Molecular Dynamics Simulation Studies on the Aggregation of Amyloid-β Peptides and Their Disaggregation by Ultrasonic Wave and Infrared Laser Irradiation
Abstract
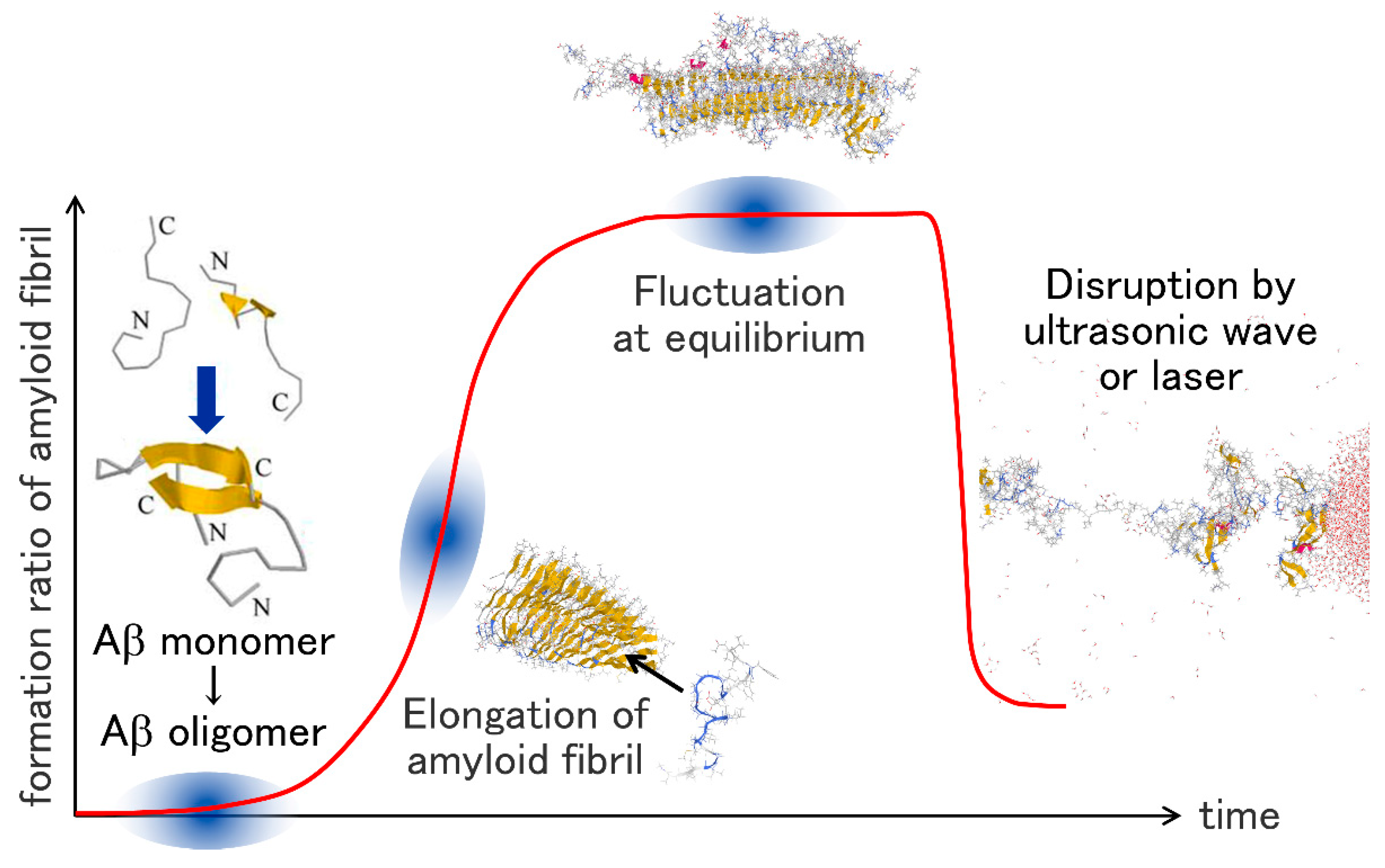
:1. Introduction
2. Aggregation of Aβ Fragments
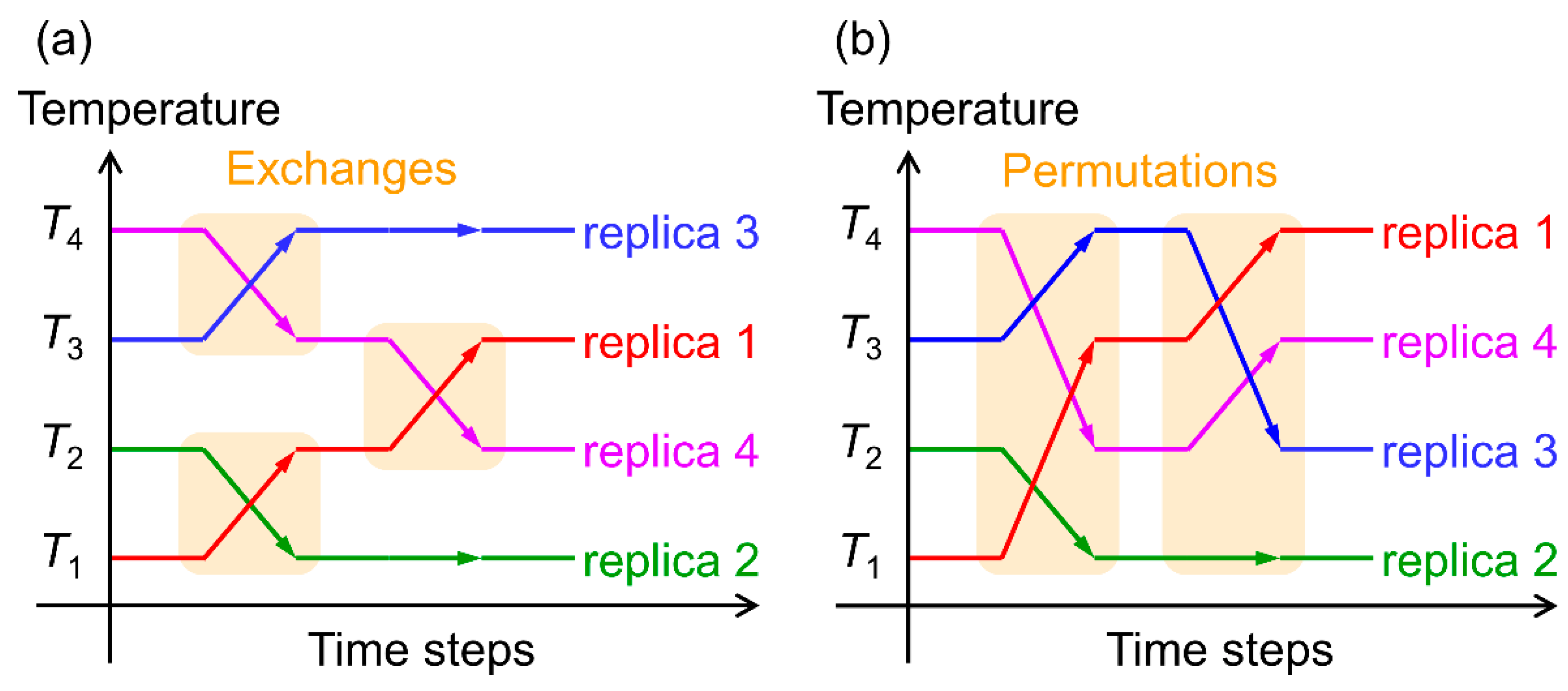
2.1. Hamiltonian Replica-Permutation Molecular Dynamics Simulation of Aβ(29–42) Peptides
2.2. Dimerization of Aβ(29–42) Peptides
3. Structure of an Aβ Peptide at an Air–Water Interface
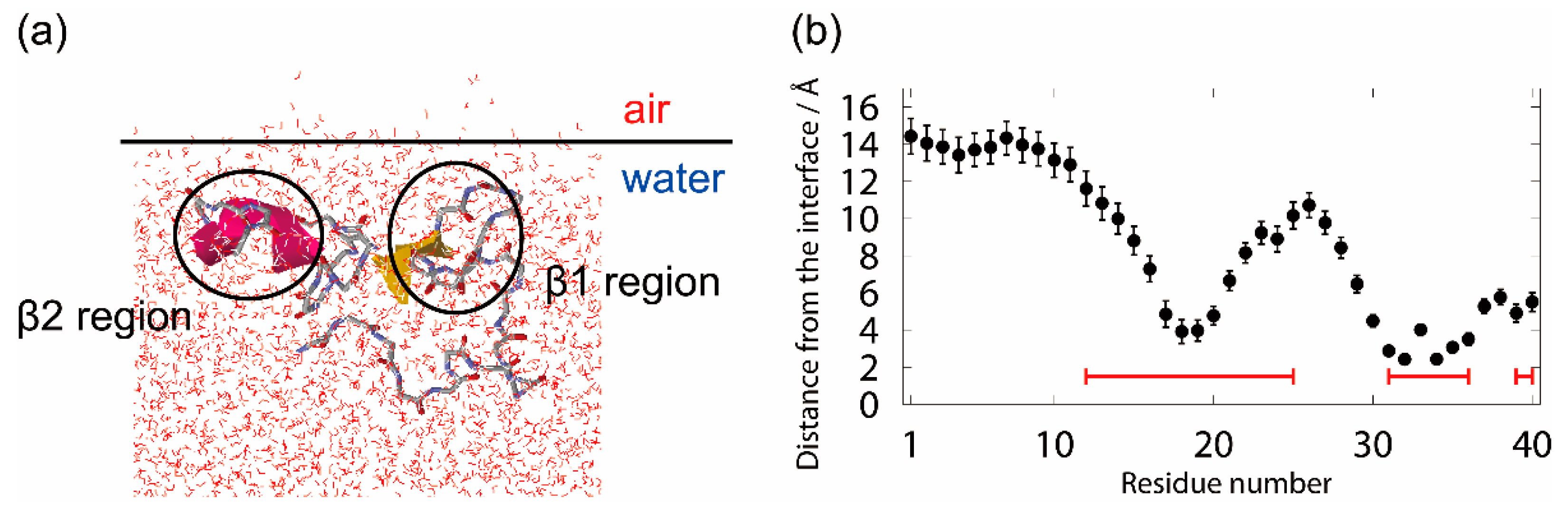
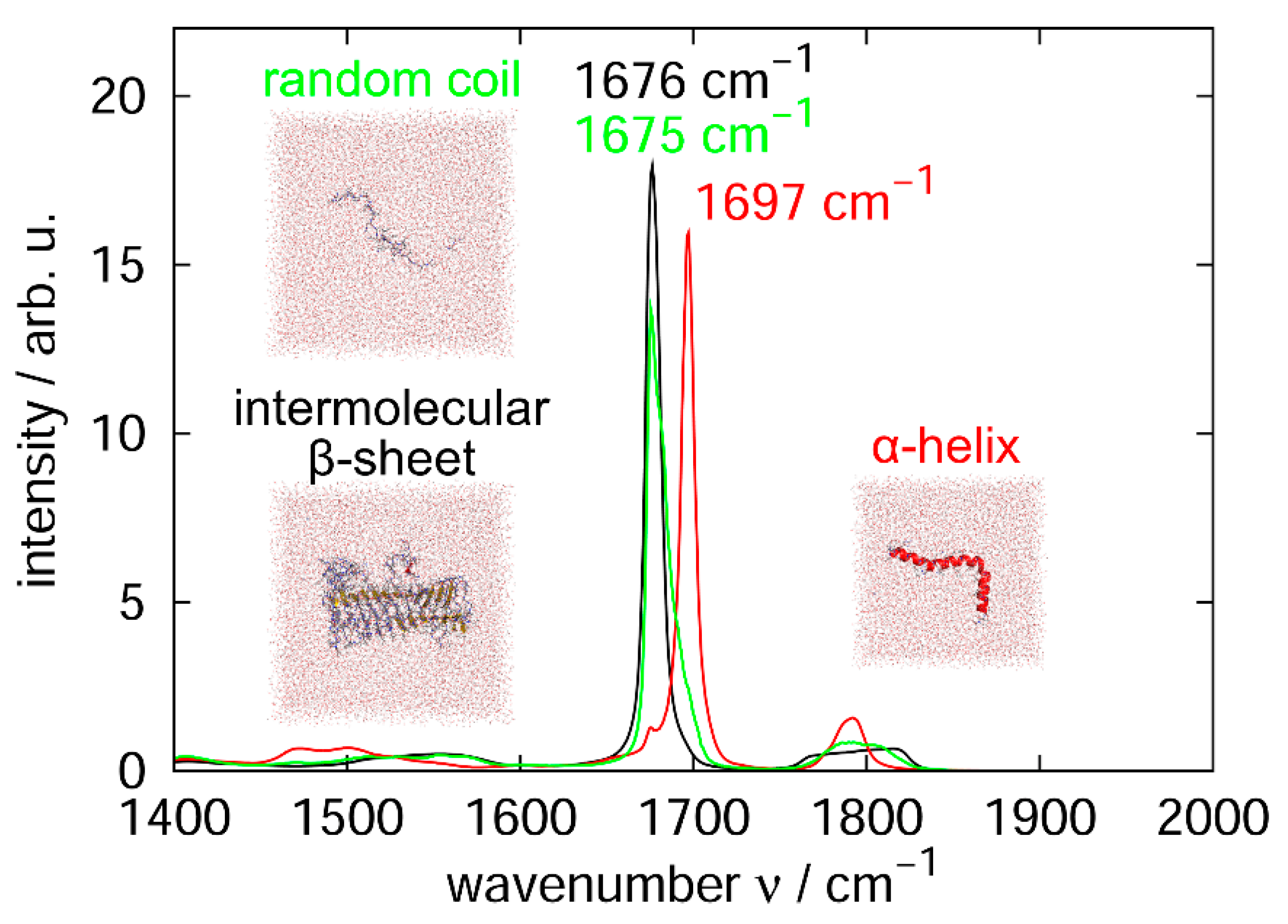
3.1. Molecular Dynamics Simulation of Aβ40 at the Air–Water Interface
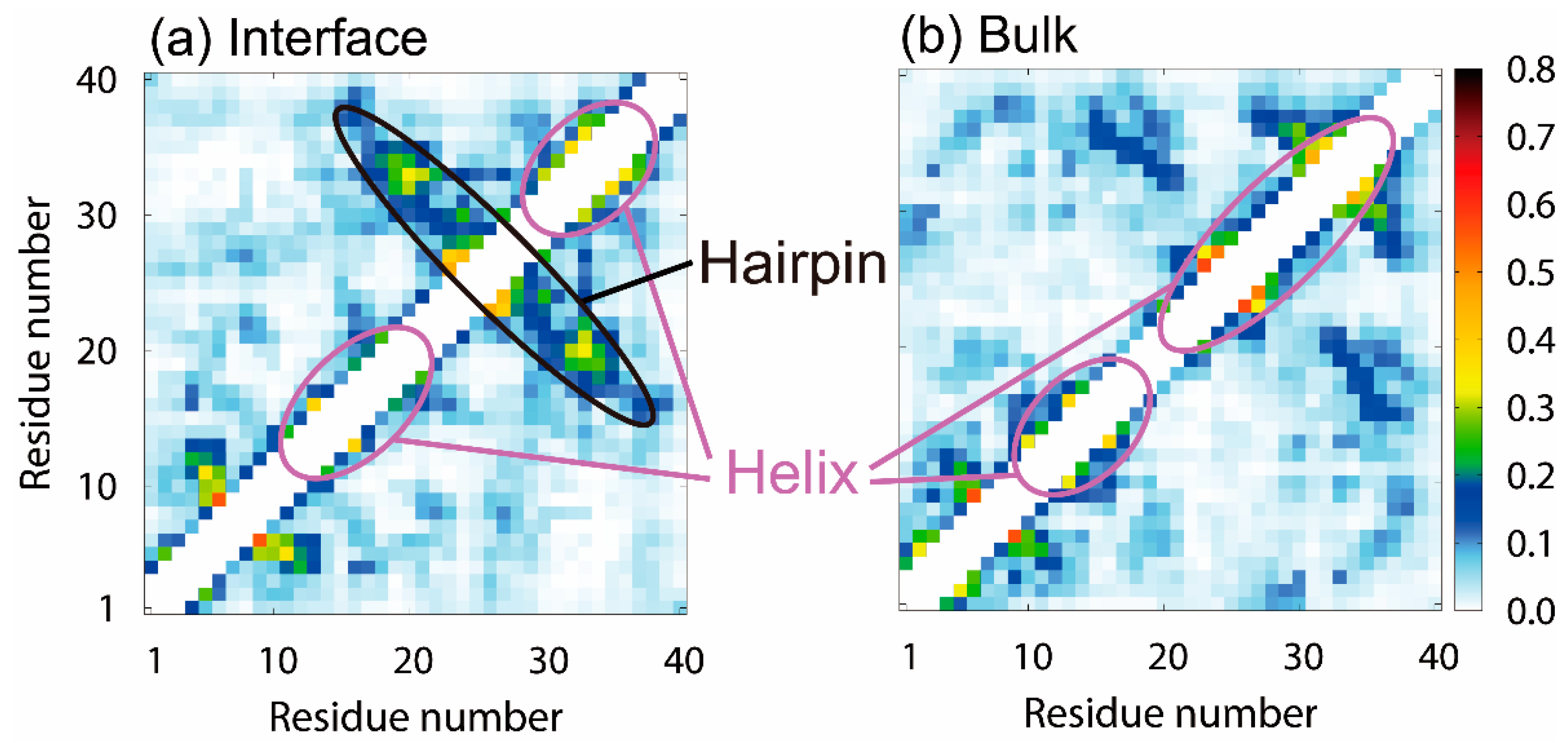
3.2. Molecular Structure of Aβ40 at the Air–Water Interface
4. Inhibitor against Aggregation of Aβ Peptides: Polyphenol
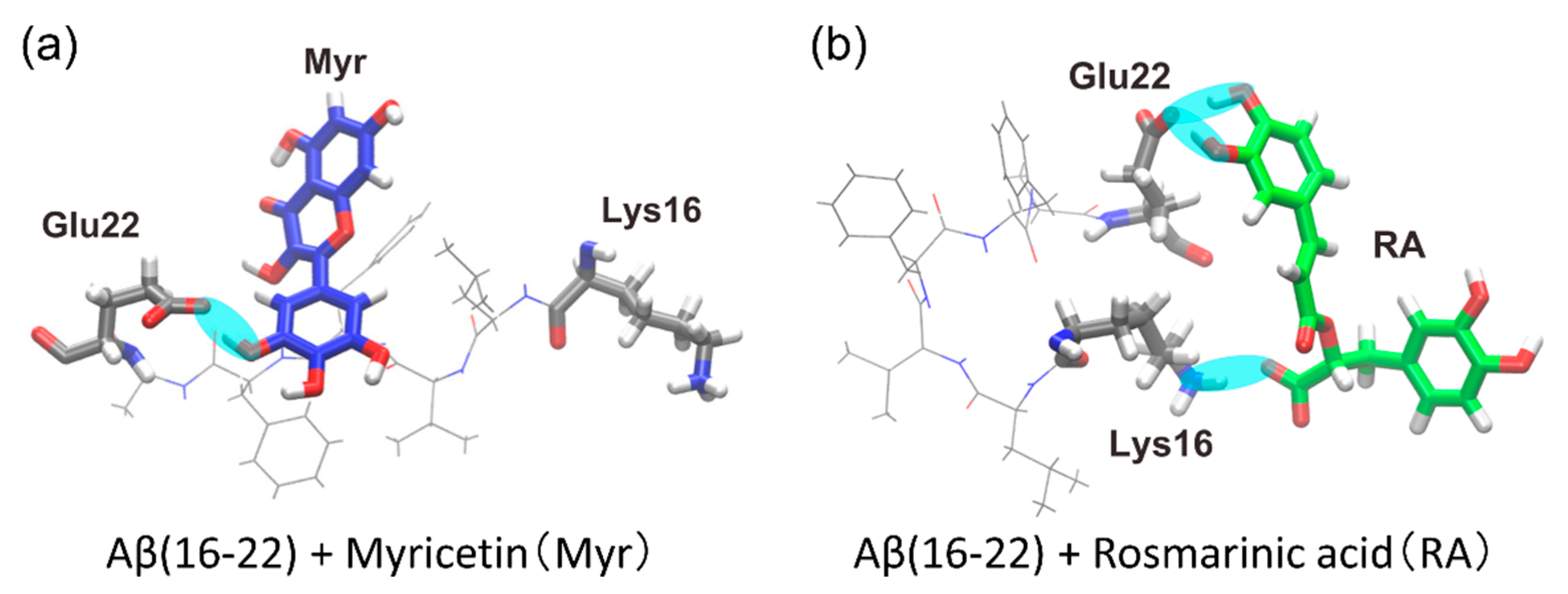
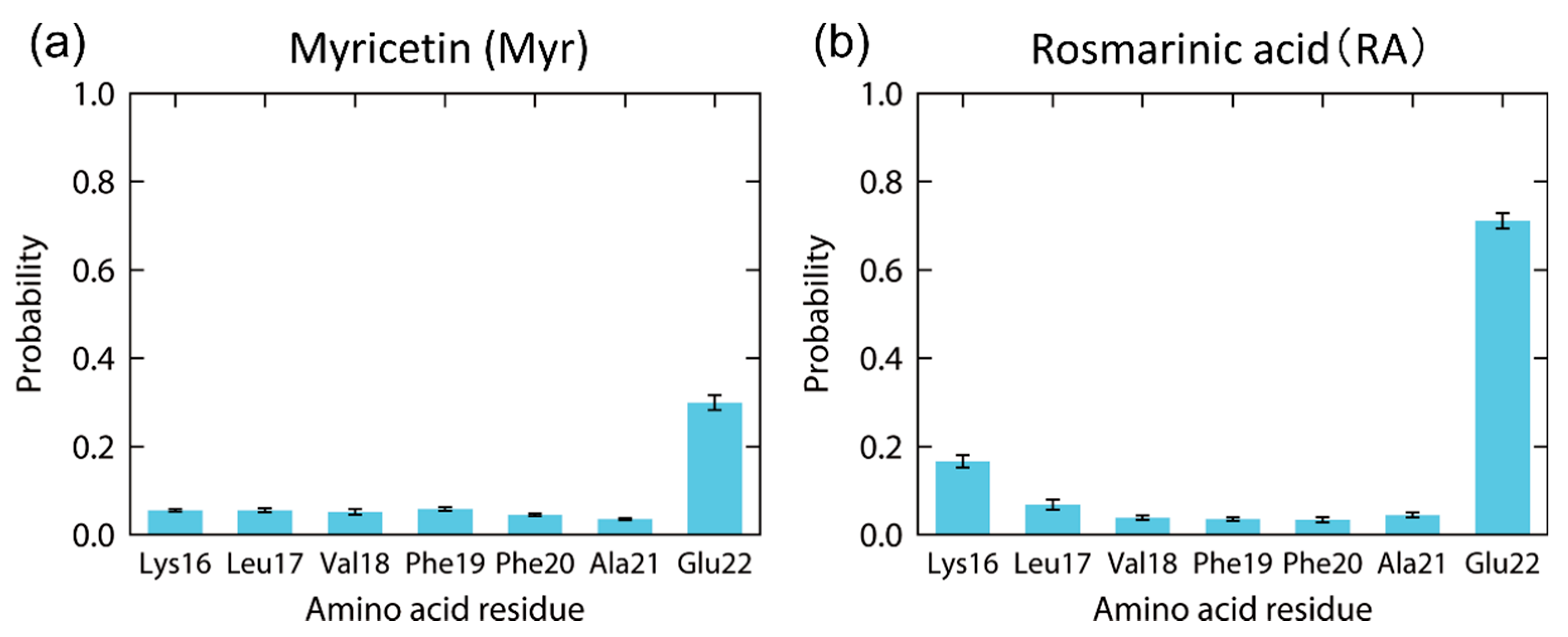
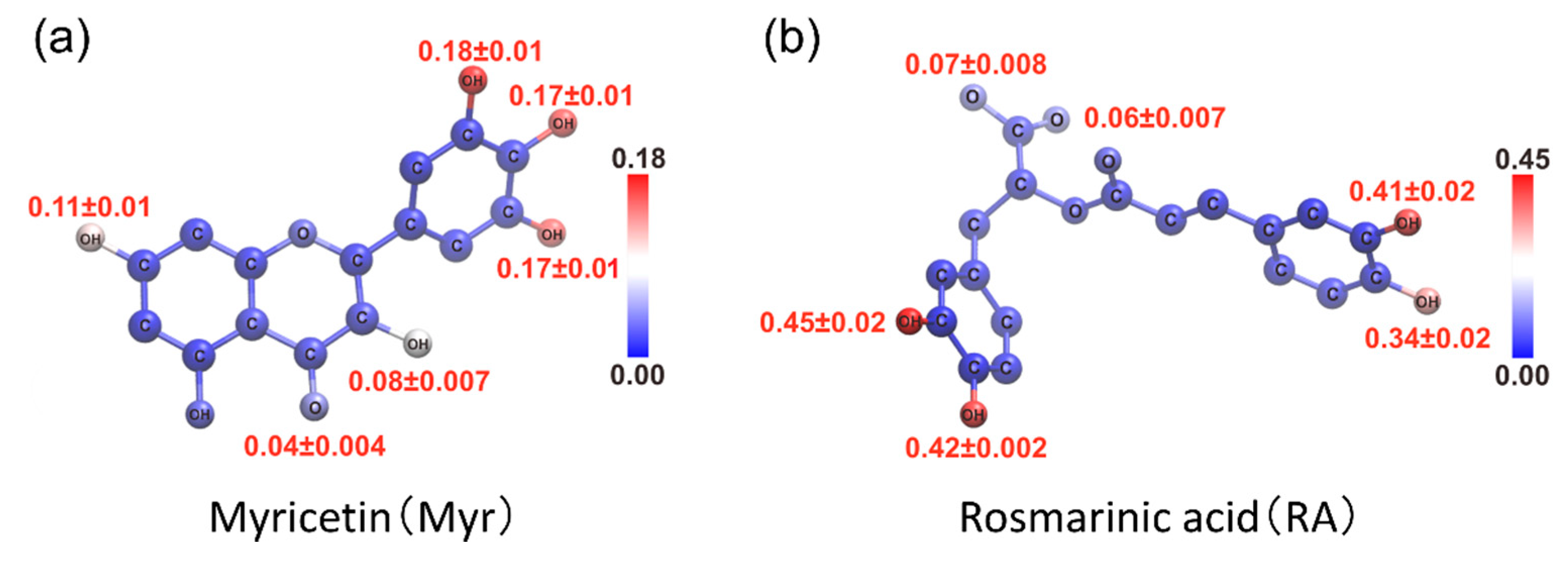
4.1. Replica-Permutation MD Simulation of an Aβ(16–22) Peptide and Polyphenols
4.2. Structure of the Complexes of an Aβ(16–22) Peptide and Polyphenols
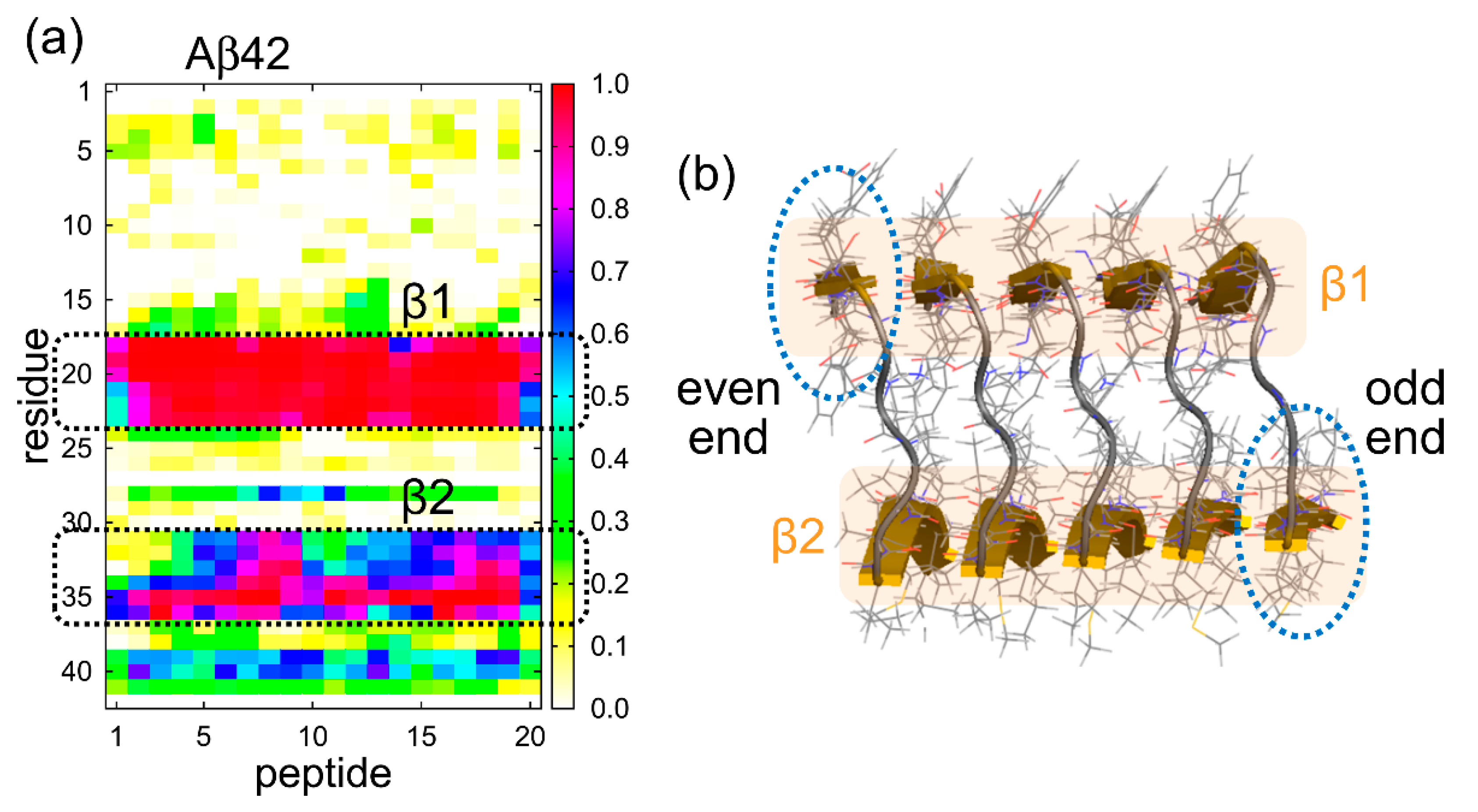
5. Structures of the Two Ends of the Aβ Amyloid Fibril
5.1. Molecular Dynamics Simulation of the Aβ Amyloid Fibril
5.2. Structure of Aβ Peptides at the Ends of the Aβ Amyloid Fibril
6. Amyloid Fibril Disruption by Ultrasonic Waves
6.1. Molecular Dynamics Simulation to Mimic Ultrasonic Waves
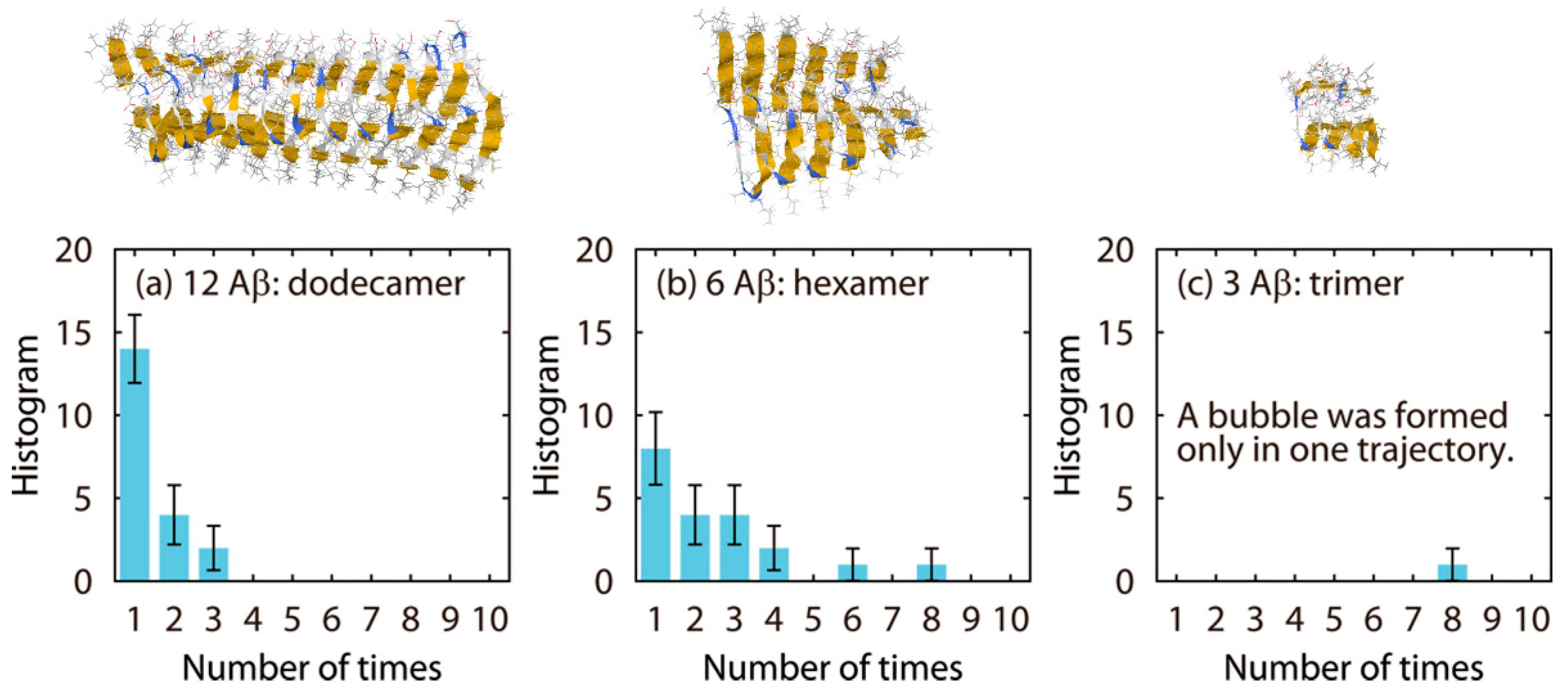
6.2. Disruption of Aβ Amyloid Fibril by Ultrasonic Waves
7. Laser-Induced Disruption of the Aβ Amyloid Fibril
7.1. Molecular Dynamics Simulation to Mimic Laser Irradiation
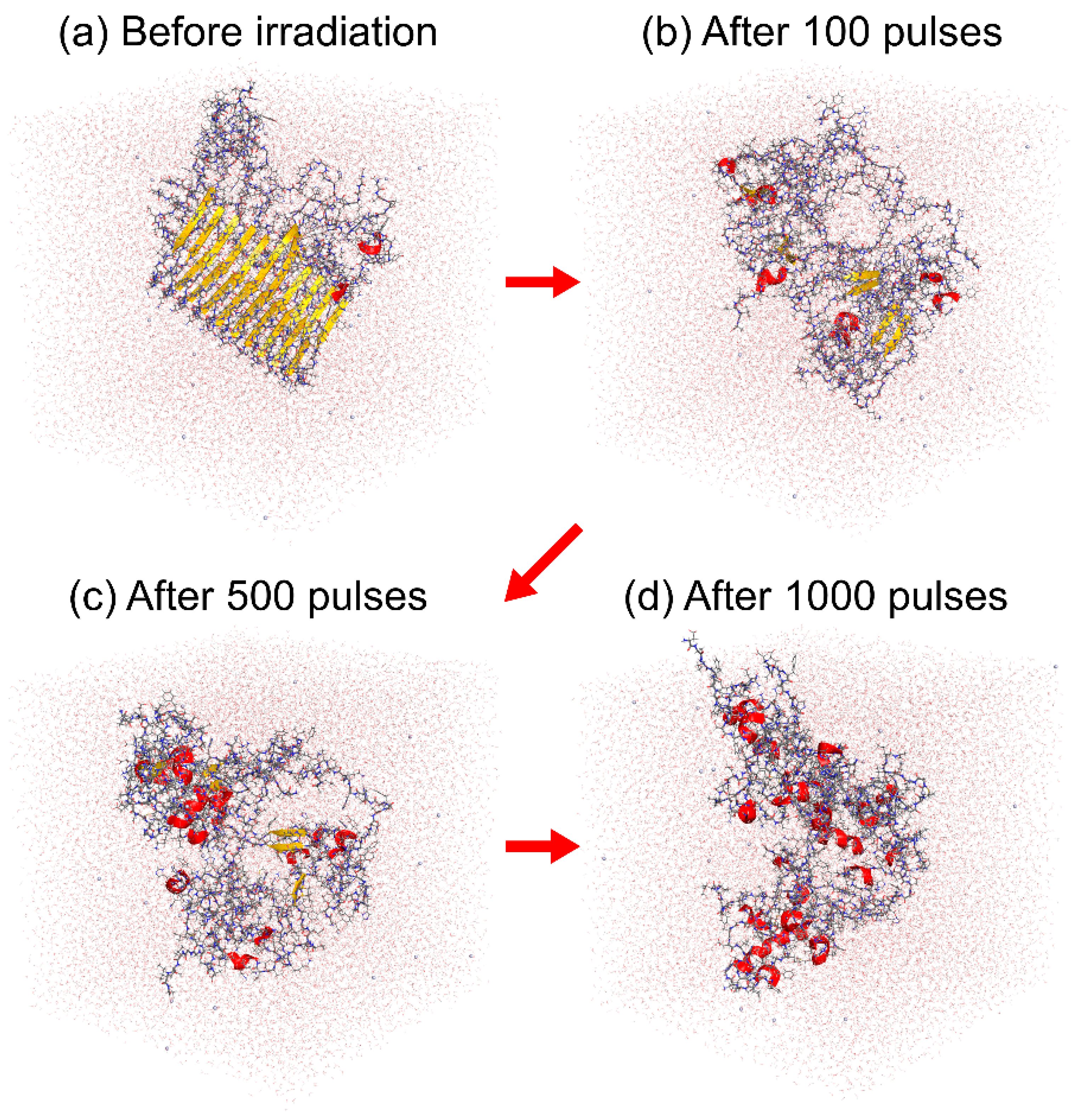
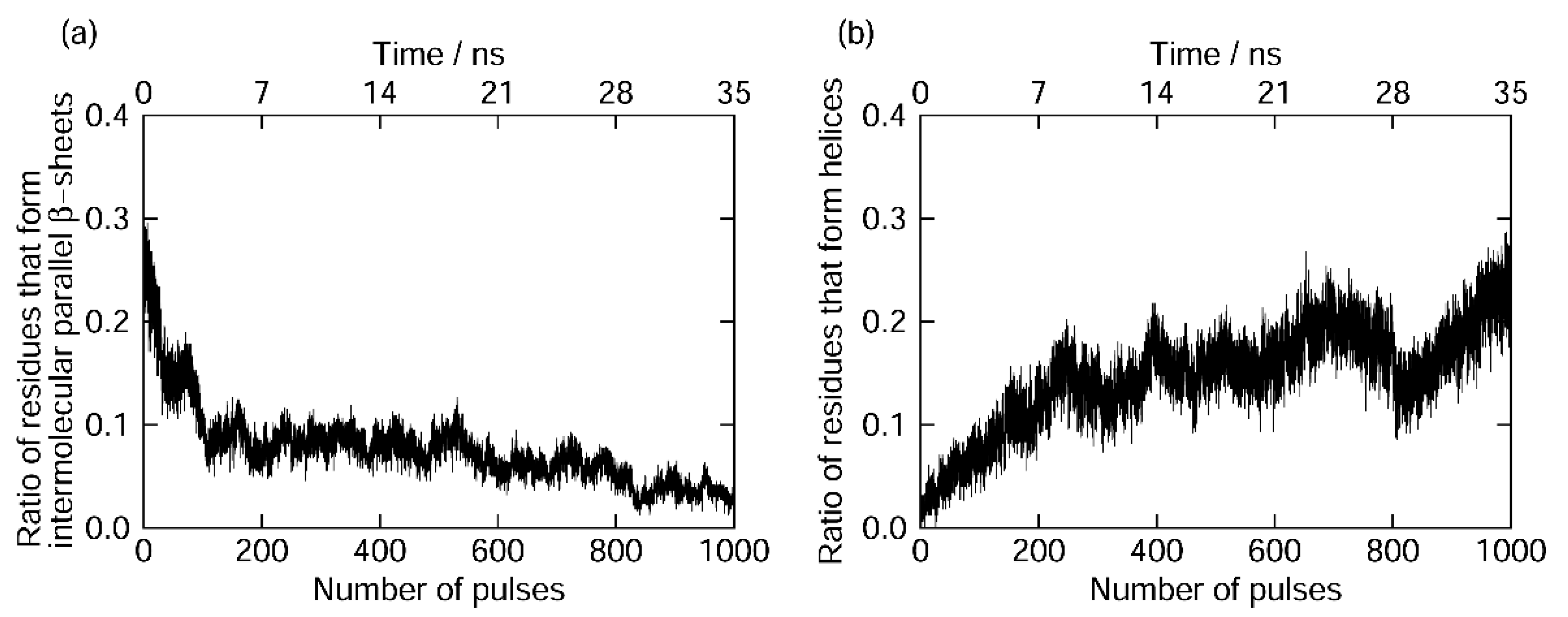
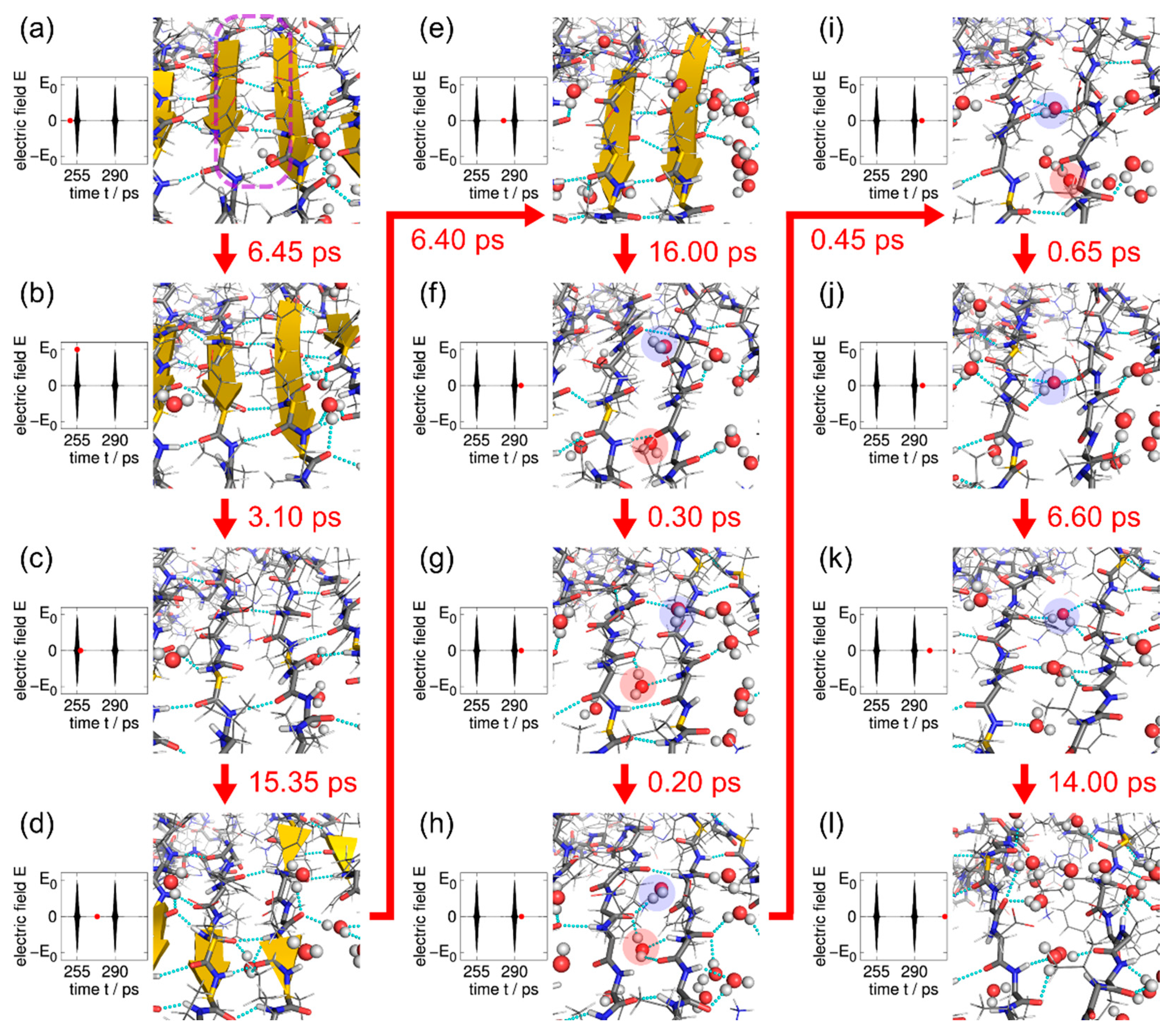
7.2. Amyloid Fibril Disruption by Laser Irradiation
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sipe, J.D.; Cohen, A.S. Review: History of the Amyloid Fibril. J. Struct. Biol. 2000, 130, 88–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tycko, R. Amyloid Polymorphism: Structural Basis and Neurobiological Relevance. Neuron 2015, 86, 632–645. [Google Scholar] [CrossRef] [Green Version]
- Post, F. The Clinical Psychiatry of Late Life; Pergamon Press: Oxford, UK, 1965. [Google Scholar]
- Tomlinson, B.; Blessed, G.; Roth, M. Observations on the brains of non-demented old people. J. Neurol. Sci. 1968, 7, 331–356. [Google Scholar] [CrossRef]
- Glenner, G.G.; Wong, C.W. Alzheimer’s disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 1984, 120, 885–890. [Google Scholar] [CrossRef]
- Masters, C.L.; Simms, G.; Weinman, N.A.; Multhaup, G.; McDonald, B.L.; Beyreuther, K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. USA 1985, 82, 4245–4249. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.; Lemaire, H.-G.; Unterbeck, A.; Salbaum, J.; Masters, C.; Grzeschik, K.-H.; Multhaup, G.; Beyreuther, K.; Müller-Hill, B. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Alzheimer Dis. Assoc. Disord. 1987, 1, 206–207. [Google Scholar] [CrossRef]
- Wetzel, R. Ideas of Order for Amyloid Fibril Structure. Structure 2002, 10, 1031–1036. [Google Scholar] [CrossRef] [Green Version]
- Petkova, A.T.; Ishii, Y.; Balbach, J.J.; Antzutkin, O.N.; Leapman, R.D.; Delaglio, F.; Tycko, R. A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. USA 2002, 99, 16742–16747. [Google Scholar] [CrossRef] [Green Version]
- Lührs, T.; Ritter, C.; Adrian, M.; Riek-Loher, D.; Bohrmann, B.; Dobeli, H.; Schubert, D.; Riek, R. 3D structure of Alzheimer’s amyloid-β(1-42) fibrils. Proc. Natl. Acad. Sci. USA 2005, 102, 17342–17347. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.X.; Qiang, W.; Yau, W.M.; Schwieters, C.D.; Meredith, S.C.; Tycko, R. Molecular Structure of β-Amyloid Fibrils in Alzheimer’s Disease Brain Tissue. Cell 2013, 154, 1257–1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.; Ma, B.; McElheny, D.; Parthasarathy, S.; Long, F.; Hoshi, M.; Nussinov, R.; Ishii, Y. Aβ(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat. Struct. Mol. Biol. 2015, 22, 499–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gremer, L.; Schölzel, D.; Schenk, C.; Reinartz, E.; Labahn, J.; Ravelli, R.B.G.; Tusche, M.; Lopez-Iglesias, C.; Hoyer, W.; Heise, H.; et al. Fibril Structure of Amyloid-β(1–42) by Cryoelectron Microscopy. Science 2017, 358, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sgourakis, N.G.; Yan, Y.; McCallum, S.A.; Wang, C.; Garcia, A.E. The Alzheimer’s Peptides Aβ40 and 42 Adopt Distinct Conformations in Water: A Combined MD / NMR Study. J. Mol. Biol. 2007, 368, 1448–1457. [Google Scholar] [CrossRef] [Green Version]
- Allison, J.R.; Varnai, P.; Dobson, C.M.; Vendruscolo, M. Determination of the Free Energy Landscape of α-Synuclein Using Spin Label Nuclear Magnetic Resonance Measurements. J. Am. Chem. Soc. 2009, 131, 18314–18326. [Google Scholar] [CrossRef]
- Vitalis, A.; Caflisch, A. Micelle-Like Architecture of the Monomer Ensemble of Alzheimer’s Amyloid-β Peptide in Aqueous Solution and Its Implications for Aβ Aggregation. J. Mol. Biol. 2010, 403, 148–165. [Google Scholar] [CrossRef]
- Sgourakis, N.G.; Merced-Serrano, M.; Boutsidis, C.; Drineas, P.; Du, Z.; Wang, C.; Garcia, A.E. Atomic-Level Characterization of the Ensemble of the Aβ(1–42) Monomer in Water Using Unbiased Molecular Dynamics Simulations and Spectral Algorithms. J. Mol. Biol. 2011, 405, 570. [Google Scholar] [CrossRef] [Green Version]
- Velez-Vega, C.; Escobedo, F.A. Characterizing the structural behavior of selected Aβ-42 monomers with different solubilities. J. Phys. Chem. B 2011, 115, 4900–4910. [Google Scholar] [CrossRef]
- Olubiyi, O.O.; Strodel, B. Structures of the amyloid β-peptides Aβ1–40 and Aβ1–42 as influenced by pH and a D-Peptide. J. Phys. Chem. B 2012, 116, 3280. [Google Scholar] [CrossRef]
- Ball, K.A.; Phillips, A.H.; Wemmer, D.E.; Head-Gordon, T. Differences in β-strand Populations of Monomeric Aβ40 and Aβ42. Biophys. J. 2013, 104, 2714–2724. [Google Scholar] [CrossRef] [Green Version]
- Itoh, S.G.; Okumura, H. Hamiltonian replica-permutation method and its applications to an alanine dipeptide and amyloid-β(29-42) peptides. J. Comput. Chem. 2013, 34, 2493–2497. [Google Scholar] [CrossRef] [PubMed]
- Itoh, S.G.; Okumura, H. Coulomb replica-exchange method: Handling electrostatic attractive and repulsive forces for biomolecules. J. Comput. Chem. 2013, 34, 622–639. [Google Scholar] [CrossRef] [PubMed]
- Rosenman, D.J.; Connors, C.R.; Chen, W.; Wang, C.; Garcia, A.E. Aβ Monomers Transiently Sample Oligomer and Fibril-Like Configurations: Ensemble Characterization Using a Combined MD/NMR Approach. J. Mol. Biol. 2013, 425, 3338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenman, D.J.; Wang, C.; García, A.E. Characterization of Aβ Monomers through the Convergence of Ensemble Properties among Simulations with Multiple Force Fields. J. Phys. Chem. B 2015, 120, 259–277. [Google Scholar] [CrossRef]
- Ilie, I.M.; Nayar, D.; den Otter, W.K.; van der Vegt, N.F.A.; Briels, W.J. Intrinsic Conformational Preferences and Interactions in α-Synuclein Fibrils: Insights from Molecular Dynamics Simulations. J. Chem. Theory Comput. 2018, 14, 3298–3310. [Google Scholar] [CrossRef]
- Mudedla, S.K.; Murugan, N.A.; Agren, H. Free Energy Landscape for α-Helix to β-Sheet Interconversion in Small Amyloid Forming Peptide under Nanoconfinement. J. Phys. Chem. B 2018, 122, 9654–9664. [Google Scholar] [CrossRef]
- Meng, F.; Bellaiche, M.M.; Kim, J.-Y.; Zerze, G.H.; Best, R.B.; Chung, H.S. Highly Disordered Amyloid-β Monomer Probed by Single-Molecule FRET and MD Simulation. Biophys. J. 2018, 114, 870–884. [Google Scholar] [CrossRef] [Green Version]
- Tachi, Y.; Okamoto, Y.; Okumura, H. Conformational Change of Amyloid-β 40 in Association with Binding to GM1-Glycan Cluster. Sci. Rep. 2019, 9, 6853. [Google Scholar] [CrossRef] [Green Version]
- Itoh, S.G.; Yagi-Utsumi, M.; Kato, K.; Okumura, H. Effects of a Hydrophilic/Hydrophobic Interface on Amyloid-β Peptides Studied by Molecular Dynamics Simulations and NMR Experiments. J. Phys. Chem. B 2018, 123, 160–169. [Google Scholar] [CrossRef]
- Ngoc, L.L.N.; Itoh, S.G.; Sompornpisut, P.; Okumura, H. Replica-permutation molecular dynamics simulations of an amyloid-β(16–22) peptide and polyphenols. Chem. Phys. Lett. 2020, 758, 137913. [Google Scholar] [CrossRef]
- Tavanti, F.; Pedone, A.; Menziani, M.C. Disclosing the Interaction of Gold Nanoparticles with Aβ(1–40) Monomers through Replica Exchange Molecular Dynamics Simulations. Int. J. Mol. Sci. 2020, 22, 26. [Google Scholar] [CrossRef] [PubMed]
- Chebaro, Y.; Mousseau, N.; Derreumaux, P. Structures and thermodynamics of Alzheimer’s amyloid-β Aβ(16-35) monomer and dimer by replica exchange molecular dynamics simulations: Implication for full-length Aβ fibrillation. J. Phys. Chem. B 2009, 113, 7668–7675. [Google Scholar] [CrossRef] [PubMed]
- Cote, S.; Laghaei, R.; Derreumaux, P.; Mousseau, N. Distinct dimerization for various alloforms of the amyloid-β protein: Aβ(1-40), Aβ(1-42), and Aβ(1-40)(D23N). J. Phys. Chem. B 2012, 116, 4043–4055. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.L.; Chen, C.J.; Okumura, H.; Hu, C.K. Transformation between α-helix and β-sheet structures of one and two polyglutamine peptides in explicit water molecules by replica-exchange molecular dynamics simulations. J. Comput. Chem. 2014, 35, 1430–1437. [Google Scholar] [CrossRef]
- Itoh, S.G.; Okumura, H. Dimerization process of amyloid-β(29-42) studied by the Hamiltonian replica-permutation molecular dynamics simulations. J. Phys. Chem. B 2014, 118, 11428–11436. [Google Scholar] [CrossRef]
- Nguyen, P.H.; Sterpone, F.; Campanera, J.M.; Nasica-Labouze, J.; Derreumaux, P. Impact of the A2V Mutation on the Heterozygous and Homozygous Aβ1–40 Dimer Structures from Atomistic Simulations. ACS Chem. Neurosci. 2016, 7, 823–832. [Google Scholar] [CrossRef]
- Tarus, B.; Tran, T.T.; Nasica-Labouze, J.; Sterpone, F.; Nguyen, P.H.; Derreumaux, P. Structures of the Alzheimer’s Wild-Type Aβ1-40 Dimer from Atomistic Simulations. J. Phys. Chem. B 2015, 119, 10478–10487. [Google Scholar] [CrossRef]
- Nguyen, P.H.; Sterpone, F.; Pouplana, R.; Derreumaux, P.; Campanera, J.M. Dimerization Mechanism of Alzheimer Aβ40 Peptides: The High Content of Intrapeptide-Stabilized Conformations in A2V and A2T Heterozygous Dimers Retards Amyloid Fibril Formation. J. Phys. Chem. B 2016, 120, 12111–12126. [Google Scholar] [CrossRef]
- Das, P.; Chacko, A.R.; Belfort, G. Alzheimer’s Protective Cross-Interaction between Wild-Type and A2T Variants Alters Aβ42 Dimer Structure. ACS Chem. Neurosci. 2016, 8, 606–618. [Google Scholar] [CrossRef]
- Man, V.H.; Nguyen, P.H.; Derreumaux, P. Conformational Ensembles of the Wild-Type and S8C Aβ1-42 Dimers. J. Phys. Chem. B 2017, 121, 2434–2442. [Google Scholar] [CrossRef] [Green Version]
- Man, V.H.; Nguyen, P.H.; Derreumaux, P. High-Resolution Structures of the Amyloid-β 1–42 Dimers from the Comparison of Four Atomistic Force Fields. J. Phys. Chem. B 2017, 121, 5977–5987. [Google Scholar] [CrossRef] [Green Version]
- Sharma, B.; Ranganathan, S.V.; Belfort, G. Weaker N-Terminal Interactions for the Protective over the Causative Aβ Peptide Dimer Mutants. ACS Chem. Neurosci. 2018, 9, 1247–1253. [Google Scholar] [CrossRef]
- Nishizawa, H.; Okumura, H. Classical Molecular Dynamics Simulation to Understand Role of a Zinc Ion for Aggregation of Amyloid-β Peptides. J. Comput. Chem. Jpn. 2018, 17, 76–79. [Google Scholar] [CrossRef] [Green Version]
- Kargar, F.; Emadi, S.; Fazli, H. Dimerization of Aβ40 inside dipalmitoylphosphatidylcholine bilayer and its effect on bilayer integrity: Atomistic simulation at three temperatures. Proteins Struct. Funct. Bioinform. 2020, 88, 1540–1552. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, M.; Okumura, H. Dimerization of α-Synuclein Fragments Studied by Isothermal–Isobaric Replica-Permutation Molecular Dynamics Simulation. J. Chem. Inf. Model. 2021, 61, 1307–1321. [Google Scholar] [CrossRef] [PubMed]
- Gsponer, J.; Haberthür, U.; Caflisch, A. The role of side-chain interactions in the early steps of aggregation: Molecular dynamics simulations of an amyloid-forming peptide from the yeast prion Sup35. Proc. Natl. Acad. Sci. USA 2003, 100, 5154–5159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbanc, B.; Betnel, M.; Cruz, L.; Bitan, G.; Teplow, D.B. Elucidation of Amyloid β-Protein Oligomerization Mechanisms: Discrete Molecular Dynamics Study. J. Am. Chem. Soc. 2010, 132, 4266–4280. [Google Scholar] [CrossRef] [Green Version]
- Pacheco, M.C.; Ismail, A.E.; Strodel, B. Oligomer Formation of Toxic and Functional Amyloid Peptides Studied with Atomistic Simulations. J. Phys. Chem. B 2015, 119, 9696–9705. [Google Scholar] [CrossRef]
- Itoh, S.G.; Okumura, H. Oligomer Formation of Amyloid-β(29-42) from Its Monomers Using the Hamiltonian Replica-Permutation Molecular Dynamics Simulation. J. Phys. Chem. B 2016, 120, 6555–6561. [Google Scholar] [CrossRef]
- Barz, B.; Liao, Q.; Strodel, B. Pathways of Amyloid-β Aggregation Depend on Oligomer Shape. J. Am. Chem. Soc. 2017, 140, 319–327. [Google Scholar] [CrossRef]
- Sun, Y.; Ge, X.; Xing, Y.; Wang, B.; Ding, F. β-barrel Oligomers as Common Intermediates of Peptides Self-Assembling into Cross-β Aggregates. Sci. Rep. 2018, 8, 10353. [Google Scholar] [CrossRef] [Green Version]
- Okumura, H.; Itoh, S.G. Molecular dynamics simulations of amyloid-β(16–22) peptide aggregation at air–water interfaces. J. Chem. Phys. 2020, 152, 95101. [Google Scholar] [CrossRef] [PubMed]
- Chrobak, W.; Pacut, D.W.; Blomgren, F.; Rodin, A.; Swenson, J.; Ermilova, I. Component of Cannabis, Cannabidiol, as a Possible Drug against the Cytotoxicity of Aβ(31–35) and Aβ(25–35) Peptides: An Investigation by Molecular Dynamics and Well-Tempered Metadynamics Simulations. ACS Chem. Neurosci. 2021, 12, 660–674. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.H.; Li, M.S.; Stock, G.; Straub, J.E.; Thirumalai, D. Monomer adds to preformed structured oligomers of Aβ-peptides by a two-stage dock–lock mechanism. Proc. Natl. Acad. Sci. USA 2007, 104, 111–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, E.P.; Okamoto, Y.; Straub, J.E.; Brooks, B.R.; Thirumalai, D. Thermodynamic Perspective on the Dock−Lock Growth Mechanism of Amyloid Fibrils. J. Phys. Chem. B 2009, 113, 14421–14430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, T.; Klimov, D.K. Probing Energetics of Aβ Fibril Elongation by Molecular Dynamics Simulations. Biophys. J. 2009, 96, 4428–4437. [Google Scholar] [CrossRef] [Green Version]
- Takeda, T.; Klimov, D.K. Replica Exchange Simulations of the Thermodynamics of Aβ Fibril Growth. Biophys. J. 2009, 96, 442–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, A.S.; Wang, L.; Singh, S.; Ling, Y.L.; Buchanan, L.; Zanni, M.T.; Skinner, J.L.; de Pablo, J.J. Stable and Metastable States of Human Amylin in Solution. Biophys. J. 2010, 99, 2208–2216. [Google Scholar] [CrossRef] [Green Version]
- Han, M.; Hansmann, U.H. Replica exchange molecular dynamics of the thermodynamics of fibril growth of Alzheimer’s Aβ42 peptide. J. Chem. Phys. 2011, 135, 65101. [Google Scholar] [CrossRef] [Green Version]
- Straub, J.E.; Thirumalai, D. Toward a Molecular Theory of Early and Late Events in Monomer to Amyloid Fibril Formation. Annu. Rev. Phys. Chem. 2011, 62, 437–463. [Google Scholar] [CrossRef] [Green Version]
- Gurry, T.; Stultz, C.M. Mechanism of Amyloid-β Fibril Elongation. Biochemistry 2014, 53, 6981–6991. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Schulten, K. Fibril Elongation by Aβ17–42: Kinetic Network Analysis of Hybrid-Resolution Molecular Dynamics Simulations. J. Am. Chem. Soc. 2014, 136, 12450–12460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwierz, N.; Frost, C.V.; Geissler, P.L.; Zacharias, M. Dynamics of Seeded Aβ40-Fibril Growth from Atomistic Molecular Dynamics Simulations: Kinetic Trapping and Reduced Water Mobility in the Locking Step. J. Am. Chem. Soc. 2016, 138, 527–539. [Google Scholar] [CrossRef]
- Sasmal, S.; Schwierz, N.; Head-Gordon, T. Mechanism of Nucleation and Growth of Aβ40 Fibrils from All-Atom and Coarse-Grained Simulations. J. Phys. Chem. B 2016, 120, 12088–12097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacci, M.; Vymětal, J.; Mihajlovic, M.; Caflisch, A.; Vitalis, A. Amyloid β Fibril Elongation by Monomers Involves Disorder at the Tip. J. Chem. Theory Comput. 2017, 13, 5117–5130. [Google Scholar] [CrossRef] [PubMed]
- Ilie, I.M.; Otter, W.K.D.; Briels, W.J. The attachment of α-synuclein to a fiber: A coarse-grain approach. J. Chem. Phys. 2017, 146, 115102. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Ding, F. αB-Crystallin Chaperone Inhibits Aβ Aggregation by Capping the β-Sheet-Rich Oligomers and Fibrils. J. Phys. Chem. B 2020, 124, 10138–10146. [Google Scholar] [CrossRef]
- Buchete, N.V.; Tycko, R.; Hummer, G. Molecular dynamics simulations of Alzheimer’s β-amyloid protofilaments. J. Mol. Biol. 2005, 353, 804–821. [Google Scholar] [CrossRef]
- Baumketner, A.; Krone, M.G.; Shea, J.-E. Role of the familial Dutch mutation E22Q in the folding and aggregation of the 15–28 fragment of the Alzheimer amyloid-β protein. Proc. Natl. Acad. Sci. USA 2008, 105, 6027–6032. [Google Scholar] [CrossRef] [Green Version]
- Lemkul, J.A.; Bevan, D.R. Assessing the Stability of Alzheimer’s Amyloid Protofibrils Using Molecular Dynamics. J. Phys. Chem. B 2010, 114, 1652–1660. [Google Scholar] [CrossRef]
- Okumura, H.; Itoh, S.G. Structural and fluctuational difference between two ends of Aβ amyloid fibril: MD simulations predict only one end has open conformations. Sci. Rep. 2016, 6, 38422. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.A.; Chen, L.Y.; Plascencia-Villa, G.; Perry, G. Thermodynamics of Amyloid-β Fibril Elongation: Atomistic Details of the Transition State. ACS Chem. Neurosci. 2017, 9, 783–789. [Google Scholar] [CrossRef] [PubMed]
- Davidson, D.S.; Brown, A.M.; Lemkul, J.A. Insights into Stabilizing Forces in Amyloid Fibrils of Differing Sizes from Polarizable Molecular Dynamics Simulations. J. Mol. Biol. 2018, 430, 3819. [Google Scholar] [CrossRef] [PubMed]
- Ilie, I.-M.; Caflisch, A. Disorder at the Tips of a Disease-Relevant Aβ42 Amyloid Fibril: A Molecular Dynamics Study. J. Phys. Chem. B 2018, 122, 11072–11082. [Google Scholar] [CrossRef] [PubMed]
- Nirmalraj, P.N.; List, J.; Battacharya, S.; Howe, G.; Xu, L.; Thompson, D.; Mayer, M. Complete aggregation pathway of amyloid β (1-40) and (1-42) resolved on an atomically clean interface. Sci. Adv. 2020, 6, eaaz6014. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Dasmahapatra, A.K. Destabilization potential of phenolics on Aβ fibrils: Mechanistic insights from molecular dynamics simulation. Phys. Chem. Chem. Phys. 2020, 22, 19643–19658. [Google Scholar] [CrossRef]
- Poma, A.B.; Thu, T.T.M.; Tri, L.T.M.; Nguyen, H.L.; Li, M.S. Nanomechanical Stability of Aβ Tetramers and Fibril-like Structures: Molecular Dynamics Simulations. J. Phys. Chem. B 2021, 125, 7628–7637. [Google Scholar] [CrossRef]
- Jahan, I.; Nayeem, S.M. Destabilization of Alzheimer’s Aβ42 protofibrils with acyclovir, carmustine, curcumin, and tetracycline: Insights from molecular dynamics simulations. New J. Chem. 2021, 45, 21031–21048. [Google Scholar] [CrossRef]
- Okumura, H.; Itoh, S.G. Amyloid Fibril Disruption by Ultrasonic Cavitation: Nonequilibrium Molecular Dynamics Simulations. J. Am. Chem. Soc. 2014, 136, 10549–10552. [Google Scholar] [CrossRef]
- Viet, M.; Derreumaux, P.; Nguyen, P.H. Nonequilibrium all-atom molecular dynamics simulation of the bubble cavitation and application to dissociate amyloid fibrils. J. Chem. Phys. 2016, 145, 174113. [Google Scholar] [CrossRef] [Green Version]
- Viet, M.; Derreumaux, P.; Li, M.S.; Roland, C.; Sagui, C.; Nguyen, P.H. Picosecond dissociation of amyloid fibrils with infrared laser: A nonequilibrium simulation study. J. Chem. Phys. 2015, 143, 155101. [Google Scholar] [CrossRef] [PubMed]
- Brogi, S.; Sirous, H.; Calderone, V.; Chemi, G. Amyloid β fibril disruption by oleuropein aglycone: Long-time molecular dynamics simulation to gain insight into the mechanism of action of this polyphenol from extra virgin olive oil. Food Funct. 2020, 11, 8122–8132. [Google Scholar] [CrossRef] [PubMed]
- Okumura, H.; Itoh, S.G.; Nakamura, K.; Kawasaki, T. Role of Water Molecules and Helix Structure Stabilization in the Laser-Induced Disruption of Amyloid Fibrils Observed by Nonequilibrium Molecular Dynamics Simulations. J. Phys. Chem. B 2021, 125, 4964–4976. [Google Scholar] [CrossRef] [PubMed]
- Nasica-Labouze, J.; Nguyen, P.H.; Sterpone, F.; Berthoumieu, O.; Buchete, N.-V.; Coté, S.; De Simone, A.; Doig, A.J.; Faller, P.; Garcia, A.; et al. Amyloid β Protein and Alzheimer’s Disease: When Computer Simulations Complement Experimental Studies. Chem. Rev. 2015, 115, 3518–3563. [Google Scholar] [CrossRef] [PubMed]
- Ilie, I.M.; Caflisch, A. Simulation Studies of Amyloidogenic Polypeptides and Their Aggregates. Chem. Rev. 2019, 119, 6956–6993. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.H.; Derreumaux, P. Structures of the intrinsically disordered Aβ, tau and α-synuclein proteins in aqueous solution from computer simulations. Biophys. Chem. 2020, 264, 106421. [Google Scholar] [CrossRef]
- Strodel, B. Amyloid aggregation simulations: Challenges, advances and perspectives. Curr. Opin. Struct. Biol. 2020, 67, 145–152. [Google Scholar] [CrossRef]
- Jarrett, J.T.; Berger, E.P.; Lansbury, P.T. The carboxy terminus of the β amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer’s disease. Biochemistry 1993, 32, 4693–4697. [Google Scholar] [CrossRef]
- Hilbich, C.; Kisterswoike, B.; Reed, J.; Masters, C.L.; Beyreuther, K. Aggregation and secondary structure of synthetic amyloid βA4 peptides of Alzheimer’s disease. J. Mol. Biol. 1991, 218, 149–163. [Google Scholar] [CrossRef]
- Barrow, C.J.; Yasuda, A.; Kenny, P.T.M.; Zagorski, M.G. Solution conformations and aggregational properties of synthetic amyloid β-peptides of Alzheimer’s disease: Analysis of circular dichroism spectra. J. Mol. Biol. 1992, 225, 1075–1093. [Google Scholar] [CrossRef]
- Serpell, L.C. Alzheimer’s amyloid fibrils: Structure and assembly. Biochim. Biophys. Acta 2000, 1502, 16–30. [Google Scholar] [CrossRef] [Green Version]
- Balbach, J.J.; Ishii, Y.; Antzutkin, O.N.; Leapman, R.D.; Rizzo, N.W.; Dyda, F.; Reed, J.; Tycko, R. Amyloid Fibril Formation by Aβ16-22, a Seven-Residue Fragment of the Alzheimer’s β-Amyloid Peptide, and Structural Characterization by Solid State NMR. Biochemistry 2000, 39, 13748–13759. [Google Scholar] [CrossRef] [PubMed]
- Haass, C.; Selkoe, D.J. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 2007, 8, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Shankar, G.M.; Li, S.M.; Mehta, T.H.; Garcia-Munoz, A.; Shepardson, N.E.; Smith, I.; Brett, F.M.; Farrell, M.A.; Rowan, M.J.; Lemere, C.A.; et al. Amyloid-β protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat. Med. 2008, 14, 837–842. [Google Scholar] [CrossRef] [Green Version]
- Itoh, S.G.; Okamoto, Y. Amyloid-β(29−42) Dimer Formations Studied by a Multicanonical−Multioverlap Molecular Dynamics Simulation. J. Phys. Chem. B 2008, 112, 2767–2770. [Google Scholar] [CrossRef]
- Itoh, S.G.; Okumura, H. Replica-Permutation Method with the Suwa–Todo Algorithm beyond the Replica-Exchange Method. J. Chem. Theory Comput. 2012, 9, 570–581. [Google Scholar] [CrossRef] [Green Version]
- Mitsutake, A.; Sugita, Y.; Okamoto, Y. Generalized-ensemble algorithms for molecular simulations of biopolymers. Biopolymers 2001, 60, 96–123. [Google Scholar] [CrossRef]
- Itoh, S.G.; Okumura, H.; Okamoto, Y. Generalized-ensemble algorithms for molecular dynamics simulations. Mol. Simul. 2007, 33, 47–56. [Google Scholar] [CrossRef]
- Yamauchi, M.; Mori, Y.; Okumura, H. Molecular simulations by generalized-ensemble algorithms in isothermal–isobaric ensemble. Biophys. Rev. 2019, 11, 457–469. [Google Scholar] [CrossRef]
- Itoh, S.G.; Okumura, H. All-Atom Molecular Dynamics Simulation Methods for the Aggregation of Protein and Peptides: Replica Exchange/Permutation and Nonequilibrium Simulations. In Computer Simulations of Aggregation of Proteins and Peptides; Li, M.S., Kloczkowski, A., Cieplak, M., Kouza, M., Eds.; Humana: New York, NY, USA, 2022; pp. 197–220. [Google Scholar]
- Hukushima, K.; Nemoto, K. Exchange Monte Carlo Method and Application to Spin Glass Simulations. J. Phys. Soc. Jpn. 1996, 65, 1604–1608. [Google Scholar] [CrossRef] [Green Version]
- Sugita, Y.; Okamoto, Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999, 314, 141–151. [Google Scholar] [CrossRef]
- Suwa, H.; Todo, S. Markov Chain Monte Carlo Method without Detailed Balance. Phys. Rev. Lett. 2010, 105, 120603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metropolis, N.; Rosenbluth, A.W.; Rosenbluth, M.N.; Teller, A.H.; Teller, E. Equation of State Calculations by Fast Computing Machines. J. Chem. Phys. 1953, 21, 1087–1092. [Google Scholar] [CrossRef] [Green Version]
- Yamauchi, M.; Okumura, H. Replica sub-permutation method for molecular dynamics and Monte Carlo simulations. J. Comput. Chem. 2019, 40, 2694–2711. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, M.; Okumura, H. Development of isothermal-isobaric replica-permutation method for molecular dynamics and Monte Carlo simulations and its application to reveal temperature and pressure dependence of folded, misfolded, and unfolded states of chignolin. J. Chem. Phys. 2017, 147, 184107. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, H.; Okumura, H. Comparison of Replica-Permutation Molecular Dynamics Simulations with and without Detailed Balance Condition. J. Phys. Soc. Jpn. 2015, 84, 74801. [Google Scholar] [CrossRef]
- Mori, Y.; Okumura, H. Simulated tempering based on global balance or detailed balance conditions: Suwa-Todo, heat bath, and Metropolis algorithms. J. Comput. Chem. 2015, 36, 2344–2349. [Google Scholar] [CrossRef]
- Fukuhara, D.; Itoh, S.G.; Okumura, H. Replica permutation with solute tempering for molecular dynamics simulation and its application to the dimerization of amyloid-β fragments. J. Chem. Phys. 2022, 156, 84109. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins Struct. Funct. Bioinform. 2006, 65, 712–725. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511–519. [Google Scholar] [CrossRef] [Green Version]
- Nosé, S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984, 52, 255–268. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 1985, 31, 1695–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abelein, A.; Abrahams, J.P.; Danielsson, J.; Gräslund, A.; Jarvet, J.; Luo, J.; Tiiman, A.; Wärmländer, S.K.T.S. The hairpin conformation of the amyloid β peptide is an important structural motif along the aggregation pathway. JBIC J. Biol. Inorg. Chem. 2014, 19, 623–634. [Google Scholar] [CrossRef]
- Maity, S.; Hashemi, M.; Lyubchenko, Y.L. Nano-assembly of amyloid β peptide: Role of the hairpin fold. Sci. Rep. 2017, 7, 2344. [Google Scholar] [CrossRef]
- Morinaga, A.; Hasegawa, K.; Nomura, R.; Ookoshi, T.; Ozawa, D.; Goto, Y.; Yamada, M.; Naiki, H. Critical role of interfaces and agitation on the nucleation of Aβ amyloid fibrils at low concentrations of Aβ monomers. Biochim. Biophys. Acta 2010, 1804, 986–995. [Google Scholar] [CrossRef]
- Jean, L.; Lee, C.F.; Vaux, D.J. Enrichment of Amyloidogenesis at an Air-Water Interface. Biophys. J. 2012, 102, 1154–1162. [Google Scholar] [CrossRef] [Green Version]
- Yagi-Utsumi, M.; Kato, K.; Nishimura, K. Membrane-Induced Dichotomous Conformation of Amyloid β with the Disordered N-Terminal Segment Followed by the Stable C-Terminal β Structure. PLoS ONE 2016, 11, e0146405. [Google Scholar] [CrossRef]
- Fantini, J.; Yahi, N. Molecular insights into amyloid regulation by membrane cholesterol and sphingolipids: Common mechanisms in neurodegenerative diseases. Expert Rev. Mol. Med. 2010, 12, e27. [Google Scholar] [CrossRef]
- Yagi-Utsumi, M.; Yamaguchi, Y.; Sasakawa, H.; Yamamoto, N.; Yanagisawa, K.; Kato, K. Up-and-down topological mode of amyloid β-peptide lying on hydrophilic/hydrophobic interface of ganglioside clusters. Glycoconj. J. 2008, 26, 999–1006. [Google Scholar] [CrossRef]
- Yagi-Utsumi, M.; Matsuo, K.; Yanagisawa, K.; Gekko, K.; Kato, K. Spectroscopic Characterization of Intermolecular Interaction of Amyloid β Promoted on GM1 Micelles. Int. J. Alzheimers Dis. 2010, 2011, 925073. [Google Scholar] [PubMed] [Green Version]
- Lemkul, J.A.; Bevan, D.R. Perturbation of membranes by the amyloid β-peptide—A molecular dynamics study. FEBS J. 2009, 276, 3060–3075. [Google Scholar] [CrossRef] [PubMed]
- Na Zhao, L.; Chiu, S.-W.; Benoit, J.; Chew, L.Y.; Mu, Y. Amyloid β Peptides Aggregation in a Mixed Membrane Bilayer: A Molecular Dynamics Study. J. Phys. Chem. B 2011, 115, 12247–12256. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Yamaguchi, T.; Fukunaga, S.; Hoshino, M.; Matsuzaki, K. Mechanism of amyloid beta-protein aggregation mediated by GM1 ganglioside clusters. Biochemistry 2011, 50, 6433–6440. [Google Scholar] [CrossRef] [PubMed]
- Poojari, C.; Kukol, A.; Strodel, B. How the amyloid-β peptide and membranes affect each other: An extensive simulation study. Biochim. Biophys. Acta-Biomembr. 2013, 1828, 327–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshino, T.; Mahmood, I.; Mori, K.; Matsuzaki, K. Binding and Aggregation Mechanism of Amyloid β-Peptides onto the GM1 Ganglioside-Containing Lipid Membrane. J. Phys. Chem. B 2013, 117, 8085–8094. [Google Scholar] [CrossRef]
- Brown, A.M.; Bevan, D.R. Molecular Dynamics Simulations of Amyloid β-Peptide (1-42): Tetramer Formation and Membrane Interactions. Biophys. J. 2016, 111, 937–949. [Google Scholar] [CrossRef] [Green Version]
- Khondker, A.; Alsop, R.J.; Rheinstädter, M.C. Membrane-Accelerated Amyloid-β Aggregation and Formation of Cross-β Sheets. Membranes 2017, 7, 49. [Google Scholar] [CrossRef] [Green Version]
- Niu, Z.; Zhang, Z.; Zhao, W.; Yang, J. Interactions between amyloid β peptide and lipid membranes. Biochim. Biophys. Acta-Biomembr. 2018, 1860, 1663–1669. [Google Scholar] [CrossRef]
- Tachi, Y.; Okamoto, Y.; Okumura, H. Conformational properties of an artificial GM1 glycan cluster based on a metal-ligand complex. J. Chem. Phys. 2018, 149, 135101. [Google Scholar] [CrossRef]
- Sato, S.; Yoshimasa, Y.; Fujita, D.; Yagi-Utsumi, M.; Yamaguchi, T.; Kato, K.; Fujita, M. A Self-Assembled Spherical Com-plex Displaying a Gangliosidic Glycan Cluster Capable of Interacting with Amyloidogenic Proteins. Angew. Chem. Int. Ed. 2015, 54, 8435–8439. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Hasegawa, K.; Naiki, H.; Yamada, M. Curcumin has potent anti-amyloidogenic effects for Alzheimer’s β-amyloid fibrils in vitro. J. Neurosci. Res. 2004, 75, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Li, L.; Takamura, Y.; Yoshiike, Y.; Zhu, L.; Han, F.; Mao, X.; Ikeda, T.; Takasaki, J.-I.; Nishijo, H.; et al. Phenolic Compounds Prevent Amyloid β-Protein Oligomerization and Synaptic Dysfunction by Site-specific Binding. J. Biol. Chem. 2012, 287, 14631–14643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klimov, D.K.; Straub, J.E.; Thirumalai, D. Aqueous urea solution destabilizes Aβ(16-22) oligomers. Proc. Natl. Acad. Sci. USA 2004, 101, 14760–14765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, P.H.; Li, M.S.; Derreumaux, P. Effects of all-atom force fields on amyloid oligomerization: Replica exchange molecular dynamics simulations of the Aβ(16-22) dimer and trimer. Phys. Chem. Chem. Phys. 2011, 13, 9778–9788. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, L.; Nguyen, P.H.; Stock, G. Construction of the Free Energy Landscape of Peptide Aggregation from Molecular Dynamics Simulations. J. Chem. Theory Comput. 2012, 8, 1471–1479. [Google Scholar] [CrossRef]
- Nguyen, P.H.; Okamoto, Y.; Derreumaux, P. Communication: Simulated tempering with fast on-the-fly weight determination. J. Chem. Phys. 2013, 138, 61102. [Google Scholar] [CrossRef] [Green Version]
- Barz, B.; Wales, D.J.; Strodel, B. A Kinetic Approach to the Sequence–Aggregation Relationship in Disease-Related Protein Assembly. J. Phys. Chem. B 2014, 118, 1003–1011. [Google Scholar] [CrossRef] [Green Version]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Okumura, H.; Okamoto, Y. Multibaric–Multithermal Molecular Dynamics Simulation of Alanine Dipeptide in Explicit Water. Bull. Chem. Soc. Jpn. 2007, 80, 1114–1123. [Google Scholar] [CrossRef]
- Okumura, H.; Itoh, S.G.; Okamoto, Y. Explicit symplectic integrators of molecular dynamics algorithms for rigid-body molecules in the canonical, isobaric-isothermal, and related ensembles. J. Chem. Phys. 2007, 126, 84103. [Google Scholar] [CrossRef] [PubMed]
- Okumura, H.; Okamoto, Y. Temperature and Pressure Dependence of Alanine Dipeptide Studied by Multibaric-Multithermal Molecular Dynamics Simulations. J. Phys. Chem. B 2008, 112, 12038–12049. [Google Scholar] [CrossRef]
- Okumura, H. Partial multicanonical algorithm for molecular dynamics and Monte Carlo simulations. J. Chem. Phys. 2008, 129, 124116. [Google Scholar] [CrossRef] [PubMed]
- Okumura, H. Optimization of partial multicanonical molecular dynamics simulations applied to an alanine dipeptide in explicit water solvent. Phys. Chem. Chem. Phys. 2011, 13, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Okumura, H. Temperature and pressure denaturation of chignolin: Folding and unfolding simulation by multibaric-multithermal molecular dynamics method. Proteins Struct. Funct. Bioinf. 2012, 80, 2397–2416. [Google Scholar] [CrossRef] [PubMed]
- Okumura, H.; Itoh, S.G. Transformation of a design peptide between the α-helix and β-hairpin structures using a helix-strand replica-exchange molecular dynamics simulation. Phys. Chem. Chem. Phys. 2013, 15, 13852–13861. [Google Scholar] [CrossRef] [PubMed]
- Mizukami, T.; Furuzawa, S.; Itoh, S.G.; Segawa, S.; Ikura, T.; Ihara, K.; Okumura, H.; Roder, H.; Maki, K. Energetics and kinetics of substrate analog-coupled staphylococcal nuclease folding revealed by a statistical mechanical approach. Proc. Natl. Acad. Sci. USA 2020, 117, 19953–19962. [Google Scholar] [CrossRef]
- Nguyen, T.H.D.; Itoh, S.G.; Okumura, H.; Tominaga, M. Structural basis for promiscuous action of monoterpenes on TRP channels. Commun. Biol. 2021, 4, 293. [Google Scholar] [CrossRef]
- Tanimoto, S.; Itoh, S.G.; Okumura, H. “Bucket brigade” using lysine residues in RNA-dependent RNA polymerase of SARS-CoV-2. Biophys. J. 2021, 120, 3615–3627. [Google Scholar] [CrossRef]
- Miyazawa, K.; Itoh, S.G.; Watanabe, H.; Uchihashi, T.; Yanaka, S.; Yagi-Utsumi, M.; Kato, K.; Arakawa, K.; Okumura, H. Tardigrade Secretory-Abundant Heat-Soluble Protein Has a Flexible β-Barrel Structure in Solution and Keeps This Structure in Dehydration. J. Phys. Chem. B 2021, 125, 9145–9154. [Google Scholar] [CrossRef] [PubMed]
- Itoh, S.G.; Tanimoto, S.; Okumura, H. Dynamic properties of SARS-CoV and SARS-CoV-2 RNA-dependent RNA polymerases studied by molecular dynamics simulations. Chem. Phys. Lett. 2021, 778, 138819. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, K.; Itoh, S.G.; Yoshida, Y.; Arakawa, K.; Okumura, H. Tardigrade Secretory-Abundant Heat-Soluble Protein Varies Entrance Propensity Depending on the Amino-Acid Sequence. J. Phys. Chem. B 2022, 126, 2361–2368. [Google Scholar] [CrossRef] [PubMed]
- Okumura, H.; Itoh, S.G.; Okamoto, Y. Generalized-Ensemble Algorithms for Simulations of Complex Molecular Systems. In Practical Aspects of Computational Chemistry II: An Overview of the Last Two Decades and Current Trends; Leszczynski, J., Shukla, M.K., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 69–101. [Google Scholar]
- Yamauchi, M.; La Penna, G.; Itoh, S.G.; Okumura, H. Implementations of replica-permutation and replica sub-permutation methods into LAMMPS. Comput. Phys. Commun. 2022, 276, 108362. [Google Scholar] [CrossRef]
- Berg, B.A.; Neuhaus, T. Multicanonical algorithms for 1st order phase-transitions. Phys. Lett. B 1991, 267, 249–253. [Google Scholar] [CrossRef]
- Berg, B.A.; Neuhaus, T. Multicanonical ensemble: A new approach to simulate first-order phase transitions. Phys. Rev. Lett. 1992, 68, 9–12. [Google Scholar] [CrossRef] [Green Version]
- Hansmann, U.H.E.; Okamoto, Y.; Eisenmenger, F. Molecular dynamics, Langevin and hybrid Monte Carlo simulations in a multicanonical ensemble. Chem. Phys. Lett. 1996, 259, 321–330. [Google Scholar] [CrossRef]
- Nakajima, N.; Nakamura, H.; Kidera, A. Multicanonical ensemble generated by molecular dynamics simulation for enhanced conformational sampling of peptides. J. Phys. Chem. B 1997, 101, 817–824. [Google Scholar] [CrossRef]
- Okumura, H.; Okamoto, Y. Monte Carlo simulations in multibaric–multithermal ensemble. Chem. Phys. Lett. 2004, 383, 391–396. [Google Scholar] [CrossRef] [Green Version]
- Okumura, H.; Okamoto, Y. Monte Carlo simulations in generalized isobaric-isothermal ensembles. Phys. Rev. E 2004, 70, 026702. [Google Scholar] [CrossRef]
- Okumura, H.; Okamoto, Y. Molecular dynamics simulations in the multibaric–multithermal ensemble. Chem. Phys. Lett. 2004, 391, 248–253. [Google Scholar] [CrossRef]
- Okumura, H.; Okamoto, Y. Multibaric-multithermal ensemble molecular dynamics simulations. J. Comput. Chem. 2005, 27, 379–395. [Google Scholar] [CrossRef] [PubMed]
- Sunde, M.; Serpell, L.; Bartlam, M.; Fraser, P.E.; Pepys, M.B.; Blake, C.C. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 1997, 273, 729–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petkova, A.T.; Yau, W.M.; Tycko, R. Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry 2006, 45, 498–512. [Google Scholar] [CrossRef] [Green Version]
- Kittel, C. Surface and interface physics. In Introduction to Solid State Physics; Wiley: New York, NY, USA, 2004; pp. 487–514. [Google Scholar]
- Buch, V.; Milet, A.; Vácha, R.; Jungwirth, P.; Devlin, J.P. Water surface is acidic. Proc. Natl. Acad. Sci. USA 2007, 104, 7342–7347. [Google Scholar] [CrossRef] [Green Version]
- Beattie, J.K.; Djerdjev, A.M.; Warr, G.G. The surface of neat water is basic. Faraday Discuss. 2008, 141, 31–39. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System; Schrödinger, LLC.: New York, NY, USA, 2015.
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Andersen, H.C. Molecular dynamics simulations at constant pressure and/or temperature. J. Chem. Phys. 1980, 72, 2384–2393. [Google Scholar] [CrossRef] [Green Version]
- Ban, T.; Hamada, D.; Hasegawa, K.; Naiki, H.; Goto, Y. Direct Observation of Amyloid Fibril Growth Monitored by Thioflavin T Fluorescence. J. Biol. Chem. 2003, 278, 16462–16465. [Google Scholar] [CrossRef] [Green Version]
- Ban, T.; Hoshino, M.; Takahashi, S.; Hamada, D.; Hasegawa, K.; Naiki, H.; Goto, Y. Direct Observation of Aβ Amyloid Fibril Growth and Inhibition. J. Mol. Biol. 2004, 344, 757–767. [Google Scholar] [CrossRef]
- Uchihashi, T.; Konno, H. Self-Assembly Dynamics of Biological Molecules Observed by High-Speed Atomic Force Microscopy. In Proceedings of the 96th Annual Meeting of the Chemical Society of Japan, No. 1S5-13. Kyotanabe, Japan, 24–27 March 2016. [Google Scholar]
- Kinjo, T.; Ohguchi, K.; Yasuoka, K.; Matsumoto, M. Computer simulation of fluid phase change: Vapor nucleation and bubble formation dynamics. Comput. Mater. Sci. 1999, 14, 138–141. [Google Scholar] [CrossRef]
- Xiao, C.; Heyes, D.M.; Powles, J.G. The collapsing bubble in a liquid by molecular dynamics simulations. Mol. Phys. 2002, 100, 3451–3468. [Google Scholar] [CrossRef]
- Okumura, H.; Ito, N. Nonequilibrium molecular dynamics simulations of a bubble. Phys. Rev. E 2003, 67, 45301. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Inaoka, H.; Ito, N. Ripening kinetics of bubbles: A molecular dynamics study. J. Chem. Phys. 2016, 145, 124707. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, H. Construction of higher order symplectic integrators. Phys. Lett. A 1990, 150, 262–268. [Google Scholar] [CrossRef]
- Miller, T.F.; Eleftheriou, M.; Pattnaik, P.; Ndirango, A.; Newns, D.; Martyna, G.J. Symplectic quaternion scheme for biophysical molecular dynamics. J. Chem. Phys. 2002, 116, 8649–8659. [Google Scholar] [CrossRef] [Green Version]
- Chatani, E.; Lee, Y.-H.; Yagi, H.; Yoshimura, Y.; Naiki, H.; Goto, Y. Ultrasonication-dependent production and breakdown lead to minimum-sized amyloid fibrils. Proc. Natl. Acad. Sci. USA 2009, 106, 11119–11124. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, T.; Ohori, G.; Chiba, T.; Tsukiyama, K.; Nakamura, K. Picosecond pulsed infrared laser tuned to amide I band dissociates polyglutamine fibrils in cells. Lasers Med. Sci. 2016, 31, 1425–1431. [Google Scholar] [CrossRef]
- Kawasaki, T.; Yaji, T.; Ohta, T.; Tsukiyama, K.; Nakamura, K. Dissociation of β-Sheet Stacking of Amyloid β Fibrils by Irradiation of Intense, Short-Pulsed Mid-infrared Laser. Cell. Mol. Neurobiol. 2018, 38, 1039–1049. [Google Scholar] [CrossRef]
- Kawasaki, T.; Tsukiyama, K.; Irizawa, A. Dissolution of a fibrous peptide by terahertz free electron laser. Sci. Rep. 2019, 9, 10636. [Google Scholar] [CrossRef] [Green Version]
- Mori, Y.; Okumura, H. Pressure-Induced Helical Structure of a Peptide Studied by Simulated Tempering Molecular Dynamics Simulations. J. Phys. Chem. Lett. 2013, 4, 2079–2083. [Google Scholar] [CrossRef]
- Mori, Y.; Okumura, H. Molecular dynamics of the structural changes of helical peptides induced by pressure. Proteins Struct. Funct. Bioinform. 2014, 82, 2970–2981. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Okumura, H. Molecular dynamics simulation study on the high-pressure behaviour of an AK16 peptide. Mol. Simul. 2015, 41, 1035–1040. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [Green Version]
- Schmid, N.; Eichenberger, A.P.; Choutko, A.; Riniker, S.; Winger, M.; Mark, A.; Van Gunsteren, W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 2011, 40, 843–856. [Google Scholar] [CrossRef]
- Watts, C.R.; Gregory, A.; Frisbie, C.; Lovas, S. Effects of force fields on the conformational and dynamic properties of amyloid β(1-40) dimer explored by replica exchange molecular dynamics simulations. Proteins Struct. Funct. Bioinform. 2018, 86, 279–300. [Google Scholar] [CrossRef]
- Man, V.H.; He, X.; Gao, J.; Wang, J. Effects of All-Atom Molecular Mechanics Force Fields on Amyloid Peptide Assembly: The Case of PHF6 Peptide of Tau Protein. J. Chem. Theory Comput. 2021, 17, 6458–6471. [Google Scholar] [CrossRef]
- Caliskan, M.; Mandaci, S.Y.; Uversky, V.N.; Coskuner-Weber, O. Secondary structure dependence of amyloid-β(1–40) on simulation techniques and force field parameters. Chem. Biol. Drug Des. 2021, 97, 1100–1108. [Google Scholar] [CrossRef]
- Still, W.C.; Tempczyk, A.; Hawley, R.C.; Hendrickson, T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 1990, 112, 6127–6129. [Google Scholar] [CrossRef]
- Dominy, B.N.; Brooks, C.L. Development of a Generalized Born Model Parametrization for Proteins and Nucleic Acids. J. Phys. Chem. B 1999, 103, 3765–3773. [Google Scholar] [CrossRef]
- Feig, M.; Im, W.; Brooks, C.L. Implicit solvation based on generalized Born theory in different dielectric environments. J. Chem. Phys. 2004, 120, 903–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davtyan, A.; Schafer, N.P.; Zheng, W.; Clementi, C.; Wolynes, P.G.; Papoian, G.A. AWSEM-MD: Protein Structure Prediction Using Coarse-Grained Physical Potentials and Bioinformatically Based Local Structure Biasing. J. Phys. Chem. B 2012, 116, 8494–8503. [Google Scholar] [CrossRef] [Green Version]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef] [Green Version]
- Monticelli, L.; Kandasamy, S.K.; Periole, X.; Larson, R.G.; Tieleman, D.P.; Marrink, S. The MARTINI Coarse-Grained Force Field: Extension to Proteins. J. Chem. Theory Comput. 2008, 4, 819–834. [Google Scholar] [CrossRef] [PubMed]
- Liwo, A.; Oldziej, S.; Pincus, M.R.; Wawak, R.J.; Rackovsky, S.; Scheraga, H.A. A united-residue force field for off-lattice protein-structure simulations. I. Functional forms and parameters of long-range side-chain interaction potentials from protein crystal data. J. Comput. Chem. 1997, 18, 849–873. [Google Scholar] [CrossRef]
- Liwo, A.; Pincus, M.R.; Wawak, R.J.; Rackovsky, S.; Ołdziej, S.; Scheraga, H.A. A united-residue force field for off-lattice protein-structure simulations. II. Parameterization of short-range interactions and determination of weights of energy terms by Z-score optimization. J. Comput. Chem. 1997, 18, 874–887. [Google Scholar] [CrossRef]
- Co, N.T.; Li, M.S.; Krupa, P. Computational Models for the Study of Protein Aggregation. In Computer Simulations of Aggregation of Proteins and Peptides; Li, M.S., Kloczkowski, A., Cieplak, M., Kouza, M., Eds.; Springer: New York, NY, USA, 2022; pp. 51–78. [Google Scholar]
- Rojas, A.V.; Maisuradze, G.G.; Scheraga, H.A.; Liwo, A. Probing Protein Aggregation Using the Coarse-Grained UNRES Force Field. In Computer Simulations of Aggregation of Proteins and Peptides; Li, M.S., Kloczkowski, A., Cieplak, M., Kouza, M., Eds.; Springer: New York, NY, USA, 2022; pp. 79–104. [Google Scholar]
- Chong, S.-H.; Ham, S. Atomic-level investigations on the amyloid-β dimerization process and its driving forces in water. Phys. Chem. Chem. Phys. 2011, 14, 1573–1575. [Google Scholar] [CrossRef]
- Masutani, K.; Yamamori, Y.; Kim, K.; Matubayasi, N. Free-energy analysis of the hydration and cosolvent effects on the β-sheet aggregation through all-atom molecular dynamics simulation. J. Chem. Phys. 2019, 150, 145101. [Google Scholar] [CrossRef]
- Li, M.S.; Klimov, D.K.; Straub, J.E.; Thirumalai, D. Probing the mechanisms of fibril formation using lattice models. J. Chem. Phys. 2008, 129, 175101. [Google Scholar] [CrossRef] [Green Version]
- Co, N.T.; Hu, C.-K.; Li, M.S. Dual effect of crowders on fibrillation kinetics of polypeptide chains revealed by lattice models. J. Chem. Phys. 2013, 138, 185101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irbäck, A.; Jónsson, S.; Linnemann, N.; Linse, B.; Wallin, S. Aggregate Geometry in Amyloid Fibril Nucleation. Phys. Rev. Lett. 2013, 110, 58101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abeln, S.; Vendruscolo, M.; Dobson, C.M.; Frenkel, D. A Simple Lattice Model That Captures Protein Folding, Aggregation and Amyloid Formation. PLoS ONE 2014, 9, e85185. [Google Scholar] [CrossRef] [PubMed]
- Hynynen, K.; McDannold, N.; Vykhodtseva, N.; Jolesz, F.A. Noninvasive MR Imaging–guided Focal Opening of the Blood-Brain Barrier in Rabbits. Radiology 2001, 220, 640–646. [Google Scholar] [CrossRef]
- Jordão, J.F.; Ayala-Grosso, C.A.; Markham, K.; Huang, Y.; Chopra, R.; McLaurin, J.; Hynynen, K.; Aubert, I. Antibodies Targeted to the Brain with Image-Guided Focused Ultrasound Reduces Amyloid-β Plaque Load in the TgCRND8 Mouse Model of Alzheimer’s Disease. PLoS ONE 2010, 5, e10549. [Google Scholar] [CrossRef] [Green Version]
- Burgess, A.; Dubey, S.; Yeung, S.; Hough, O.; Eterman, N.; Aubert, I.; Hynynen, K. Alzheimer Disease in a Mouse Model: MR Imaging–guided Focused Ultrasound Targeted to the Hippocampus Opens the Blood-Brain Barrier and Improves Pathologic Abnormalities and Behavior. Radiology 2014, 273, 736–745. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.-H.; Lin, Y.-T.; Chung, Y.-H.; Lin, K.-J.; Yang, L.-Y.; Yen, T.-C.; Liu, H.-L. Focused Ultrasound-Induced Blood-Brain Barrier Opening Enhances GSK-3 Inhibitor Delivery for Amyloid-β Plaque Reduction. Sci. Rep. 2018, 8, 12882. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okumura, H.; Itoh, S.G. Molecular Dynamics Simulation Studies on the Aggregation of Amyloid-β Peptides and Their Disaggregation by Ultrasonic Wave and Infrared Laser Irradiation. Molecules 2022, 27, 2483. https://doi.org/10.3390/molecules27082483
Okumura H, Itoh SG. Molecular Dynamics Simulation Studies on the Aggregation of Amyloid-β Peptides and Their Disaggregation by Ultrasonic Wave and Infrared Laser Irradiation. Molecules. 2022; 27(8):2483. https://doi.org/10.3390/molecules27082483
Chicago/Turabian StyleOkumura, Hisashi, and Satoru G. Itoh. 2022. "Molecular Dynamics Simulation Studies on the Aggregation of Amyloid-β Peptides and Their Disaggregation by Ultrasonic Wave and Infrared Laser Irradiation" Molecules 27, no. 8: 2483. https://doi.org/10.3390/molecules27082483





