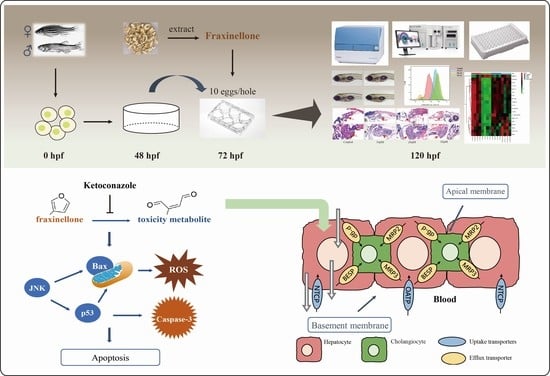
Fraxinellone Induces Hepatotoxicity in Zebrafish through Oxidative Stress and the Transporters Pathway
Abstract
:1. Introduction
2. Results
2.1. Effects of FRA on Survival of Zebrafish Larvae
2.2. Hepatotoxic Effects of FRA on Zebrafish Larvae
2.3. Histopathological Evaluation of FRA-Induced Hepatotoxicity
2.4. Effects of FRA on ROS Levels of Zebrafish Larvae
2.5. Effects of FRA on the Expression of Bile Acid Transporters and JNK/p53 Pathway
2.6. Inhibitory Effects of KCZ on FRA-Induced Hepatotoxicity
2.7. KCZ Altered the Expression Patterns of Transporter and Apoptotic Proteins Inhibited by FRA
2.8. KCZ Restored Amino Acid Metabolism Altered by FRA
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Animals
4.3. Drug Administration
4.4. Determination of Survival Rate
4.5. Microscopic Evaluation of Hepatotoxicity
4.5.1. Liver Gray Value Assay
4.5.2. AO Staining
4.6. Liver Biochemical Assay
4.7. Histopathological Analysis
4.8. ROS Assay
4.9. qRT-PCR Analysis
4.10. Western Blot Analysis
4.11. Targeted Amino Acid Metabolomics Test
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- European Association for the Study of the Liver. Electronic address eee, Clinical Practice Guideline Panel C, Panel m, representative EGB. EASL Clinical Practice Guidelines: Drug-induced liver injury. J. Hepatol. 2019, 70, 1222–1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, J.; Teschke, R. Traditional Chinese Medicine and Herb-induced Liver Injury: Comparison with Drug-induced Liver Injury. J. Clin. Transl. Hepatol. 2018, 6, 57–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrade, R.J.; Chalasani, N.; Björnsson, E.S.; Suzuki, A.; Kullak-Ublick, G.A.; Watkins, P.B.; Devarbhavi, H.; Merz, M.; Lucena, M.I.; Kaplowitz, N.; et al. Drug-induced liver injury. Nat. Rev. Dis. Primers 2019, 5, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Real, M.; Barnhill, M.S.; Higley, C.; Rosenberg, J.; Lewis, J.H. Drug-Induced Liver Injury: Highlights of the Recent Literature. Drug Saf. 2019, 42, 365–387. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ruan, J.; Sun, F.; Wang, H.; Yang, S.; Zhang, Y.; Yan, J.; Yu, H.; Guo, Y.; Zhang, Y.; et al. Anti-inflammatory Limonoids From Cortex Dictamni. Front Chem. 2020, 8, 73. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Xian, Y.F.; Loo, S.; Chan, W.Y.; Liu, L.; Lin, Z.X. Anti-atopic dermatitis effects of dictamni cortex: Studies on in vitro and in vivo experimental models. Phytomedicine 2021, 82, 153453. [Google Scholar] [CrossRef]
- Li, Z.-Q.; Jiang, L.-L.; Zhao, D.-S.; Zhou, J.; Wang, L.-L.; Wu, Z.-T.; Zheng, X.; Shi, Z.-Q.; Li, P.; Li, H.-J. The Modulatory Role of CYP3A4 in Dictamnine-Induced Hepatotoxicity. Front. Pharmacol. 2018, 9, 1033. [Google Scholar] [CrossRef] [Green Version]
- Shi, F.; Pan, H.; Cui, B.; Li, Y.; Huang, L.; Lu, Y. Dictamnine-induced hepatotoxicity in mice: The role of metabolic activation of furan. Toxicol. Appl. Pharmacol. 2019, 364, 68–76. [Google Scholar] [CrossRef]
- Wang, R.; Qi, X.; Yoshida, E.M.; Mendez-Sanchez, N.; Teschke, R.; Sun, M.; Liu, X.; Su, C.; Deng, J.; Deng, H.; et al. Clinical characteristics and outcomes of traditional Chinese medicine-induced liver injury: A systematic review. Expert Rev. Gastroenterol. Hepatol. 2018, 12, 425–434. [Google Scholar] [CrossRef]
- Huang, L.; Li, Y.; Pan, H.; Lu, Y.; Zhou, X.; Shi, F. Cortex dictamni-induced liver injury in mice: The role of P450-mediated metabolic activation of furanoids. Toxicol Lett. 2020, 330, 41–52. [Google Scholar] [CrossRef]
- Lang, X.; Zhang, X.; Wang, D.; Zhou, W. In Vitro and In Vivo Metabolic Activation of Obacunone, A Bioactive and Potentially Hepatotoxic Constituent of Dictamni Cortex. Planta Med. 2020, 86, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liu, Q.; Zahang, J.; Yao, J.; Wang, C.; Zhang, Y.; Li, Y.; Zhang, X.; Zhang, L. Cytochrome P450-Mediated Bioactivation: Implication for the Liver Injury Induced by Fraxinellone, A Bioactive Constituent from Dictamni Cortex. Chem. Res. Toxicol. 2020, 33, 1960–1968. [Google Scholar] [CrossRef]
- Li, H.-Y.; Yang, J.-B.; Li, W.-F.; Qiu, C.-X.; Hu, G.; Wang, S.-T.; Song, Y.-F.; Gao, H.-Y.; Liu, Y.; Wang, Q.; et al. In vivo hepatotoxicity screening of different extracts, components, and constituents of Polygoni Multiflori Thunb. in zebrafish (Danio rerio) larvae. Biomed. Pharmacother. 2020, 131, 110524. [Google Scholar] [CrossRef] [PubMed]
- De Souza Anselmo, C.; Sardela, V.F.; De Sousa, V.P.; Pereira, H.M.G. Zebrafish (Danio rerio): A valuable tool for predicting the metabolism of xenobiotics in humans? Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2018, 212, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.; Cavanaugh, K.; Verbueken, E.; Pype, C.; Casteleyn, C.; Van Ginneken, C.; Van Cruchten, S. Xenobiotic metabolism in the zebrafish: A review of the spatiotemporal distribution, modulation and activity of Cytochrome P450 families 1 to 3. J. Toxicol. Sci. 2016, 41, 1. [Google Scholar] [CrossRef] [Green Version]
- Pham, D.H.; Yin, C. Zebrafish as a Model to Study Cholestatic Liver Diseases. Methods Mol. Biol. 2019, 1981, 273–289. [Google Scholar] [CrossRef]
- Foulkes, M.J.; Henry, K.M.; Rougeot, J.; Hooper-Greenhill, E.; Loynes, C.A.; Jeffrey, P.; Fleming, A.; Savage, C.O.; Meijer, A.H.; Jones, S.; et al. Expression and regulation of drug transporters in vertebrate neutrophils. Sci. Rep. 2017, 7, 4967. [Google Scholar] [CrossRef] [Green Version]
- Nigam, S.K. What do drug transporters really do? Nat. Rev. Drug Discov. 2015, 14, 29–44. [Google Scholar] [CrossRef] [Green Version]
- Telbisz, A.; Homolya, L. Recent advances in the exploration of the bile salt export pump (BSEP/ABCB11) function. Expert Opin. Ther. Targets 2016, 20, 501–514. [Google Scholar] [CrossRef] [Green Version]
- International Transporter Consortium; Giacomini, K.M.; Huang, S.M.; Tweedie, D.J.; Benet, L.Z.; Brouwer, K.L.R.; Chu, X.; Dahlin, A.; Evers, R.; Fischer, V.; et al. Membrane transporters in drug development. Nat. Rev. Drug. Discov. 2010, 9, 215–236. [Google Scholar] [CrossRef]
- Choi, J.M.; Oh, S.J.; Lee, S.Y.; Im, J.H.; Oh, J.M.; Ryu, C.S.; Kwak, H.C.; Lee, J.-Y.; Kang, K.W.; Kim, S.K. HepG2 cells as an in vitro model for evaluation of cytochrome P450 induction by xenobiotics. Arch. Pharm. Res. 2015, 38, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Xuan, J.; Chen, S.; Ning, B.; Tolleson, W.H.; Guo, L. Development of HepG2-derived cells expressing cytochrome P450s for assessing metabolism-associated drug-induced liver toxicity. Chem. Biol. Interact. 2016, 255, 63–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.M.; Oh, S.J.; Lee, J.-Y.; Jeon, J.S.; Ryu, C.S.; Kim, Y.-M.; Lee, K.; Kim, S.K. Prediction of Drug-Induced Liver Injury in HepG2 Cells Cultured with Human Liver Microsomes. Chem. Res. Toxicol. 2015, 28, 872–885. [Google Scholar] [CrossRef]
- Gerets, H.H.J.; Tilmant, K.; Gerin, B.; Chanteux, H.; Depelchin, B.O.; Dhalluin, S.; Atienzar, F.A. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol. Toxicol. 2012, 28, 69–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banales, J.M.; Iñarrairaegui, M.; Arbelaiz, A.; Milkiewicz, P.; Muntane, J.; Muñoz-Bellvis, L.; La Casta, A.; Gonzalez, L.M.; Arretxe, E.; Alonso, C.; et al. Serum Metabolites as Diagnostic Biomarkers for Cholangiocarcinoma, Hepatocellular Carcinoma, and Primary Sclerosing Cholangitis. Hepatology 2019, 70, 47–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kou, F.; Zhu, B.; Zhou, W.; Lv, C.; Cheng, Y.; Wei, H. Targeted metabolomics in the cell culture media reveals increased uptake of branched amino acids by breast cancer cells. Anal. Biochem. 2021, 624, 114192. [Google Scholar] [CrossRef]
- Kakisaka, K.; Yoshida, Y.; Suzuki, Y.; Sato, T.; Kuroda, H.; Miyasaka, A.; Takikawa, Y. Serum markers for mitochondrial dysfunction and cell death are possible predictive indicators for drug-induced liver injury by direct acting antivirals. Hepatol. Res. 2018, 48, 78–86. [Google Scholar] [CrossRef] [Green Version]
- Bando, K.; Kunimatsu, T.; Sakai, J.; Kimura, J.; Funabashi, H.; Seki, T.; Bamba, T.; Fukusaki, E. GC-MS-based metabolomics reveals mechanism of action for hydrazine induced hepatotoxicity in rats. J. Appl. Toxicol. 2011, 31, 524–535. [Google Scholar] [CrossRef]
- Yu, M.; Zhu, Y.; Cong, Q.; Wu, C. Metabonomics Research Progress on Liver Diseases. Can. J. Gastroenterol. Hepatol. 2017, 2017, 8467192. [Google Scholar] [CrossRef]
- An, Z.; Hu, T.; Lv, Y.; Li, P.; Liu, L. Targeted amino acid and related amines analysis based on iTRAQ(R)-LC-MS/MS for discovering potential hepatotoxicity biomarkers. J. Pharm. Biomed. Anal. 2020, 178, 112812. [Google Scholar] [CrossRef]
- Troisi, J.; Masarone, M.; Caruso, R.; Persico, M. Metabolomics in the progression of fatty liver disease. J. Hepatol. 2018, 68, S568–S569. [Google Scholar] [CrossRef]
- Li, H.; Yang, H.; Li, P.; Li, M.; Yao, Q.; Min, L.; Zhang, Y.; Wang, J.; Zheng, N. Maillard reaction products with furan ring, like furosine, cause kidney injury through triggering ferroptosis pathway. Food Chem. 2020, 319, 126368. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.A. Reactive metabolites in the biotransformation of molecules containing a furan ring. Chem. Res. Toxicol. 2013, 26, 6–25. [Google Scholar] [CrossRef] [Green Version]
- Pei, J.; Xiao, W.; Zhu, D.; Ji, X.; Shi, L.; Deng, X. Cytochrome P450 Enzyme-Mediated Bioactivation as an Underlying Mechanism of Columbin-Induced Hepatotoxicity. Chem. Res. Toxicol. 2020, 33, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Pauli-Magnus, C.; Meier, P.J. Hepatocellular transporters and cholestasis. J. Clin. Gastroenterol. 2005, 39, S103. [Google Scholar] [CrossRef]
- Liu, X. ABC Family Transporters. Adv. Exp. Med. Biol. 2019, 1141, 13–100. [Google Scholar] [CrossRef]
- Chen, H.L.; Wu, S.H.; Hsu, S.H.; Liou, B.Y.; Chen, H.L.; Chang, M.H. Jaundice revisited: Recent advances in the diagnosis and treatment of inherited cholestatic liver diseases. J. Biomed. Sci. 2018, 25, 75. [Google Scholar] [CrossRef] [Green Version]
- Roma, M.G.; Barosso, I.R.; Miszczuk, G.S.; Crocenzi, F.A.; Pozzi, E.J.S. Dynamic Localization of Hepatocellular Transporters: Role in Biliary Excretion and Impairment in Cholestasis. Curr. Med. Chem. 2019, 26, 1113–1154. [Google Scholar] [CrossRef]
- Dietrich, C.G.; Geier, A. Effect of drug transporter pharmacogenetics on cholestasis. Expert Opin. Drug Metab. Toxicol. 2014, 10, 1533–1551. [Google Scholar] [CrossRef]
- Amirneni, S.; Haep, N.; Gad, M.A.; Soto-Gutierrez, A.; Squires, J.E.; Florentino, R.M. Molecular overview of progressive familial intrahepatic cholestasis. World J. Gastroenterol. 2020, 26, 7470–7484. [Google Scholar] [CrossRef]
- Lowjaga, K.; Kirstgen, M.; Müller, S.; Goldmann, N.; Geyer, J. Long-term trans-inhibition of the hepatitis B and D virus receptor NTCP by taurolithocholic acid. AJP Gastrointest. Liver Physiol. 2020, 320, G66–G80. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.-L.; Cen, J.; Wang, J.-B.; Zhang, F.; Xia, Q.; Wang, X.; Chen, X.-Q.; Wang, R.-C.; Hsiao, C.-D.; Liu, K.-C.; et al. Mechanism of isoniazid-induced hepatotoxicity in zebrafish larvae: Activation of ROS-mediated ERS, apoptosis and the Nrf2 pathway. Chemosphere 2019, 227, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, Y.; Liu, Y.; Yin, K.; Wang, D.; Li, B.; Yu, H.; Xing, M. ROS-Induced Hepatotoxicity under Cypermethrin: Involvement of the Crosstalk between Nrf2/Keap1 and NF-kappaB/ikappaB-alpha Pathways Regulated by Proteasome. Environ. Sci. Technol. 2021, 55, 6171–6183. [Google Scholar] [CrossRef]
- Yang, S.; Lian, G. ROS and diseases: Role in metabolism and energy supply. Mol. Cell. Biochem. 2020, 467, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Yi, J.; Lu, J.; Nie, M.; Huang, M.; Rong, J.; Zhu, Z.; Chen, J.; Zhou, X.; Li, B.; et al. N-Acetylcysteine Reduces ROS-Mediated Oxidative DNA Damage and PI3K/Akt Pathway Activation Induced by Helicobacter pylori Infection. Oxid. Med. Cell. Longev. 2018, 2018, 1874985. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Nikulenkov, F.; Zawacka-Pankau, J.; Li, H.; Gabdoulline, R.; Xu, J.; Eriksson, S.; Hedström, E.; Issaeva, N.; Kel, A.; et al. ROS-dependent activation of JNK converts p53 into an efficient inhibitor of oncogenes leading to robust apoptosis. Cell Death Differ. 2014, 21, 612–623. [Google Scholar] [CrossRef] [Green Version]
- Banankhah, P.S.; Garnick, K.A.; Greenblatt, D.J. Ketoconazole-Associated Liver Injury in Drug-Drug Interaction Studies in Healthy Volunteers. J. Clin. Pharmacol. 2016, 56, 1196–1202. [Google Scholar] [CrossRef]
- Loerracher, A.K.; Braunbeck, T. Cytochrome P450-dependent biotransformation capacities in embryonic, juvenile and adult stages of zebrafish (Danio rerio)-a state-of-the-art review. Arch. Toxicol. 2021, 95, 2299–2334. [Google Scholar] [CrossRef]
- Saad, M.; Matheeussen, A.; Bijttebier, S.; Verbueken, E.; Pype, C.; Casteleyn, C.; Van Ginneken, C.; Apers, S.; Maes, L.; Cos, P.; et al. In vitro CYP-mediated drug metabolism in the zebrafish (embryo) using human reference compounds. Toxicol. Vitr. 2017, 42, 329–336. [Google Scholar] [CrossRef]
- Mak, T.W.; Grusdat, M.; Duncan, G.S.; Dostert, C.; Nonnenmacher, Y.; Cox, M.; Binsfeld, C.; Hao, Z.; Bruestle, A.; Itsumi, M.; et al. Glutathione Primes T Cell Metabolism for Inflammation. Immunity 2017, 46, 675–689. [Google Scholar] [CrossRef] [Green Version]
- Bansal, A.; Simon, M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ullah, R.; Jo, M.H.; Riaz, M.; Alam, S.I.; Saeed, K.; Ali, W.; Rehman, I.U.; Ikram, M.; Kim, M.O. Glycine, the smallest amino acid, confers neuroprotection against D-galactose-induced neurodegeneration and memory impairment by regulating c-Jun N-terminal kinase in the mouse brain. J. Neuroinflamm. 2020, 17, 303. [Google Scholar] [CrossRef] [PubMed]
- Holecek, M. Branched-chain amino acids and ammonia metabolism in liver disease: Therapeutic implications. Nutrition 2013, 29, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Ishigami, M.; Luo, F.; Lingyun, M.; Ishizu, Y.; Kuzuya, T.; Hayashi, K.; Nakano, I.; Ishikawa, T.; Feng, G.-G.; et al. Branched-chain amino acids alleviate hepatic steatosis and liver injury in choline-deficient high-fat diet induced NASH mice. Metabolism 2017, 69, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Heidari, R.; Mohammadi, H.; Ghanbarinejad, V.; Ahmadi, A.; Ommati, M.M.; Niknahad, H.; Jamshidzadeh, A.; Azarpira, N.; Abdoli, N. Proline supplementation mitigates the early stage of liver injury in bile duct ligated rats. J. Basic Clin. Physiol. Pharmacol. 2018, 30, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Wrzosek, L.; Ciocan, D.; Hugot, C.; Spatz, M.; Dupeux, M.; Houron, C.; Moal, V.L.-L.; Puchois, V.; Ferrere, G.; Trainel, N.; et al. Microbiota tryptophan metabolism induces aryl hydrocarbon receptor activation and improves alcohol-induced liver injury. Gut 2021, 70, 1299–1308. [Google Scholar] [CrossRef]
- Maezono, K.; Kajiwara, K.; Mawatari, K.; Shinkai, A.; Torii, K.; Maki, T. Alanine protects liver from injury caused by D-galactosamine and CCl4. Hepatology 1996, 24, 185–191. [Google Scholar]
- Katoch, S.; Patial, V. Zebrafish: An emerging model system to study liver diseases and related drug discovery. J. Appl. Toxicol. 2021, 41, 33–51. [Google Scholar] [CrossRef]
- MacRae, C.A.; Peterson, R.T. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015, 14, 721–731. [Google Scholar] [CrossRef]
- Wong, W.; Huang, Y.; Wu, Z.; Kong, Y.; Luan, J.; Zhang, Q.; Pan, J.; Yan, K.; Zhang, Z. Mvda is required for zebrafish early development. Biol. Res. 2021, 54, 17. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Q.; Li, W.; Li, H.; Bao, J.; Yang, C.; Wang, A.; Wei, J.; Chen, S.; Jin, H. Role of Nrf2 in the antioxidation and oxidative stress induced developmental toxicity of honokiol in zebrafish. Toxicol. Appl. Pharmacol. 2019, 373, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Stevens, A.P.; Fagerer, S.R.; Kaspar, H.; Oefner, P.J. Amino Acid Analysis in Physiological Samples by GC-MS with Propyl Chloroformate Derivatization and iTRAQ-LC-MS/MS. Methods Mol. Biol. 2019, 2030, 173–190. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics by Gas Chromatography-Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30.4.1–30.4.32. [Google Scholar] [CrossRef] [PubMed]









| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Gene ID |
|---|---|---|---|
| P-gp | TCGTCATCCTCGCTGTTAGC | ATGACAGCACTTCCTCTGCC | 100136865 |
| Bsep | ATTTCCGCAGCAAAGAAGGC | GTTTTTGACCCCGGGAAAGC | 571189 |
| Ntcp | TGGTCATCCGCTTGGCTTTA | GAAGGCAGTGAAGGGCATGA | 562260 |
| Cyp3a65 | AAACCCTGATGAGCATGGAC | CAAGTCTTTGGGGATGAGGA | 553969 |
| JNK1 | GAGAACTGGTCCTGATGA | TCACCAGATAAACATCCT | 65236 |
| p53 | GGGCAATCAGCGAGCAAA | ACTGACCTTCCTGAGTCTCCA | 30590 |
| Caspase-3 | TCAGGCTTGTCGAGGAAC | CTGCCATACTTTGTCATCATTT | 140621 |
| GAPDH | ACAGCAACACAGAAGACCGT | GGCAGGTTTCTCAAGACGGA | 317743 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Bao, J.; Li, J.; Li, W.; Tian, M.; Qiu, C.; Pang, F.; Li, X.; Yang, J.; Hu, Y.; et al. Fraxinellone Induces Hepatotoxicity in Zebrafish through Oxidative Stress and the Transporters Pathway. Molecules 2022, 27, 2647. https://doi.org/10.3390/molecules27092647
Wang S, Bao J, Li J, Li W, Tian M, Qiu C, Pang F, Li X, Yang J, Hu Y, et al. Fraxinellone Induces Hepatotoxicity in Zebrafish through Oxidative Stress and the Transporters Pathway. Molecules. 2022; 27(9):2647. https://doi.org/10.3390/molecules27092647
Chicago/Turabian StyleWang, Shuting, Jie Bao, Jie Li, Wanfang Li, Mengyin Tian, Caixia Qiu, Fei Pang, Xin Li, Jianbo Yang, Yuchi Hu, and et al. 2022. "Fraxinellone Induces Hepatotoxicity in Zebrafish through Oxidative Stress and the Transporters Pathway" Molecules 27, no. 9: 2647. https://doi.org/10.3390/molecules27092647
APA StyleWang, S., Bao, J., Li, J., Li, W., Tian, M., Qiu, C., Pang, F., Li, X., Yang, J., Hu, Y., Wang, S., & Jin, H. (2022). Fraxinellone Induces Hepatotoxicity in Zebrafish through Oxidative Stress and the Transporters Pathway. Molecules, 27(9), 2647. https://doi.org/10.3390/molecules27092647