Compositional Analysis of Four Kinds of Citrus Fruits with an NMR-Based Method for Understanding Nutritional Value and Rational Utilization: From Pericarp to Juice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Citrus Sample Collection and Preparation
2.2. 1H-NMR Spectroscopy and Spectral Preprocessing
2.3. Univariate and Multivariate Statistical Analyses
3. Results and Discussion
3.1. The Comparison of 1H-NMR Spectral Profiles and Chemical Composition of Different Varieties of Citrus Juice and Pericarp
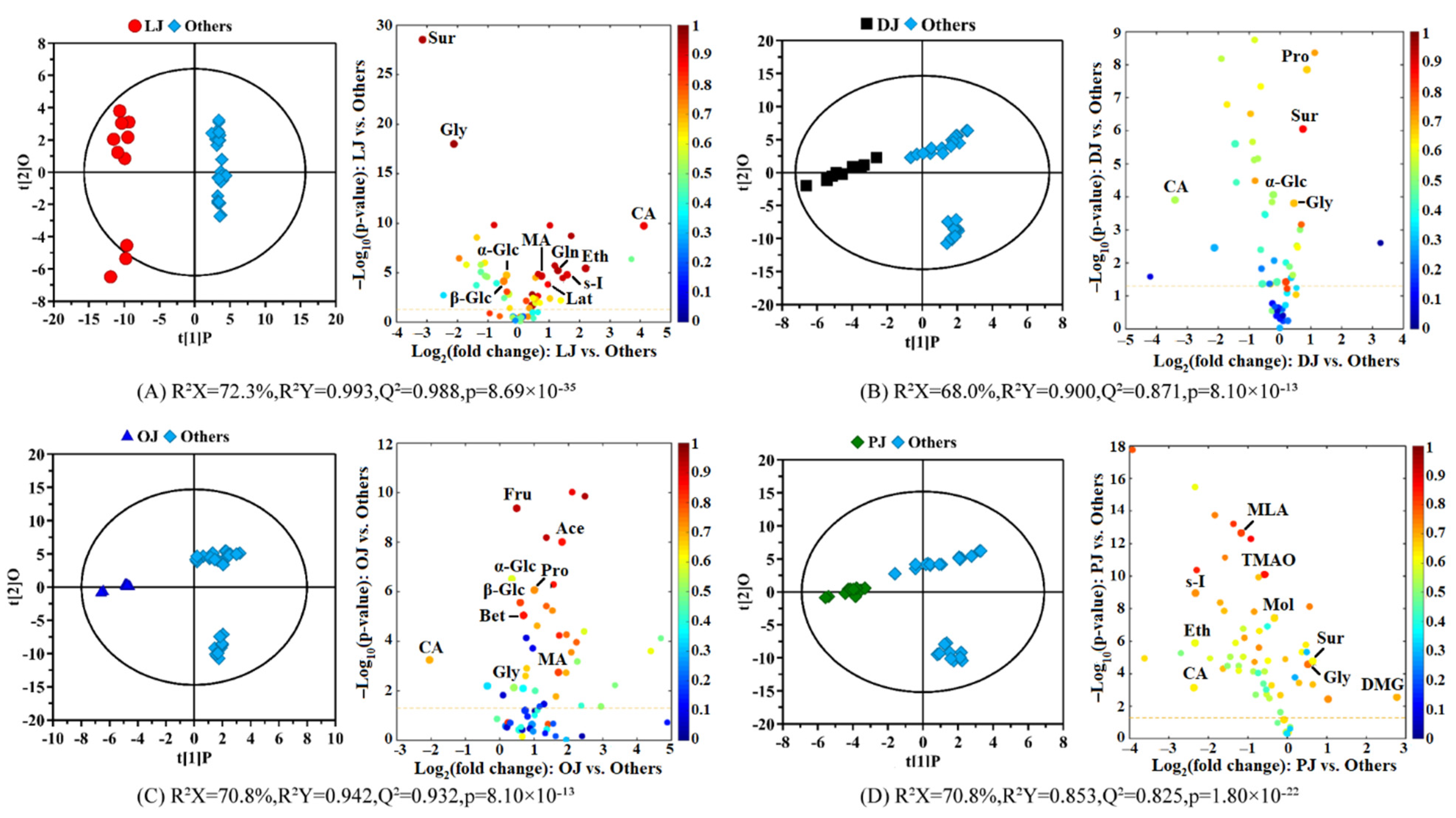
3.2. Compositional Differences between the Citrus Juices in the Different Varieties
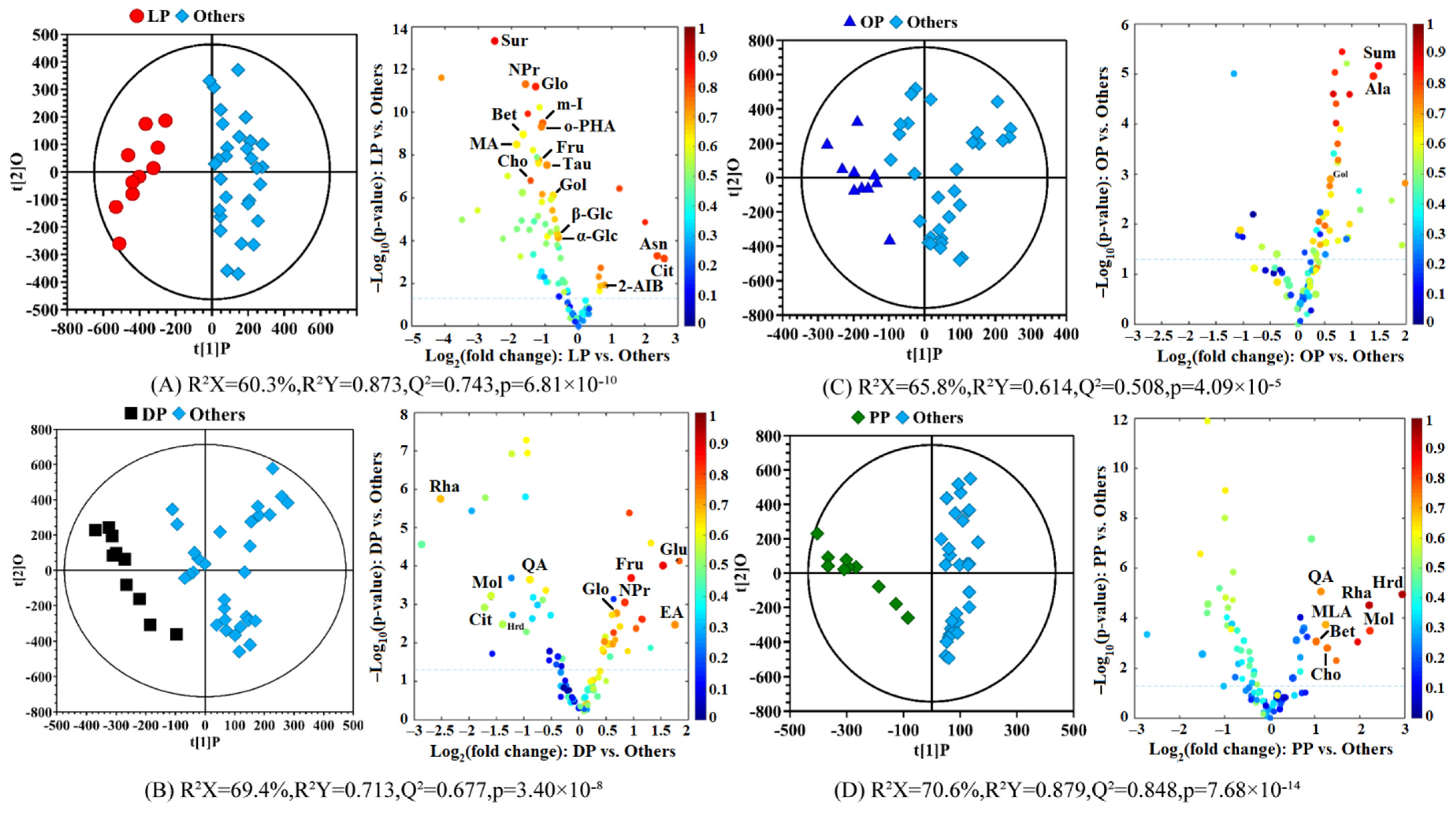
3.3. Compositional Differences between the Citrus Pericarps in the Different Varieties
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Saini, R.K.; Ranjit, A.; Sharma, K.; Prasad, P.; Shang, X.; Gowda, K.G.M.; Keum, Y.-S. Bioactive compounds of citrus fruits: A review of composition and health benefits of carotenoids, flavonoids, limonoids, and terpenes. Antioxidants 2022, 11, 239. [Google Scholar] [CrossRef] [PubMed]
- Villa-Ruano, N.; Perez-Hernandez, N.; Gerardo Zepeda-Vallejo, L.; Quiroz-Acosta, T.; Mendieta-Moctezuma, A.; Montoya-Garcia, C.; Garcia-Nava, M.L.; Becerra-Martinez, E. H-1-NMR based metabolomics profiling of citrus juices produced in Veracruz, Mexico. Chem. Biodivers. 2019, 16, e1800479. [Google Scholar] [CrossRef] [PubMed]
- Gavahian, M.; Chu, Y.-H.; Khaneghah, A.M. Recent advances in orange oil extraction: An opportunity for the valorisation of orange peel waste a review. Int. J. Food Sci. Technol. 2019, 54, 925–932. [Google Scholar] [CrossRef]
- Karen Tovar, A.; Godinez, L.A.; Espejel, F.; Ramirez-Zamora, R.-M.; Robles, I. Optimization of the integral valorization process for orange peel waste using a design of experiments approach: Production of high-quality pectin and activated carbon. Waste Manag. 2019, 85, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Zhang, R.; Zicari, S. (Eds.) Integrated Processing Technologies for Food and Agricultural By-Products; Academic Press: Cambridge, MA, USA, 2019; pp. 439–444. [Google Scholar]
- Yadav, V.; Sarker, A.; Yadav, A.; Miftah, A.O.; Bilal, M.; Iqbal, H.M.N. Integrated biorefinery approach to valorize citrus waste: A sustainable solution for resource recovery and environmental management. Chemosphere 2022, 293, 133459. [Google Scholar] [CrossRef]
- Phucharoenrak, P.; Muangnoi, C.; Trachootham, D. A green extraction method to achieve the highest yield of limonin and hesperidin from lime peel powder (Citrus aurantifolia). Molecules 2022, 27, 820. [Google Scholar] [CrossRef]
- Brits, M.; Naessens, T.; Theunis, M.; Taktak, O.; Allouche, N.; Pieters, L.; Foubert, K. Identification and quantification of polymethoxylated flavonoids in different citrus species using UPLC-QTOF-MS/MS and HPLC-DAD (#). Planta Med. 2021, 87, 1080–1088. [Google Scholar]
- Chen, X.; Chen, H.; Xiao, J.; Liu, J.; Tang, N.; Zhou, A. Variations of volatile flavour compounds in finger citron (Citrus medico L. var. sarcodactylis) pickling process revealed by E-nose, HS-SPME-GC-MS and HS-GC-IMS. Food Res. Int. 2020, 138, 109717. [Google Scholar]
- Md Nor, S.; Ding, P.; Abas, F.; Mediani, A. H-1 NMR reveals dynamic changes of primary metabolites in purple passion fruit (Passiflora edulis Sims) juice during maturation and ripening. Agriculture 2022, 12, 156. [Google Scholar] [CrossRef]
- Wang, S.; Tu, H.; Wan, J.; Chen, W.; Liu, X.; Luo, J.; Xu, J.; Zhang, H. Spatio-temporal distribution and natural variation of metabolites in citrus fruits. Food Chem. 2016, 199, 8–17. [Google Scholar] [CrossRef]
- Markley, J.L.; Bruschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S. NMR metabolomics: A look ahead. J. Magn. Reson. 2019, 306, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Eisenmann, P.; Ehlers, M.; Weinert, C.H.; Tzvetkova, P.; Silber, M.; Rist, M.J.; Luy, B.; Muhle-Goll, C. Untargeted NMR spectroscopic analysis of the metabolic variety of new apple cultivars. Metabolites 2016, 6, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Zhao, Y.; Yang, J.; Jiang, Y.; Lu, F.; Jia, Y.; Yang, B. Metabolomic analyses of banana during postharvest senescence by H-1-high resolution-NMR. Food Chem. 2017, 218, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Hu, J.; Huang, Y.; Wang, H.; Adeleye, A.; Ortiz, C.; Keller, A.A. H-1 NMR and GC-MS based metabolomics reveal nano-Cu altered cucumber (Cucumis sativus) fruit nutritional supply. Plant Physiol. Biochem. 2017, 110, 138–146. [Google Scholar] [CrossRef] [Green Version]
- Koda, M.; Furihata, K.; Wei, F.; Miyakawa, T.; Tanokura, M. Metabolic discrimination of mango juice from various cultivars by band-selective NMR spectroscopy. J. Agric. Food Chem. 2012, 60, 1158–1166. [Google Scholar] [CrossRef]
- Migues, I.; Hodos, N.; Moltini, A.I.; Gambaro, A.; Rivas, F.; Moyna, G.; Heinzen, H. H-1 NMR metabolic profiles as selection tools of new mandarin cultivars based on fruit acceptability. Sci. Hortic. 2021, 287, 110262. [Google Scholar] [CrossRef]
- Salvino, R.A.; Colella, M.F.; De Luca, G. NMR-based metabolomics analysis of Calabrian citrus fruit juices and its application to industrial process quality control. Food Control 2021, 121, 107619. [Google Scholar] [CrossRef]
- Lin, H.; He, C.; Liu, H.; Shen, G.; Xia, F.; Feng, J. NMR-based quantitative component analysis and geographical origin identification of China’s sweet orange. Food Control 2021, 130, 108292. [Google Scholar] [CrossRef]
- Le Mao, I.; Martin-Pernier, J.; Bautista, C.; Lacampagne, S.; Richard, T.; Da Costa, G. 1H-NMR metabolomics as a tool for Winemaking monitoring. Molecules 2021, 26, 6771. [Google Scholar] [CrossRef]
- Mucci, A.; Parenti, F.; Righi, V.; Schenetti, L. Citron and lemon under the lens of HR-MAS NMR spectroscopy. Food Chem. 2013, 141, 3167–3176. [Google Scholar] [CrossRef] [PubMed]
- Spraul, M.; Schuetz, B.; Humpfer, E.; Moertter, M.; Schaefer, H.; Koswig, S.; Rinke, P. Mixture analysis by NMR as applied to fruit juice quality control. Magn. Reson. Chem. 2009, 47, S130–S137. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, G.; Puaud, M.; Colquhoun, I.J. Discrimination between orange juice and pulp wash by H-1 nuclear magnetic resonance spectroscopy: Identification of marker compounds. J. Agric. Food Chem. 2001, 49, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, E.L.; Akutsu, H.; Doreleijers, J.F.; Harano, Y.; Ioannidis, Y.E.; Lin, J.; Livny, M.; Mading, S.; Maziuk, D.; Miller, Z.; et al. BioMagResBank. Nucleic Acids Res. 2008, 36, D402–D408. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.J.; Lee, S.Y.; Kang, N.G.; Jin, M. H A multi-faceted comparison of phytochemicals in seven citrus peels and improvement of chemical composition and antioxidant activity by steaming. LWT Food Sci. Technol. 2022, 160, 113297. [Google Scholar] [CrossRef]
- Yu, L.; Chen, M.; Liu, J.; Huang, X.; He, W.; Qing, Z.; Zeng, J. Systematic detection and identification of bioactive ingredients from Citrus aurantium L. var. amara using HPLC-Q-TOF-MS combined with a screening method. Molecules 2020, 25, 357. [Google Scholar]
- Chen, J.; Wang, Y.; Zhu, T.; Yang, S.; Cao, J.; Li, X.; Wang, L.-S.; Sun, C. Beneficial regulatory effects of polymethoxyflavone-rich fraction from ougan (Citrus reticulata cv. Suavissima) fruit on gut microbiota and identification of its intestinal metabolites in mice. Antioxidants 2020, 9, 831. [Google Scholar]
- Qi, H.; Ding, S.; Pan, Z.; Li, X.; Fu, F. Characteristic volatile fingerprints and odor activity values in different citrus-tea by HS-GC-IMS and HS-SPME-GC-MS. Molecules 2020, 25, 6027. [Google Scholar] [CrossRef]
- Li, K.; Xia, Y.; Li, F.; Fan, Y.-L.; Liu, T.-X. Comparative analysis of volative compounds in five citrus cultivars with HS-SPME-GC-MS. Pak. J. Agric. Sci. 2020, 57, 1203–1209. [Google Scholar]
- Hanganu, A.; Todasca, M.-C.; Chira, N.-A.; Rosca, S. Influence of common and selected yeasts on wine composition studied using H-1-NMR spectroscopy. Rev. Chim. 2011, 62, 689–692. [Google Scholar]
- Sulaiman, F.; Ahmad Azam, A.; Ahamad Bustamam, M.S.; Fakurazi, S.; Abas, F.; Lee, Y.X.; Ismail, A.A.; Mohd Faudzi, S.M.; Ismail, I.S. Metabolite profiles of red and yellow watermelon (Citrullus lanatus) cultivars using a(1)H-NMR metabolomics approach. Molecules 2020, 25, 3235. [Google Scholar] [CrossRef] [PubMed]
- Villa-Ruano, N.; Velasquez-Valle, R.; Gerardo Zepeda-Vallejo, L.; Perez-Hernandez, N.; Velazquez-Ponce, M.; Arcos-Adame, V.M.; Becerra-Martinez, E. H-1 NMR-based metabolomic profiling for identification of metabolites in Capsicum annuum cv. mirasol infected by beet mild curly top virus (BMCTV). Food Res. Int. 2018, 106, 870–877. [Google Scholar] [CrossRef] [PubMed]
- Doddipalla, R.; Rendedula, D.; Ganneru, S.; Kaliyaperumal, M.; Mudiam, M.K.R. Understanding metabolic perturbations in palm wine during storage using multi-platform metabolomics. LWT Food Sci. Technol. 2022, 155, 112889. [Google Scholar] [CrossRef]
- Michon, C.; Kang, C.-M.; Karpenko, S.; Tanaka, K.; Ishikawa, S.; Yoshida, K.-I. A bacterial cell factory converting glucose into scyllo-inositol, a therapeutic agent for Alzheimer’s disease. Commun. Biol. 2020, 3, 93. [Google Scholar] [CrossRef] [Green Version]
- Fichman, Y.; Gerdes, S.Y.; Kovacs, H.; Szabados, L.; Zilberstein, A.; Csonka, L.N. Evolution of proline biosynthesis: Enzymology, bioinformatics, genetics, and transcriptional regulation. Biol. Rev. 2015, 90, 1065–1099. [Google Scholar] [CrossRef]
- Miller, R.A.; Harrison, D.E.; Astle, C.M.; Bogue, M.A.; Brind, J.; Fernandez, E.; Flurkey, K.; Javors, M.; Ladiges, W.; Leeuwenburgh, C.; et al. Glycine supplementation extends lifespan of male and female mice. Aging Cell 2019, 18, e12953. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhang, Y.; Fan, J. Acetic acid in aged vinegar affects molecular targets for thrombus disease management. Food Funct. 2015, 6, 2845–2853. [Google Scholar]
- Knoepp, F.; Ashley, Z.; Barth, D.; Baldin, J.-P.; Jennings, M.; Kazantseva, M.; Saw, E.L.; Katare, R.; Alvarez de la Rosa, D.; Weissmann, N.; et al. Shear force sensing of epithelial Na+ channel (ENaC) relies on N-glycosylated asparagines in the palm and knuckle domains of alpha ENaC. Proc. Natl. Acad. Sci. USA 2020, 117, 717–726. [Google Scholar] [CrossRef] [Green Version]
- Hamerlinck, F.F.V. Neopterin: A review. Exp. Dermatol. 1999, 8, 167–176. [Google Scholar] [CrossRef]
- Su, C.-Y.; Tseng, C.-L.; Wu, S.-H.; Shih, B.-W.; Chen, Y.-Z.; Fang, H.-W. Poly-gamma-glutamic acid functions as an effective lubricant with antimicrobial activity in multipurpose contact lens care solutions. Polymers 2019, 11, 1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Yue, J.; Ji, X.; Nian, M.; Kang, K.; Qiao, H.; Zheng, X. Research progress in biological activities of succinimide derivatives. Bioorganic Chem. 2021, 108, 104557. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Dong, Y.; Liu, H.; Ren, H.; Cui, Z. Hesperetin protects against H2O2- triggered oxidative damage via upregulation of the Keap1-Nrf2/ HO-1 signal pathway in ARPE-19 cells. Biomed. Pharmacother. 2017, 88, 124–133. [Google Scholar] [CrossRef] [PubMed]
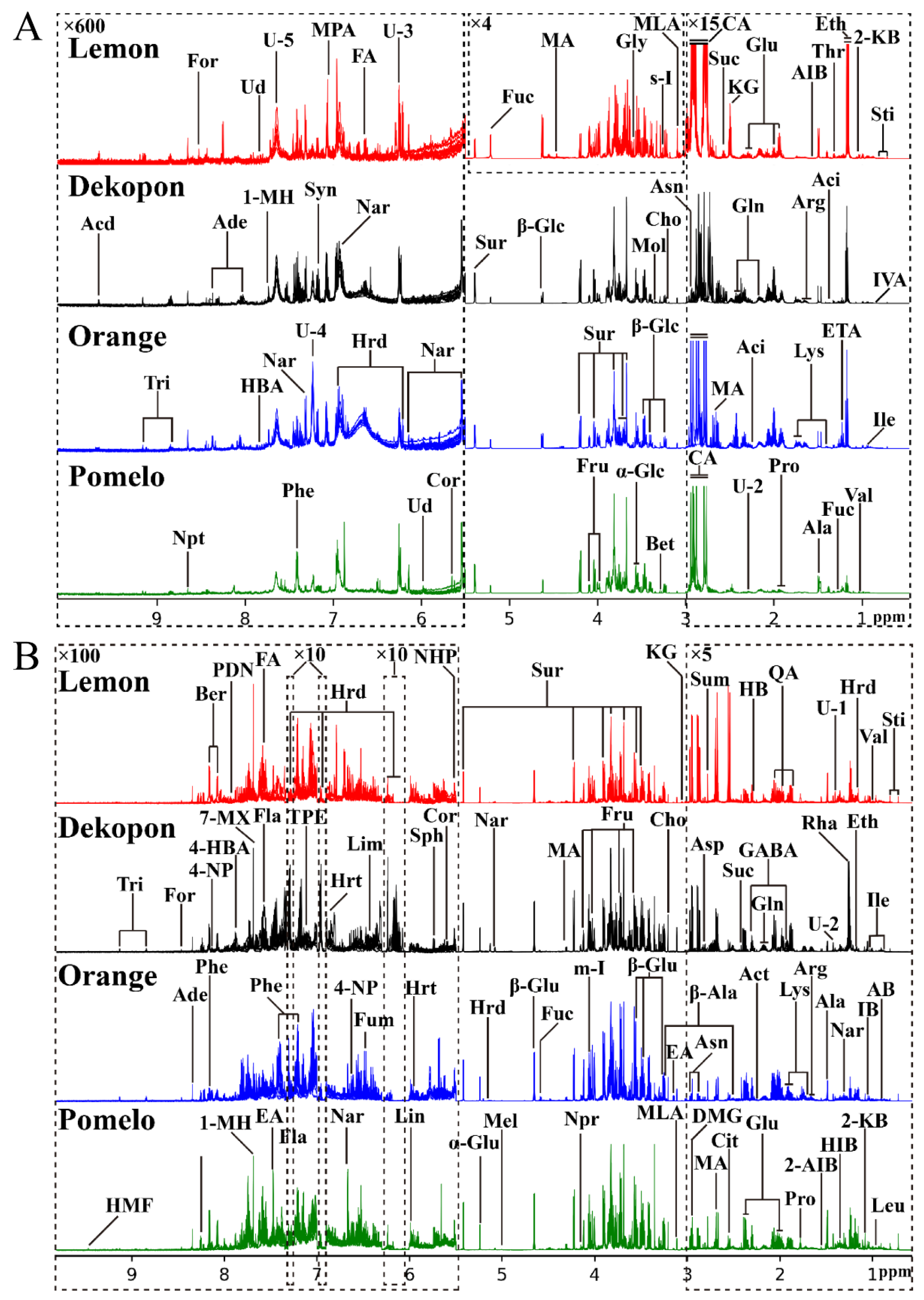

| Components | Abbr. | Chemical Shift (ppm) (Multiplicity) | Source | Ref. |
|---|---|---|---|---|
| 1-Methylhistidine | 1-MH | 7.08(s a), 7.73(s)(1) b | Juice | [20,25,26] |
| 1-Methylhistidine | 1-MH | 7.08(s), 7.68(s)(1) | Pericarp | [20,25,26] |
| 2-Aminoisobutyrate | 2-AIB | 1.55(s)(6) | Pericarp | [25,26] |
| 2-Furoic acid | FA | 6.54(dd), 6.65(d), 7.57(dd)(1) | All c | [20,25,26] |
| 2-Ketobutyric acid | 2-KB | 1.07(t)(3) | All | [20,25,26] |
| 3-Hydroxybutyric acid | HB | 1.21(d)(3), 2.28(dd) | Pericarp | [22,25,26] |
| 3-Methylphenylacetic acid | MPA | 7.07(m)(2) | All | [20,25,26] |
| 4-Hydroxybenzoic acid | HBA | 7.82(d)(2) | All | [20,25,26] |
| 4-Nitrophenol | 4-NP | 6.70(m), 8.13(m)(2) | Pericarp | [25,26] |
| 7-Methylxanthine | 7-MX | 7.93(s)(1) | Pericarp | [25,26] |
| Acetaldehyde | Acd | 2.23(d), 9.67(q)(1) | Juice | [20,25,26] |
| Acetic acid | Ace | 1.91(s)(3) | Juice | [20,21,22,23,24,30,32,33,34,35] |
| Acetoin | Aci | 1.37(d), 2.21(s)(3) | All | [2,20,21] |
| Acetone | Act | 2.25(s)(6) | Pericarp | [25,26] |
| Adenosine | Ade | 6.11(d), 8.09(s), 8.36(s)(1) | All | [20,28,34] |
| Alanine | Ala | 1.47(d)(3) | All | [2,10,20,21,22,23,24,33,34] |
| Arginine | Arg | 1.68(m)(2) | All | [2,20,21,22,24,28,33] |
| Ascorbic acid | Asc | 4.50(d)(1) | Juice | [2,20,33,35] |
| Asparagine | Asn | 2.88(dd), 2.95(dd)(1) | All | [20,22,28,34] |
| Aspartic acid | Asp | 2.67(dd)(1), 2.73(dd) | All | [2,10,20,33,34] |
| Berberine | Ber | 4.91(t), 8.08(d), 8.16(d)(1) | Pericarp | [25,26] |
| Betaine | Bet | 3.27(s)(9), 3.90 (s) | All | [20,22] |
| Choline | Cho | 3.19(s)(9) | All | [2,10,20,21,22,34] |
| Citric acid | CA | 2.75(1/2AB)(2), 2.85(1/2AB) | Juice | [2,10,20,21,22,23,34,35] |
| Citric acid | Cit | 2.55(1/2AB)(2), 2.73(1/2AB) | Pericarp | [2,10,20,22,33,34,35] |
| Corticosterone | Cor | 5.67(d)(1) | All | [20,25,26] |
| Cuminaldehyde | Cum | 9.98(s)(1) | Pericarp | [25,26] |
| Ellagic acid | EA | 7.47(s)(2) | Pericarp | [25,26] |
| Erlose | Erl | 5.36(d)(1) | Juice | [25,26] |
| Ethanol | Eth | 1.17(t)(3), 3.64(q) | All | [2,10,20,21,22,23,32,33,34,35] |
| Ethanolamine | ELA | 3.15(t)(2) | Pericarp | [22,34] |
| Ethyl acetate | ETA | 1.23(t)(3) | Juice | [20,21,30] |
| Flavone | Fla | 7.55(m), 7.62(m), 7.74(m), 7.88(m), 8.24(dd)(1) | Pericarp | [2] |
| Formic acid | For | 8.45(s)(1) | All | [20,23,33,34] |
| Fructose | Fru | 3.58(m), 3.70(m), 3.72(m), 3.80(m), 3.88(dd), 3.99(m), 4.01(m), 4.10(d)(2) | All | [2,10,20,21,22,23,24,33,34,35] |
| Fucose | Fuc | 1.26(t), 4.59(d), 5.26(d)(1) | All | [20,25,26] |
| Fumaric acid | Fum | 6.52(s)(2) | Pericarp | [2,21,23,33,34] |
| Galactitol | Gol | 3.96(t)(1) | Pericarp | [25,26] |
| Glutamic acid | Glu | 2.00(m), 2.07(m), 2.34(m)(2) | All | [20,22,24] |
| Glutamine | Gln | 2.16(m), 2.45(m)(2) | All | [2,20,22,24,28,33,34] |
| Glycerol | Glo | 3.67(dd)(2) | Pericarp | [21,22,32,34,35] |
| Glycine | Gly | 3.57(s)(2) | Juice | [20,25,26] |
| Gulonolactone | GNA | 4.52(s), 4.55(s)(1) | All | [20,25,26] |
| Hesperedin | Hrd | 1.16(m), 3.35(m), 4.52(s), 4.61(m), 4.76(m), 5.13(s), 5.32(dd), 6.16(m), 6.23(d)(2), 6.40(m), 6.80(m), 6.96(d), 7.34(m) | All | [20,27,28] |
| Hesperitin | Hrt | 5.44(q)(1), 5.95(dd), 6.85(m), 6.91(m) | All | [27,28,29] |
| Histamine | Him | 8.00(s)(1) | Pericarp | [25,26] |
| Hydroxymethylfurfural | HMF | 9.47(s)(1) | Pericarp | [25,26] |
| Isobutyric acid | IB | 1.05(d)(6) | Pericarp | [25,26,34] |
| Isoleucine | Ile | 0.93(t), 1.00(d)(3) | All | [2,10,20,21,33,34] |
| Isovaleric acid | IVA | 0.90(t)(6) | All | [20,25,26] |
| Lactose | Lat | 4.46(d)(1),5.17(d) | All | [20,35] |
| Leucine | Leu | 0.95(t)(6) | All | [2,10,20,21,33,34] |
| Limonin | Lim | 6.49(d)(1), 7.53(d),7.71(d) | All | [20,22,28,30,31] |
| Linalool | Lin | 5.97(dd)(1) | Pericarp | [30] |
| Lysine | Lys | 1.42(m), 1.75(m)(2), 1.90(m),3.04(m) | All | [10,22] |
| Malic acid | MA | 2.60(dd), 4.43(dd)(1) | All | [2,10,20,21,22,23,33,34] |
| Malonic acid | MLA | 3.10(s)(2) | All | [2,10,20,34] |
| Maltotriose | Mal | 5.38(m)(1) | Pericarp | [25,26] |
| Melezitose | Mel | 4.99(d)(1) | Pericarp | [25,26] |
| Methanol | Mol | 3.35(s)(3) | All | [2,20,21,22,23,34] |
| myo-Inositol | m-I | 3.63(t), 4.08(t)(1) | Pericarp | [2,21,22,34] |
| N,N-Dimethylglycine | DMG | 2.94(s)(6) | All | [20,25,26] |
| Naringenin | Nan | 5.49(d)(2), 7.41(d) | Juice | [20,27,28] |
| Naringin | Nar | 1.28(dd), 5.06(d), 5.54(d), 6.15(t), 6.79(d), 6.94(d), 7.32(d)(2) | All | [20,27,28] |
| Neohesperidin | NHP | 5.51(dd)(1), 5.62(dd) | Pericarp | [29] |
| Neopterin | NPr | 4.13(dd)(1) | Pericarp | [25,26] |
| Neopterin | NPt | 8.65(s)(1) | Juice | [20,25,26] |
| ortho-Hydroxyphenylacetic acid | o-HPA | 3.61(s)(2) | Pericarp | [25,26] |
| para-Aminobenzoic acid | p-ABA | 6.57(d)(2), 7.66(d) | Pericarp | [25,26] |
| p-Cresol | Cre | 7.12(d)(2) | Juice | [20,25,26] |
| Phenindione | PDN | 7.86(d), 8.04(d)(2) | Pericarp | [25,26] |
| Phenylalanine | Phe | 7.28(m), 7.38(m), 7.45(m)(2) | All | [2,10,20,24,28,33,34] |
| Proline | Pro | 1.94(m), 4.13(dd)(1) | All | [2,10,20,21,22,23,24] |
| Quinic acid | QA | 1.88(dd)(1), 1.97(dt), 2.06(m), 3.94(m), 4.15(q) | Pericarp | [22,23,24] |
| Raffinose | Raf | 4.97(d)(1) | All | [20,25,26] |
| Rhamnose | Rha | 1.20(d), 5.08(d)(1) | All | [20,25,26] |
| scyllo-Inositol | s-I | 3.33(s)(6) | Juice | [20,22] |
| Sebacic acid | Seb | 1.31(s)(8) | Pericarp | [25,26] |
| Sphingosine | Sph | 5.72(m)(2) | Pericarp | [25,26] |
| Stigmasterol | Sti | 0.71(s)(6), 0.79(m), 0.81(s), 1.78(m), 5.04(m) | All | [20,25,26] |
| Succinic acid | Suc | 2.63(s)(4) | All | [2,10,20,21,22,23,24,32,35] |
| Succinimide | Sum | 2.78(s)(3) | Pericarp | [25,26] |
| Sucrose | Sur | 3.47(t),3.54(dd), 3.67(s), 3.75(t), 3.81(t), 3.84(m), 3.90(m), 4.05(t), 4.21(d), 5.40(d)(1) | All | [2,10,20,23,24,33,34,35] |
| Synephrine | Syn | 6.89(d), 7.18(d)(2) | Juice | [20,28] |
| Taurine | Tau | 3.28(t)(2) | Pericarp | [25,26] |
| Threonine | Thr | 1.32(d)(3), 4.28(m) | All | [2,10,20,21,22,24,33,34] |
| Tiglic acid | Tig | 1.75(m)(2), 1.76(s) | Pericarp | [25,26] |
| Trehalose | Tre | 5.18(d)(2) | All | [10,20] |
| Trigonelline | Tri | 4.36(s), 8.08(m), 8.83(t), 9.12(s)(1) | All | [2,20,21,22] |
| Trimethylamine | TMA | 2.92(s)(9) | Pericarp | [25,26] |
| Trimethylamine N-oxide | TMAO | 3.29(s)(9) | Juice | [20,25,26] |
| Uridine | Ud | 5.89(d), 5.90(d), 7.87(d)(1) | Juice | [20,34] |
| Valine | Val | 0.98(d), 1.03(d)(3) | All | [2,10,20,21,22,24,33,34] |
| Vitamin C | VC | 4.48(d)(1) | Pericarp | [2,25,26] |
| α-Amino-N-butyric acid | AB | 0.96(t)(3) | Pericarp | [20,25,26] |
| α-Aminoisobutyrate | AIB | 1.54(s)(6) | Juice | [25,26] |
| α-Glucose | α-Glc | 3.41(t), 3.53(dd), 3.70(m), 3.84(m), 5.23(d)(1) | All | [2,10,20,21,22,23,24,33,34,35] |
| α-Hydroxyisobutyric acid | HIB | 1.35(s)(6) | Pericarp | [25,26] |
| α-Ketoglutaric acid | KG | 2.46(t)(2), 3.04(m) | All | [20,25,26] |
| β-Alanine | β-Ala | 2.51(t)(2), 3.23(t) | Pericarp | [25,26] |
| β-Glucose | β-Glc | 3.23(dd), 3.39(t), 3.45(dd), 3.47(t), 3.72(m), 3.88(dd), 4.64(d)(1) | All | [2,10,20,21,22,23,24,33,34,35] |
| γ-Aminobutyric acid | GABA | 1.91(m), 2.30(t), 3.02(t)(2) | Pericarp | [2,10,21,22,24,33,34] |
| γ-Terpinene | TPE | 2.20(m), 2.32(s), 7.15(d)(1), 7.19(m) | Pericarp | [30,31] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pei, Y.; He, C.; Liu, H.; Shen, G.; Feng, J. Compositional Analysis of Four Kinds of Citrus Fruits with an NMR-Based Method for Understanding Nutritional Value and Rational Utilization: From Pericarp to Juice. Molecules 2022, 27, 2579. https://doi.org/10.3390/molecules27082579
Pei Y, He C, Liu H, Shen G, Feng J. Compositional Analysis of Four Kinds of Citrus Fruits with an NMR-Based Method for Understanding Nutritional Value and Rational Utilization: From Pericarp to Juice. Molecules. 2022; 27(8):2579. https://doi.org/10.3390/molecules27082579
Chicago/Turabian StylePei, Yong, Chenxi He, Huili Liu, Guiping Shen, and Jianghua Feng. 2022. "Compositional Analysis of Four Kinds of Citrus Fruits with an NMR-Based Method for Understanding Nutritional Value and Rational Utilization: From Pericarp to Juice" Molecules 27, no. 8: 2579. https://doi.org/10.3390/molecules27082579
APA StylePei, Y., He, C., Liu, H., Shen, G., & Feng, J. (2022). Compositional Analysis of Four Kinds of Citrus Fruits with an NMR-Based Method for Understanding Nutritional Value and Rational Utilization: From Pericarp to Juice. Molecules, 27(8), 2579. https://doi.org/10.3390/molecules27082579







