torniainen, J.; Brommer, H.; Nippolainen, E.; Shaikh, R.; Mäkelä, J.; Korhonen, R.K.; et al. Machine learning augmented near-infrared spectroscopy: In vivo follow-up of cartilage defects. Osteoarthr. Cartil. 2021, 29, 423–432. [Google Scholar] [CrossRef]
- Afara, I.; Sahama, T.; Oloyede, A. Near infrared for non-destructive testing of articular cartilage. In Nondestructive Testing of Materials and Structures; Springer: Berlin/Heidelberg, Germany, 2013; pp. 399–404. [Google Scholar]
- Afara, I.; Singh, S.; Oloyede, A. Application of near infrared (NIR) spectroscopy for determining the thickness of articular cartilage. Med. Eng. Phys. 2013, 35, 88–95. [Google Scholar] [CrossRef]
- Prakash, M.; Joukainen, A.; Torniainen, J.; Honkanen, M.; Rieppo, L.; Afara, I.; Kröger, H.; Töyräs, J.; Sarin, J. Near-infrared spectroscopy enables quantitative evaluation of human cartilage biomechanical properties during arthroscopy. Osteoarthr. Cartil. 2019, 27, 1235–1243. [Google Scholar] [CrossRef]
- Li, G.; Thomson, M.; Dicarlo, E.; Yang, X.; Nestor, B.; Bostrom, M.P.; Camacho, N.P. A chemometric analysis for evaluation of early-stage cartilage degradation by infrared fiber-optic probe spectroscopy. Appl. Spectrosc. 2005, 59, 1527–1533. [Google Scholar] [CrossRef]
- Hanifi, A.; McCarthy, H.; Roberts, S.; Pleshko, N. Fourier transform infrared imaging and infrared fiber optic probe spectroscopy identify collagen type in connective tissues. PLoS ONE 2013, 8, e64822. [Google Scholar] [CrossRef] [PubMed]
- Mackanos, M.A.; Contag, C.H. Fiber-optic probes enable cancer detection with FTIR spectroscopy. Trends Biotechnol. 2010, 28, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Gonzalez, E.; Hitchins, V.; Ilev, I. Detecting bacteria contamination on medical device surfaces using an integrated fiber-optic mid-infrared spectroscopy sensing method. Sens. Actuators B Chem. 2016, 231, 646–654. [Google Scholar] [CrossRef]
- Ozaki, Y.; Matsunaga, T.; Miura, T. Nondestructive and noninvasive monitoring of deoxyhemoglobin in the vein by use of a near-infrared reflectance spectrometer with a fiber-optic probe. Appl. Spectrosc. 1992, 46, 180–182. [Google Scholar] [CrossRef]
- Bibikova, O.A.; Mironovich, V.; Usenov, I.; Feliksberger, E.; Bocharnikov, A.; Surkova, A.; Belikova, V.; Nippolainen, E.; Afara, I.; Haas, J.; et al. Mid-infrared fiber spectroscopy for detection of cartilage degeneration in osteoarthritis (Conference Presentation). In Optical Fibers and Sensors for Medical Diagnostics and Treatment Applications XX; International Society for Optics and Photonics: Bellingham, WA, USA, 2020; Volume 11233, p. 112330J. [Google Scholar]
- Venn, M.; Maroudas, A. Chemical composition and swelling of normal and osteoarthrotic femoral head cartilage. I. Chemical composition. Ann. Rheum. Dis. 1977, 36, 121–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwaighofer, A.; Brandstetter, M.; Lendl, B. Quantum cascade lasers (QCLs) in biomedical spectroscopy. Chem. Soc. Rev. 2017, 46, 5903–5924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasahara, R.; Kino, S.; Soyama, S.; Matsuura, Y. Noninvasive glucose monitoring using mid-infrared absorption spectroscopy based on a few wavenumbers. Biomed. Opt. Express 2018, 9, 289–302. [Google Scholar] [CrossRef] [Green Version]
- Kasyutich, V.L.; Ibrahim, R.R.; Martin, P.A. Stability of widely tuneable, continuous wave external-cavity quantum cascade laser for absorption spectroscopy. Infrared Phys. Technol. 2010, 53, 381–387. [Google Scholar] [CrossRef]
- Miracleproject. Horizon 2020 Photonics21 Project MIRACLE. 2022. Available online: https://miracleproject.eu/ (accessed on 29 December 2021).
- Tafintseva, V.; Lintvedt, T.A.; Solheim, J.H.; Zimmermann, B.; Rehman, H.U.; Virtanen, V.; Shaikh, R.; Nippolainen, E.; Afara, I.; Saarakkala, S.; et al. Preprocessing Strategies for Sparse Infrared Spectroscopy: A Case Study on Cartilage Diagnostics. Molecules 2022, 27, 873. [Google Scholar] [CrossRef]
- Shaikh, R.; Tafintseva, V.; Nippolainen, E.; Virtanen, V.; Solheim, J.; Zimmermann, B.; Rieppo, L.; Saarakkala, S.; Kröger, H.; Kohler, A.; et al. Identification of cartilage damage by data fusion of mid-infrared, near-infrared and Raman spectroscopies. Ann. Biomed. Eng. 2021; in press. [Google Scholar]
- Virtanen, V.; Tafintseva, V.; Shaikh, R.; Nippolainen, E.; Haas, J.; Afara, I.; Töyräs, J.; Kröger, H.; Solheim, J.; Zimmermann, B.; et al. Infrared spectroscopy as objective tool for assessing articular cartilage tissue health. Osteoarthr. Cartil. Open 2021, in press. [Google Scholar]
- Kohler, A.; Solheim, J.; Tafintseva, V.; Zimmermann, B.; Shapaval, V. Model-Based Pre-Processing in Vibrational Spectroscopy. 2020. Available online: https://www.sciencedirect.com/science/article/pii/B9780124095472146074?via%3Dihub (accessed on 15 February 2022).
- Lasch, P. Spectral pre-processing for biomedical vibrational spectroscopy and microspectroscopic imaging. Chemom. Intell. Lab. Syst. 2012, 117, 100–114. [Google Scholar] [CrossRef] [Green Version]
- Liland, K.H.; Almøy, T.; Mevik, B.H. Optimal choice of baseline correction for multivariate calibration of spectra. Appl. Spectrosc. 2010, 64, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Purvine, S.; Kolker, N.; Kolker, E. Spectral quality assessment for high-throughput tandem mass spectrometry proteomics. OMICS J. Integr. Biol. 2004, 8, 255–265. [Google Scholar] [CrossRef]
- Tafintseva, V.; Shapaval, V.; Smirnova, M.; Kohler, A. Extended multiplicative signal correction for FTIR spectral quality test and pre-processing of infrared imaging data. J. Biophotonics 2020, 13, e201960112. [Google Scholar] [CrossRef]
- Petibois, C.; Déléris, G. Chemical mapping of tumor progression by FT-IR imaging: Towards molecular histopathology. Trends Biotechnol. 2006, 24, 455–462. [Google Scholar] [CrossRef]
- Servaty, R.; Schiller, J.; Binder, H.; Arnold, K. Hydration of polymeric components of cartilage—An infrared spectroscopic study on hyaluronic acid and chondroitin sulfate. Int. J. Biol. Macromol. 2001, 28, 121–127. [Google Scholar] [CrossRef]
- Martens, H.; Stark, E. Extended multiplicative signal correction and spectral interference subtraction: New preprocessing methods for near infrared spectroscopy. J. Pharm. Biomed. Anal. 1991, 9, 625–635. [Google Scholar] [CrossRef]
- Rasskazov, I.L.; Singh, R.; Carney, P.S.; Bhargava, R. Extended Multiplicative Signal Correction for Infrared Microspectroscopy of Heterogeneous Samples with Cylindrical Domains. Appl. Spectrosc. 2019, 73, 859–869. [Google Scholar] [CrossRef]
- Afseth, N.K.; Kohler, A. Extended multiplicative signal correction in vibrational spectroscopy, a tutorial. Chemom. Intell. Lab. Syst. 2012, 117, 92–99. [Google Scholar] [CrossRef]
- Sophia Fox, A.J.; Bedi, A.; Rodeo, S.A. The basic science of articular cartilage: Structure, composition, and function. Sports Health 2009, 1, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Ilari, J.L.; Martens, H.; Isaksson, T. Determination of Particle Size in Powders by Scatter Correction in Diffuse Near-Infrared Reflectance. Appl. Spectrosc. 1988, 42, 722–728. [Google Scholar] [CrossRef]
- Kohler, A.; Kristian Afseth, N.; Martens, H. Chemometrics in biospectroscopy. In Handbook of Vibrational Spectroscopy; Wiley: Hoboken, NJ, USA, 2006. [Google Scholar]
- Tafintseva, V.; Shapaval, V.; Blazhko, U.; Kohler, A. Correcting replicate variation in spectroscopic data by machine learning and model-based pre-processing. Chemom. Intell. Lab. Syst. 2021, 215, 104–350. [Google Scholar] [CrossRef]
- Diehn, S.; Zimmermann, B.; Tafintseva, V.; Bagcioglu, M.; Kohler, A.; Ohlson, M.; Fjellheim, S.; Kneipp, J. Discrimination of grass pollen of different species by FTIR spectroscopy of individual pollen grains. Anal. Bioanal. Chem. 2021, 412, 6459–6474. [Google Scholar] [CrossRef] [PubMed]
- Trukhan, S.; Tafintseva, V.; Tøndel, K.; Großerueschkamp, F.; Mosig, A.; Kovalev, V.; Gerwert, K.; Kohler, A. Grayscale representation of infrared microscopy images by extended multiplicative signal correction for registration with histological images. J. Biophotonics 2020, 13, e201960223. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehman, H.U.; Tafintseva, V.; Zimmermann, B.; Solheim, J.H.; Virtanen, V.; Shaikh, R.; Nippolainen, E.; Afara, I.; Saarakkala, S.; Rieppo, L.; et al. Preclassification of Broadband and Sparse Infrared Data by Multiplicative Signal Correction Approach. Molecules 2022, 27, 2298. https://doi.org/10.3390/molecules27072298
Rehman HU, Tafintseva V, Zimmermann B, Solheim JH, Virtanen V, Shaikh R, Nippolainen E, Afara I, Saarakkala S, Rieppo L, et al. Preclassification of Broadband and Sparse Infrared Data by Multiplicative Signal Correction Approach. Molecules. 2022; 27(7):2298. https://doi.org/10.3390/molecules27072298
Chicago/Turabian StyleRehman, Hafeez Ur, Valeria Tafintseva, Boris Zimmermann, Johanne Heitmann Solheim, Vesa Virtanen, Rubina Shaikh, Ervin Nippolainen, Isaac Afara, Simo Saarakkala, Lassi Rieppo, and et al. 2022. "Preclassification of Broadband and Sparse Infrared Data by Multiplicative Signal Correction Approach" Molecules 27, no. 7: 2298. https://doi.org/10.3390/molecules27072298






