In Silico Prediction of Cross-Reactive Epitopes of Tropomyosin from Shrimp and Other Arthropods Involved in Allergy
Abstract
:1. Introduction
2. Results
2.1. Sequence Retrieval and Analysis
2.2. Identification of Possible Cross-Reactive Linear B Cell Epitopes
2.3. Identification of Cross-Reactive T Cell Epitope Candidates
2.4. Structural Modeling, Refinement and Validation
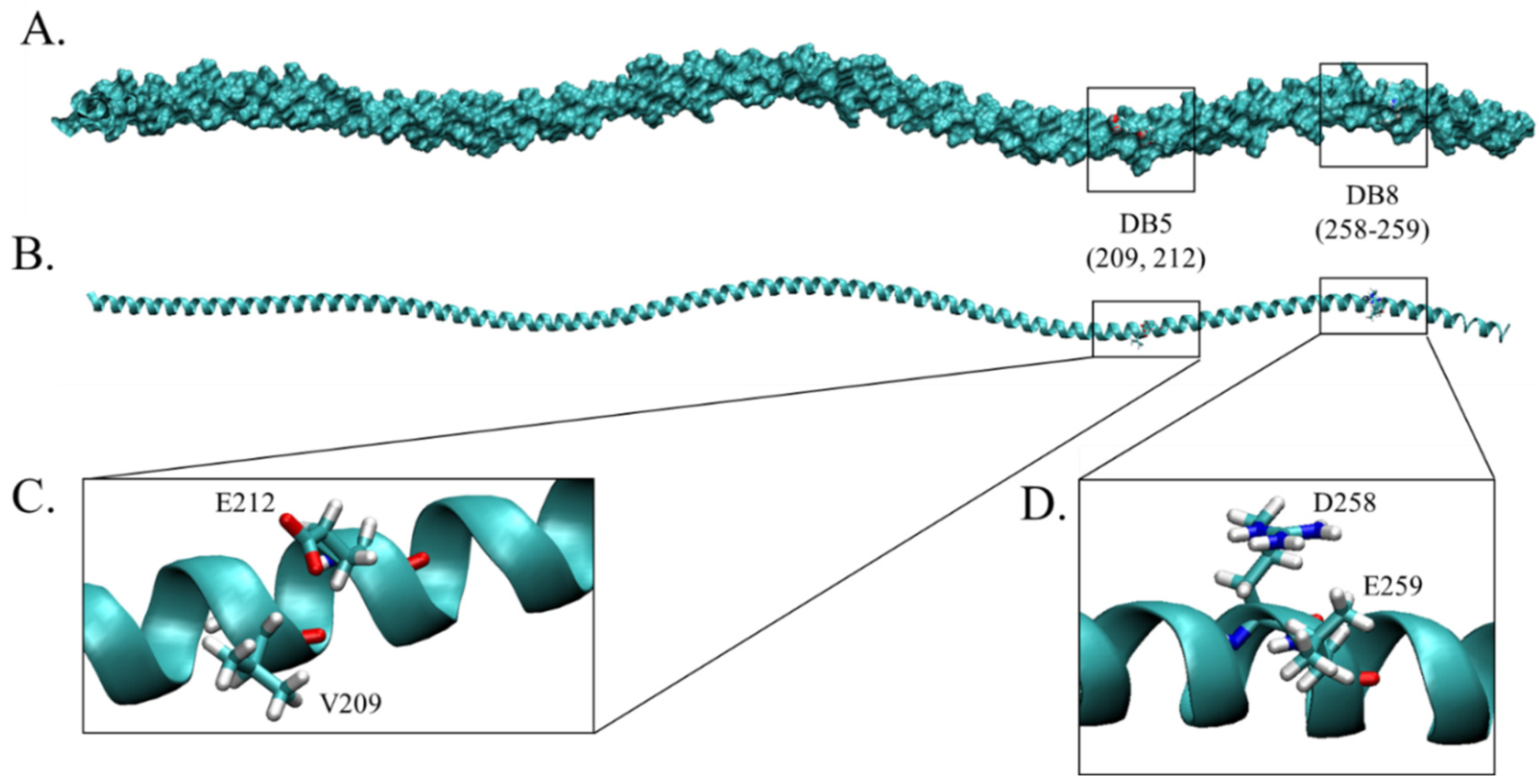
2.5. Discontinuous B Cell Epitope Prediction
3. Discussion
4. Materials and Methods
4.1. Sequence Retrieval and Analysis
4.2. Prediction of Linear B Cell Epitopes and IgE Epitopes
4.3. Prediction of T Cell Epitope
4.4. Structure Retrieval and Homology Modeling
4.5. Structural Refinement and Validation
4.6. Discontinuous Epitope Prediction
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Renz, H.; Allen, K.J.; Sicherer, S.H.; Sampson, H.A.; Lack, G.; Beyer, K.; Oettgen, H.C. Food allergy. Nat. Rev. Dis. Prim. 2018, 4, 17098. [Google Scholar] [CrossRef]
- Ruethers, T.; Taki, A.C.; Johnston, E.B.; Nugraha, R.; Le, T.T.K.; Kalic, T.; McLean, T.R.; Kamath, S.D.; Lopata, A.L. Seafood allergy: A comprehensive review of fish and shellfish allergens. Mol. Immunol. 2018, 100, 28–57. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Lau, C.H.; Sita, E.E.; Smith, B.; Greenhawt, M.J. Factors associated with reported food allergy tolerance among US children. Ann. Allergy Asthma Immunol. 2013, 111, 194–198.e4. [Google Scholar] [CrossRef] [PubMed]
- Alonso, L.L.; Armstrong, L.; Warrington, S.J. Shellfish Allergy. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Thalayasingam, M.; Gerez, I.F.A.; Yap, G.C.; Llanora, G.V.; Chia, I.P.; Chua, L.; Lee, C.J.A.O.; Ta, L.D.H.; Cheng, Y.K.; Thong, B.; et al. Clinical and immunochemical profiles of food challenge proven or anaphylactic shrimp allergy in tropical Singapore. Clin. Exp. Allergy 2015, 45, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Pascal, M.; Grishina, G.; Yang, A.C.; Sánchez-García, S.; Lin, J.; Towle, D.; Ibañez, M.D.; Sastre, J.; Sampson, H.A.; Ayuso, R. Molecular Diagnosis of Shrimp Allergy: Efficiency of Several Allergens to Predict Clinical Reactivity. J. Allergy Clin. Immunol. Pract. 2015, 3, 521–529.e10. [Google Scholar] [CrossRef]
- Kamath, S.D.; Johnston, E.B.; Iyer, S.; Schaeffer, P.M.; Koplin, J.; Allen, K.; Lopata, A.L. IgE reactivity to shrimp allergens in infants and their cross-reactivity to house dust mite. Pediatr. Allergy Immunol. 2017, 28, 703–707. [Google Scholar] [CrossRef]
- Ayuso, R.; Sánchez-Garcia, S.; Lin, J.; Fu, Z.; Ibáñez, M.D.; Carrillo, T.; Blanco, C.; Goldis, M.; Bardina, L.; Sastre, J.; et al. Greater epitope recognition of shrimp allergens by children than by adults suggests that shrimp sensitization decreases with age. J. Allergy Clin. Immunol. 2010, 125, 1286–1293.e3. [Google Scholar] [CrossRef]
- Gamez, C.; Sánchez-García, S.; Ibáñez, M.D.; López, R.; Aguado, E.; López, E.; Sastre, B.; Sastre, J.; Del Pozo, V. Tropomyosin IgE-positive results are a good predictor of shrimp allergy. Allergy 2011, 66, 1375–1383. [Google Scholar] [CrossRef]
- Bauermeister, K.; Wangorsch, A.; Garoffo, L.P.; Reuter, A.; Conti, A.; Taylor, S.L.; Lidholm, J.; De Witt, Å.M.; Enrique, E.; Vieths, S.; et al. Generation of a comprehensive panel of crustacean allergens from the North Sea Shrimp Crangon crangon. Mol. Immunol. 2011, 48, 1983–1992. [Google Scholar] [CrossRef]
- Wai, C.Y.Y.; Leung, N.Y.H.; Chu, K.H.; Leung, P.S.C.; Leung, A.S.Y.; Wong, G.W.K.; Leung, T.F. Overcoming Shellfish Allergy: How Far Have We Come? Int. J. Mol. Sci. 2020, 21, 2234. [Google Scholar] [CrossRef] [Green Version]
- Nugraha, R.; Kamath, S.D.; Johnston, E.; Karnaneedi, S.; Ruethers, T.; Lopata, A.L. Conservation Analysis of B-Cell Allergen Epitopes to Predict Clinical Cross-Reactivity Between Shellfish and Inhalant Invertebrate Allergens. Front. Immunol. 2019, 10, 2676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faber, M.A.; Pascal, M.; El Kharbouchi, O.; Sabato, V.; Hagendorens, M.M.; Decuyper, I.I.; Bridts, C.H.; Ebo, D.G. Shellfish allergens: Tropomyosin and beyond. Allergy 2017, 72, 842–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asero, R.; Pravettoni, V.; Scala, E.; Villalta, D. House Dust Mite-Shrimp Allergen Interrelationships. Curr. Allergy Asthma Rep. 2020, 20, 9. [Google Scholar] [CrossRef] [PubMed]
- Chiang, W.C.; Kidon, M.I.; Liew, W.K.; Goh, A.; Tang, J.P.L.; Chay, O.M. The changing face of food hypersensitivity in an Asian community. Clin. Exp. Allergy 2007, 37, 1055–1061. [Google Scholar] [CrossRef]
- Wang, J.; Calatroni, A.; Visness, C.M.; Sampson, H.A. Correlation of specific IgE to shrimp with cockroach and dust mite exposure and sensitization in an inner-city population. J. Allergy Clin. Immunol. 2011, 128, 834–837. [Google Scholar] [CrossRef]
- Boquete, M.; Iraola, V.; Morales, M.; Pinto, H.; Francisco, C.; Carballás, C.; Carnes, J. Seafood hypersensitivity in mite sensitized individuals: Is tropomyosin the only responsible allergen? Ann. Allergy Asthma Immunol. 2011, 106, 223–229. [Google Scholar] [CrossRef]
- Arlian, L.G.; Morgan, M.S.; Vyszenski-Moher, D.L.; Sharra, D. Cross-reactivity between storage and dust mites and between mites and shrimp. Exp. Appl. Acarol. 2009, 47, 159–172. [Google Scholar] [CrossRef]
- Wong, L.H.; Huang, C.H.; Lee, B.W. Shellfish and House Dust Mite Allergies: Is the Link Tropomyosin? Allergy Asthma Immunol. Res. 2016, 8, 101–106. [Google Scholar] [CrossRef] [Green Version]
- Gelis, S.; Rueda, M.; Valero, A.; Fernández, E.A.; Moran, M.; Fernández-Caldas, E. Shellfish Allergy: Unmet Needs in Diagnosis and Treatment. J. Investig. Allergy Clin. Immunol. 2020, 30, 409–420. [Google Scholar] [CrossRef]
- Saidova, A.; Hershkop, A.M.; Ponce, M.; Eiwegger, T. Allergen-Specific T Cells in IgE-Mediated Food Allergy. Arch. Immunol. Ther. Exp. 2018, 66, 161–170. [Google Scholar] [CrossRef]
- Pereira, J.; Simpkin, A.J.; Hartmann, M.D.; Rigden, D.J.; Keegan, R.M.; Lupas, A.N. High-accuracy protein structure prediction in CASP14. Proteins Struct. Funct. Bioinform. 2021, 89, 1687–1699. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.R.; Won, J.; Heo, L.; Seok, C. GalaxyRefine2: Simultaneous refinement of inaccurate local regions and overall protein structure. Nucleic Acids Res. 2019, 47, W451–W455. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo—distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef] [Green Version]
- Shek, L.; Cabrera-Morales, E.A.; Soh, S.E.; Gerez, I.; Ng, P.Z.; Yi, F.C.; Ma, S.; Lee, B.W. A population-based questionnaire survey on the prevalence of peanut, tree nut, and shellfish allergy in 2 Asian populations. J. Allergy Clin. Immunol. 2010, 126, 324–331.e7. [Google Scholar] [CrossRef]
- Savage, J.; Johns, C.B. Food Allergy: Epidemiology and natural history. Immunol. Allergy Clin. N. Am. 2015, 35, 45–59. [Google Scholar] [CrossRef] [Green Version]
- McGowan, E.C.; Keet, C.A. Prevalence of self-reported food allergy in the National Health and Nutrition Examination Survey (NHANES) 2007–2010. J. Allergy Clin. Immunol. 2013, 132, 1216–1219.e5. [Google Scholar] [CrossRef] [Green Version]
- Rosenfield, L.; Tsoulis, M.W.; Milio, K.; Schnittke, M.; Kim, H. High rate of house dust mite sensitization in a shrimp allergic southern Ontario population. Allergy Asthma Clin. Immunol. 2017, 13, 5. [Google Scholar] [CrossRef] [Green Version]
- Diez, S.; Puerta, L.; Martínez, D.; Muñoz, M.; Hernández, K.; Sánchez, J. Clinical Relevance of Shrimp Sensitization in Patients with Allergic Rhinitis: Anti-Der p 10 IgE as Predictor. Int. Arch. Allergy Immunol. 2021, 182, 971–979. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2018: Meeting the Sustainable Development Goals; the State of World Fisheries and Aquaculture (SOFIA); FAO: Rome, Italy, 2018; ISBN 978-92-5-130562-1. [Google Scholar]
- Neudecker, T.; Damm, U. The by-catch situation in German brown shrimp (Crangon crangonL.) fisheries with particular reference to plaice (Pleuronectes platessa L.). J. Appl. Ichthyol. 2010, 26, 67–74. [Google Scholar] [CrossRef]
- Thomas, W.R.; Hales, B.J.; Smith, W.-A. House dust mite allergens in asthma and allergy. Trends Mol. Med. 2010, 16, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Pomés, A.; Mueller, G.A.; Randall, T.A.; Chapman, M.D.; Arruda, L.K. New Insights into Cockroach Allergens. Curr. Allergy Asthma Rep. 2017, 17, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.-Y.; Mei, X.-J.; Hu, M.-J.; Yang, Y.; Liu, M.; Li, M.-S.; Zhang, M.-L.; Cao, M.-J.; Liu, G.-M. Analysis of the Allergenic Epitopes of Tropomyosin from Mud Crab Using Phage Display and Site-Directed Mutagenesis. J. Agric. Food Chem. 2018, 66, 9127–9137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, X.-M.; Xiao, H.; Nowak-Wegrzyn, A.; Zhou, P. IgE-binding epitope mapping of tropomyosin allergen (Exo m 1) from Exopalaemon modestus, the freshwater Siberian prawn. Food Chem. 2020, 309, 125603. [Google Scholar] [CrossRef]
- Ayuso, R.; Lehrer, S.B.; Reese, G. Identification of Continuous, Allergenic Regions of the Major Shrimp Allergen Pen a 1 (Tropomyosin). Int. Arch. Allergy Immunol. 2002, 127, 27–37. [Google Scholar] [CrossRef]
- Fu, L.; Wang, J.; Ni, S.; Wang, C.; Wang, Y. Identification of Allergenic Epitopes and Critical Amino Acids of Major Allergens in Chinese Shrimp (Penaeus chinensis) by Immunoinformatics Coupled with Competitive-Binding Strategy. J. Agric. Food Chem. 2018, 66, 2944–2953. [Google Scholar] [CrossRef]
- Beezhold, D.H.; Hickey, V.L.; Slater, J.E.; Sussman, G.L. Human IgE-binding epitopes of the latex allergen Hev b 5. J. Allergy Clin. Immunol. 1999, 103, 1166–1172. [Google Scholar] [CrossRef]
- Sereda, M.J.; Hartmann, S.; Lucius, R. Helminths and allergy: The example of tropomyosin. Trends Parasitol. 2008, 24, 272–278. [Google Scholar] [CrossRef]
- Ravkov, E.V.; Pavlov, I.Y.; Martins, T.B.; Gleich, G.J.; Wagner, L.A.; Hill, H.R.; Delgado, J.C. Identification and validation of shrimp-tropomyosin specific CD4 T cell epitopes. Hum. Immunol. 2013, 74, 1542–1549. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Ponomarenko, J.; Zhu, Z.; Tamang, D.; Wang, P.; Greenbaum, J.; Lundegaard, C.; Sette, A.; Lund, O.; Bourne, P.E.; et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012, 40, W525–W530. [Google Scholar] [CrossRef] [Green Version]
- Tan, Z.; Chan, C.H.; Maleska, M.; Jara, B.B.; Lohman, B.K.; Ricks, N.J.; Bond, D.R.; Hammond, M.C. The Signaling Pathway That cGAMP Riboswitches Found: Analysis and Application of Riboswitches to Study cGAMP Signaling in Geobacter sulfurreducens. Int. J. Mol. Sci. 2022, 23, 1183. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Enríquez, A.; Herrera-Camacho, I.; Millán-Pérez-Peña, L.; Reyes-Leyva, J.; Santos-López, G.; Rivera-Benítez, J.F.; Rosas-Murrieta, N.H. Predicted 3D model of the M protein of Porcine Epidemic Diarrhea Virus and analysis of its immunogenic potential. PLoS ONE 2022, 17, e0263582. [Google Scholar] [CrossRef] [PubMed]
- Nitanai, Y.; Minakata, S.; Maeda, K.; Oda, N.; Maéda, Y. Crystal Structures of Tropomyosin: Flexible Coiled-Coil. Adv. Exp. Med. Biol. 2007, 592, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.H.; Zhou, Z.; Reshetnikova, L.; Robinson, H.; Yammani, R.D.; Tobacman, L.S.; Cohen, C. Structure of the mid-region of tropomyosin: Bending and binding sites for actin. Proc. Natl. Acad. Sci. USA 2005, 102, 18878–18883. [Google Scholar] [CrossRef] [Green Version]
- Ochiai, Y.; Ozawa, H. Biochemical and physicochemical characteristics of the major muscle proteins from fish and shellfish. Fish. Sci. 2020, 86, 729–740. [Google Scholar] [CrossRef]
- Broekman, H.C.H.P.; Knulst, A.C.; De Jong, G.; Gaspari, M.; Jager, C.F.D.H.; Houben, G.F.; Verhoeckx, K.C.M. Is mealworm or shrimp allergy indicative for food allergy to insects? Mol. Nutr. Food Res. 2017, 61, 1601061. [Google Scholar] [CrossRef]
- de Gier, S.; Verhoeckx, K. Insect (food) allergy and allergens. Mol. Immunol. 2018, 100, 82–106. [Google Scholar] [CrossRef]
- Kamemura, N.; Sugimoto, M.; Tamehiro, N.; Adachi, R.; Tomonari, S.; Watanabe, T.; Mito, T. Cross-allergenicity of crustacean and the edible insect Gryllus bimaculatus in patients with shrimp allergy. Mol. Immunol. 2019, 106, 127–134. [Google Scholar] [CrossRef]
- Eddy, S.R. Where did the BLOSUM62 alignment score matrix come from? Nat. Biotechnol. 2004, 22, 1035–1036. [Google Scholar] [CrossRef]
- Sharma, N.; Patiyal, S.; Dhall, A.; Pande, A.; Arora, C.; Raghava, G.P.S. AlgPred 2.0: An improved method for predicting allergenic proteins and mapping of IgE epitopes. Brief. Bioinform. 2021, 22, bbaa294. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynisson, B.; Barra, C.; Kaabinejadian, S.; Hildebrand, W.H.; Peters, B.; Nielsen, M. Improved Prediction of MHC II Antigen Presentation through Integration and Motif Deconvolution of Mass Spectrometry MHC Eluted Ligand Data. J. Proteome Res. 2020, 19, 2304–2315. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [Green Version]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [Green Version]
- Ponomarenko, J.V.; Bui, H.-H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef] [Green Version]
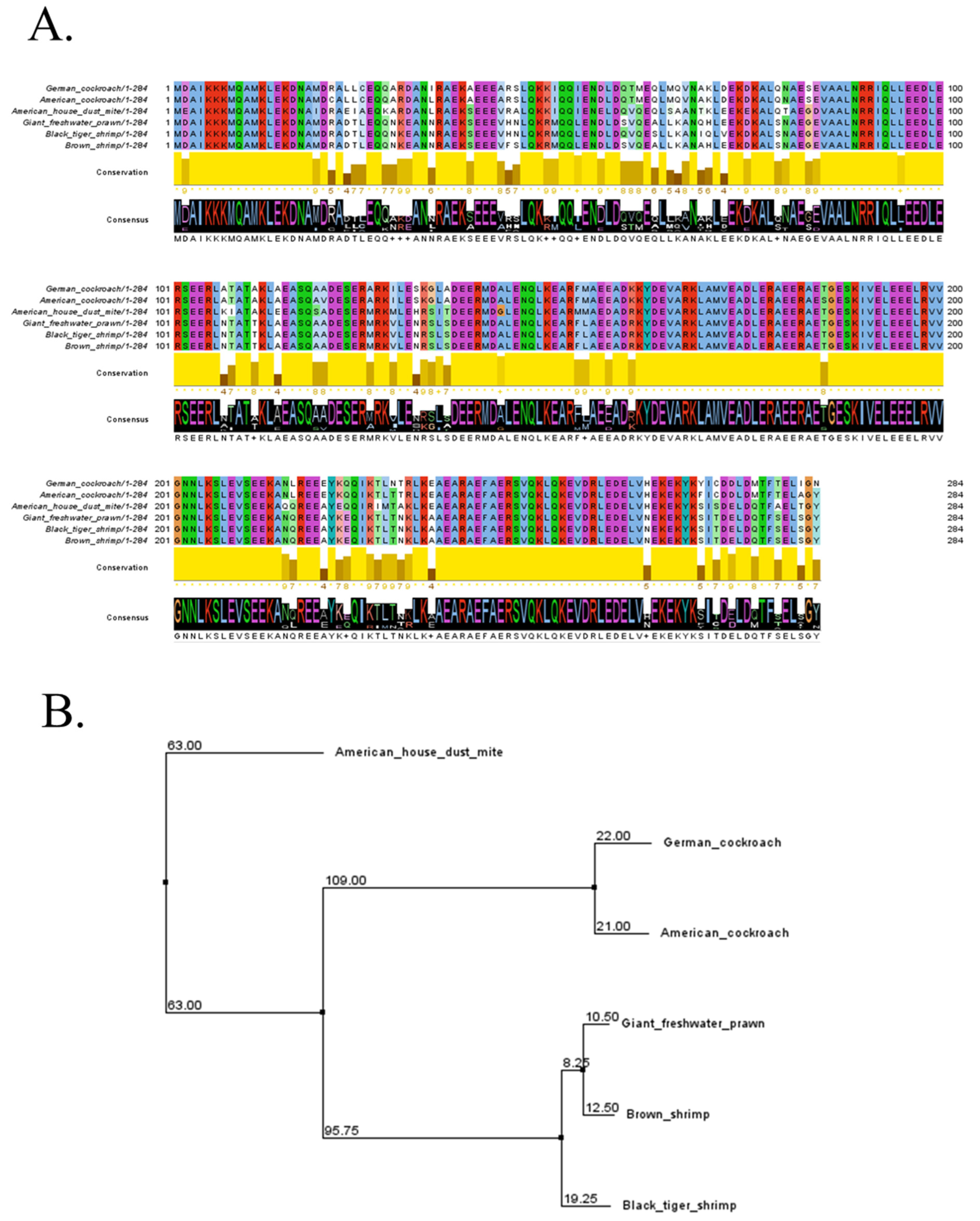


| Allergen | Biological Source | Common Name | Uniprot Acession No. | Structure Sources |
|---|---|---|---|---|
| Mac r 1 | Macrobrachium rosenbergii | Giant freshwater prawn | D3XNR9 | Robetta |
| Pen m 1 | Penaeus monodon | Black tiger shrimp | A1KYZ2 | AlphaFold |
| Pen a 1 | Penaeus aztecus | Brown shrimp | Q3Y8M6 | Robetta |
| Der f 1 | Dermatophagoides farinae | American house dust mite | Q23939 | AlphaFold |
| Bla g 7 | Blattella germanica | German cockroach | Q9NG56 | Robetta |
| Per a 7 | Periplaneta americana | American cockroach | Q9UB83 | AlphaFold |
| AlgPRED (IgE Epitope) | BepiPRED-2.0 (Linear B-Cell Epitopes) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Residue | Peptide ID | Epitope Sequence | Species Consensus | AHDM | AC | BTS | BS | GC | GFP | |
| Group 1 overlapped peptides | 238–252 | B1 | RAEFAERSVQKLQKE | All | / | / | / | / | / | / |
| 241–255 | B2 | FAERSVQKLQKEVDR | All | / | / | / | / | / | / | |
| 243–259 | B3 | ERSVQKLQKEVDRLEDE | All | / | / | / | / | / | / | |
| 244–258 | B4 | RSVQKLQKEVDRLED | All | / | / | / | / | / | / | |
| 251–259 | B5 | KEVDRLEDE | All | / | / | / | / | / | / | |
| Group 2 overlapped peptides | 187–197 | B6 | ESKIVELEEEL | All | / | / | / | / | / | / |
| 190–204 | B7 | IVELEEELRVVGNNL | All | / | / | / | / | / | / | |
| 193–207 | B8 | LEEELRVVGNNLKSL | All | / | / | / | / | / | / | |
| 196–210 | B9 | ELRVVGNNLKSLEVS | All | / | / | / | / | / | / | |
| 199–206 | B10 | VVGNNLKS | All | / | / | / | / | / | / | |
| 199–213 | B11 | VVGNNLKSLEVSEEK | All | / | / | / | / | / | / | |
| Unique 1 | 85–91 | B12 | VAALNRR | All | / | / | / | / | / | / |
| Unique 2 | 137–141 | B13 | DEERM | All | / | / | / | / | / | / |
| Unique 3 | 144–151 | B14 | LENQLKEA | All | / | / | / | / | / | / |
| Unique 4 | 169–183 | B15 | LAMVEADLERAEERA | All | / | / | / | / | / | X |
| Epitope Prediction | MHCII Alleles | Peptide ID | Residues | Consensus Core Epitope | Binding Type |
|---|---|---|---|---|---|
| HLA-DQ | DQA1*101–DQB1*501 | - | - | ||
| DQA1*102–DQB1*602 | T01 | 166–174 | ARKLAMVEA | Weak binder | |
| DQA1*301–DQB1*302 | T02 | 188–196 | SKIVELEEE | Strong binder | |
| T03 | 187–195 | ESKIVELEE | Strong binder | ||
| T04 | 167–175 | RKLAMVEAD | Weak binder | ||
| T05 | 169–177 | LAMVEADLE | Weak binder | ||
| T06 | 165–173 | VARKLAMVE | Weak binder | ||
| T07 | 173–181 | EADLERAEE | Weak binder | ||
| T08 | 186–194 | GESKIVELE | Weak binder | ||
| DQA1*401–DQB1*402 | T05 | 169–177 | LAMVEADLE | Strong binder | |
| T07 | 186–194 | EADLERAEE | Strong binder | ||
| T02 | 188–196 | SKIVELEEE | Weak binder | ||
| T06 | 165–173 | VARKLAMVE | Weak binder | ||
| T03 | 187–195 | ESKIVELEE | Weak binder | ||
| T09 | 204–212 | LKSLEVSEE | Weak binder | ||
| DQA1*501–DQB1*201 | T05 | 169–177 | LAMVEADLE | Strong binder | |
| T02 | 188–196 | SKIVELEEE | Strong binder | ||
| T05 | 169–177 | LAMVEADLE | Weak binder | ||
| T10 | 189–197 | KIVELEEEL | Weak binder | ||
| T11 | 253–261 | VDRLEDELV | Weak binder | ||
| DQA1*501–DQB1*301 | - | - | |||
| HLA-DR | DRB1*0101 | - | - | ||
| DRB1*0301 | T12 | 144–152 | LENQLKEAR | Weak binder | |
| T13 | 190–198 | IVELEEELR | Weak binder | ||
| T14 | 172–180 | VEADLERAE | Weak binder | ||
| DRB3*0101 | - | - | |||
| DRB4*0101 | T05 | 169–177 | LAMVEADLE | Strong binder | |
| T15 | 171–179 | MVEADLERA | Strong binder | ||
| T16 | res8–16 | MQAMKLEKD | Strong binder | ||
| T09 | 204–212 | LKSLEVSEE | Weak binder | ||
| T11 | 253–261 | VDRLEDELV | Weak binder | ||
| DRB5*0101 | T17 | 197–205 | LRVVGNNLK | Strong binder | |
| T13 | 190–198 | IVELEEELR | Weak binder |
| Ramachandran Plot (%) | ERRAT Overall Quality Factor | QMEAN Score | ProSA Z-Score | ||||
|---|---|---|---|---|---|---|---|
| Residues in Favorable Regions | Residues in Allowed Regions | Residues in Generally Allowed Regions | Residues in Disallowed Regions | ||||
| Black tiger shrimp | 100 | 0 | 0 | 0 | 100 | 0.78 | −2.91 |
| Brown shrimp | 98.9 | 0.7 | 0 | 0.4 | 100 | 0.68 | −4.94 |
| Giant Freshwater prawn | 99.3 | 0.4 | 0.4 | 0 | 100 | 0.66 | −4.97 |
| American cockroach | 100 | 0 | 0 | 0 | 100 | 0.82 | −2.5 |
| American house dust mite | 100 | 0 | 0 | 0 | 100 | 0.81 | −3.01 |
| German cockroach | 100 | 0 | 0 | 0 | 100 | 0.82 | −2.63 |
| ID | Sequences | Residues | Linear B Cell Epitopes (IgE Epitope) | Linear T Cell Epitopes |
|---|---|---|---|---|
| DB1 | M [D or E] AIKK, M | 1–7, 8 | No | Yes (8; strong binder) |
| DB2 | QAMKLEKDNA[M or I]D[R or C] | 9–21 | No | Yes (9–16; strong binder) |
| DB3 | [L or N or I]R, E | 34–35, 37 | No | No |
| DB4 | [A or I or T], [DE or EE or VE], K, [S or G] | 69, 72–73, 76, 83 | No | No |
| DB5 | V, EK | 209, 212–213 | Yes | Yes (209, 212; weak) |
| DB6 | [T or I], [TR or AK or NK] | 227, 230–231 | No | No |
| DB7 | AR, E, RS, KL, E, RL | 237–238, 240, 244–245, 248–249, 252, 255–256 | Yes (238, 240, 244–245, 248–249, 252, 255–256) | Yes (255–256; weak) |
| DB8 | Vary | 258–262, 264–265 | Yes (258–259) | Yes (258–261; weak) |
| DB9 | Vary | 268–284 | No | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saetang, J.; Tipmanee, V.; Benjakul, S. In Silico Prediction of Cross-Reactive Epitopes of Tropomyosin from Shrimp and Other Arthropods Involved in Allergy. Molecules 2022, 27, 2667. https://doi.org/10.3390/molecules27092667
Saetang J, Tipmanee V, Benjakul S. In Silico Prediction of Cross-Reactive Epitopes of Tropomyosin from Shrimp and Other Arthropods Involved in Allergy. Molecules. 2022; 27(9):2667. https://doi.org/10.3390/molecules27092667
Chicago/Turabian StyleSaetang, Jirakrit, Varomyalin Tipmanee, and Soottawat Benjakul. 2022. "In Silico Prediction of Cross-Reactive Epitopes of Tropomyosin from Shrimp and Other Arthropods Involved in Allergy" Molecules 27, no. 9: 2667. https://doi.org/10.3390/molecules27092667







