Synthesis, X-ray Structure and Biological Studies of New Self-Assembled Cu(II) Complexes Derived from s-Triazine Schiff Base Ligand
Abstract
:1. Introduction
2. Results and Discussion
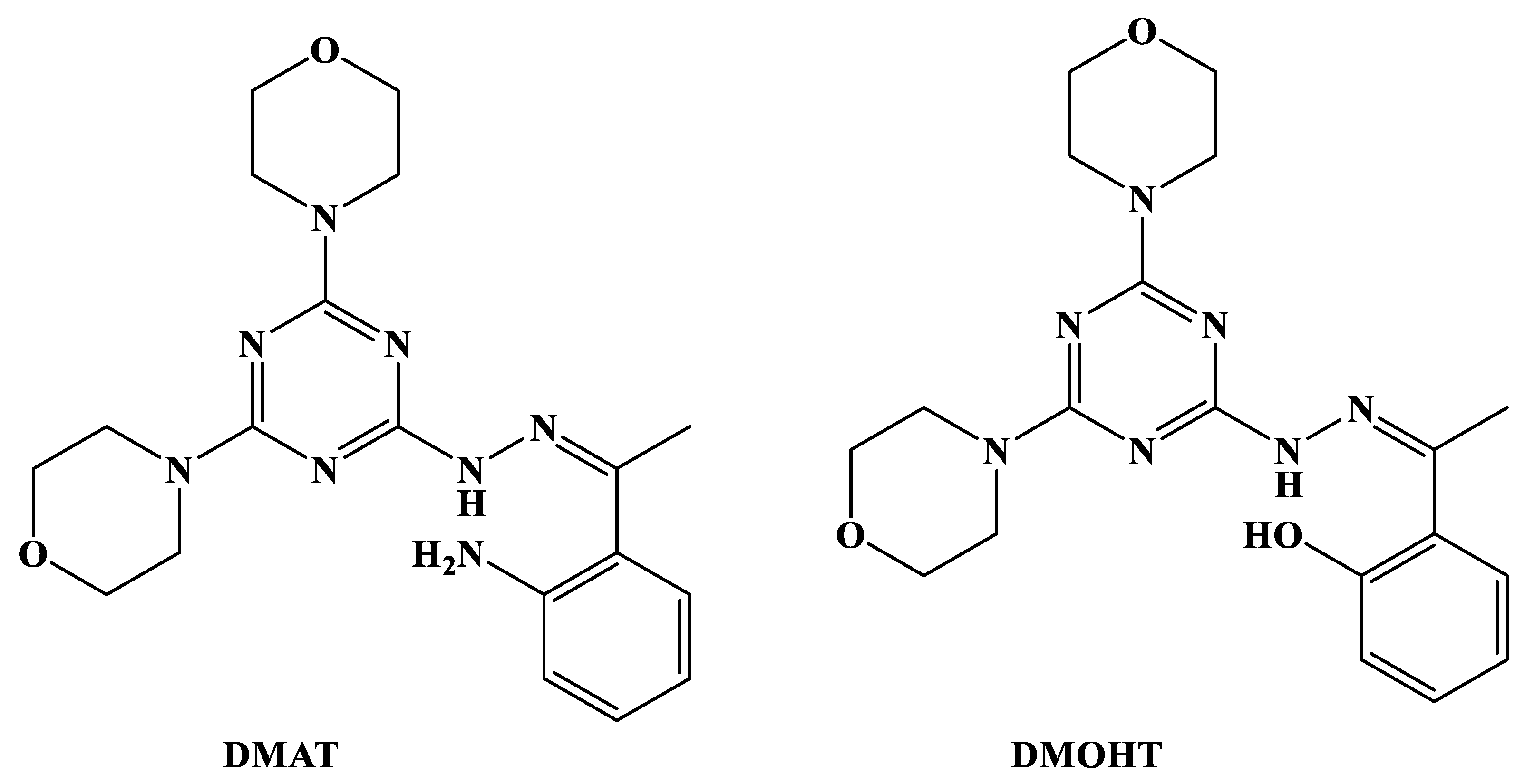
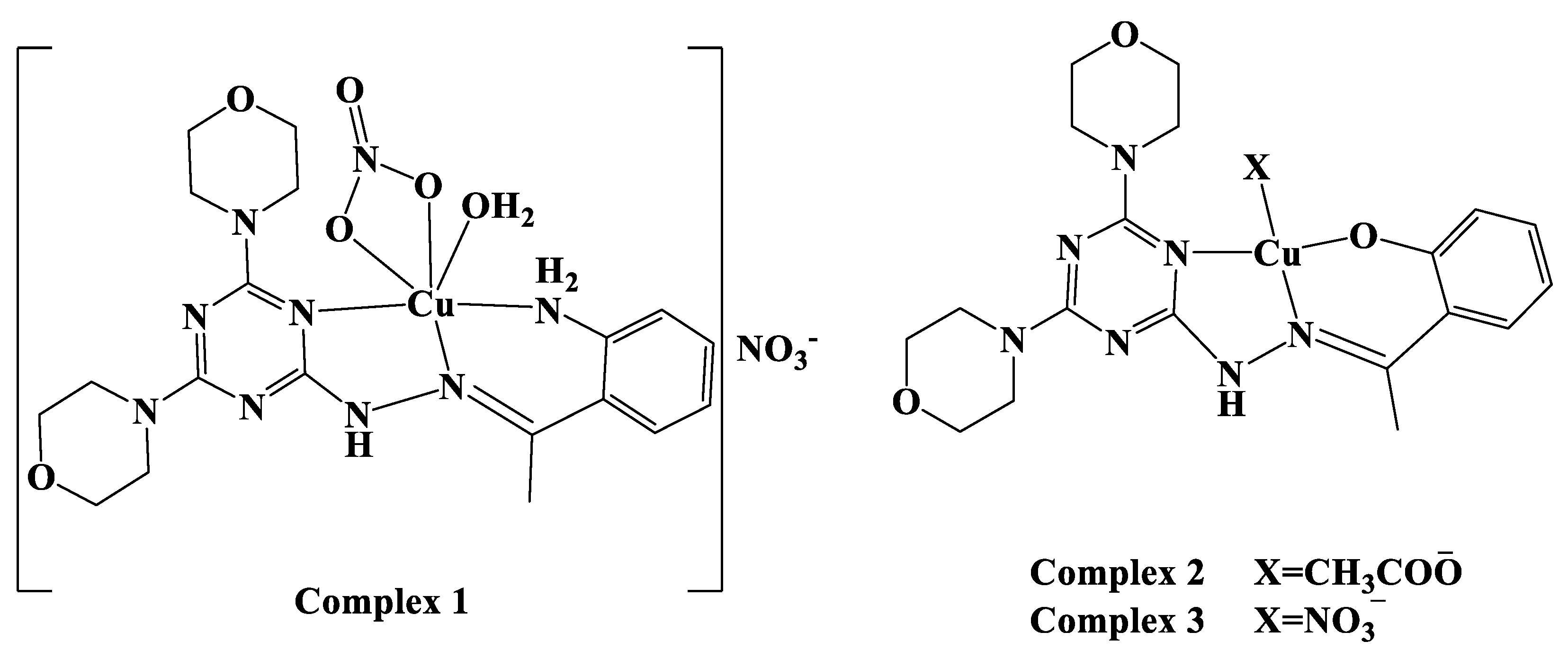
2.1. Synthesis and Characterizations
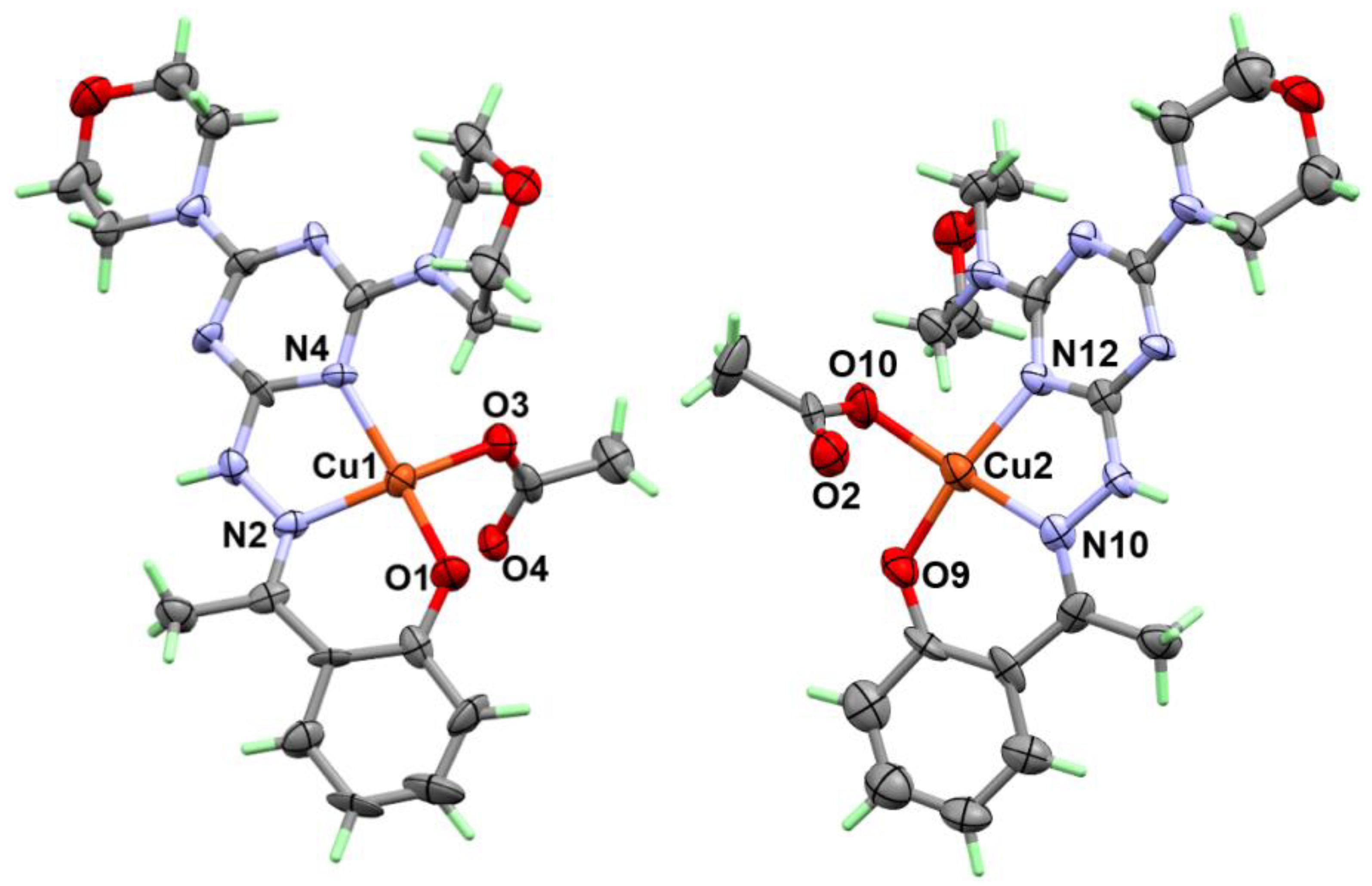
2.2. X-ray Structure
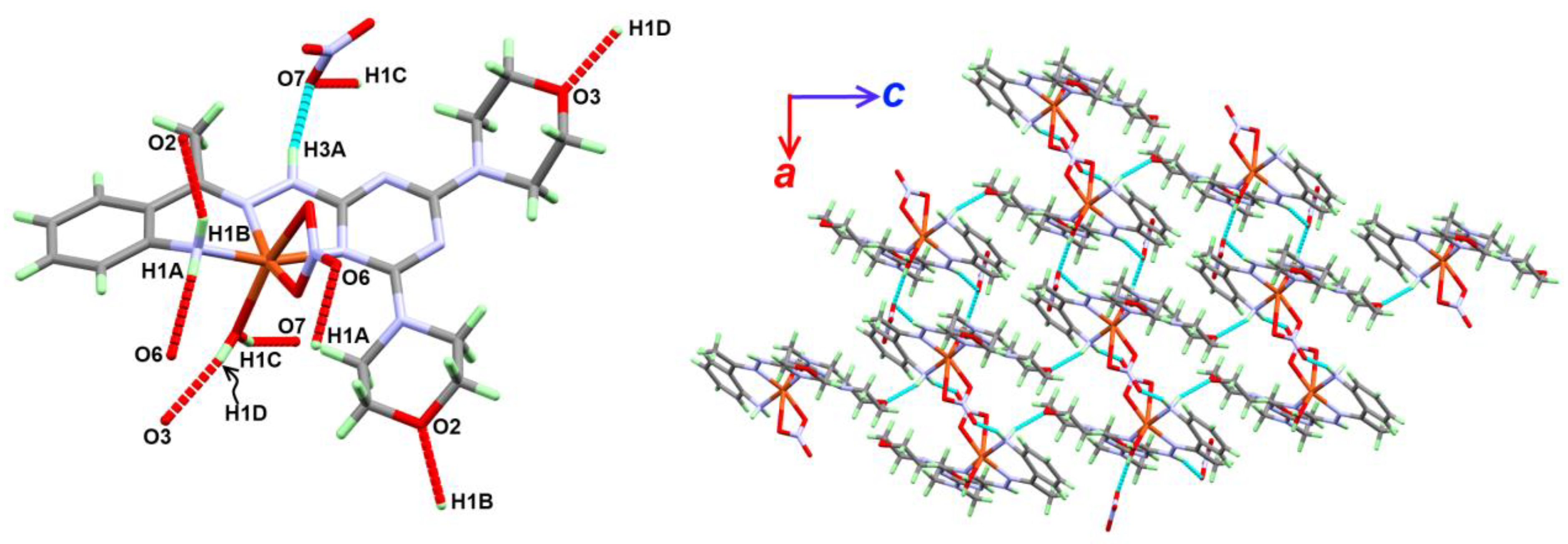
2.2.1. Structure Description of Complex 1
2.2.2. Structure Description of Complex 2
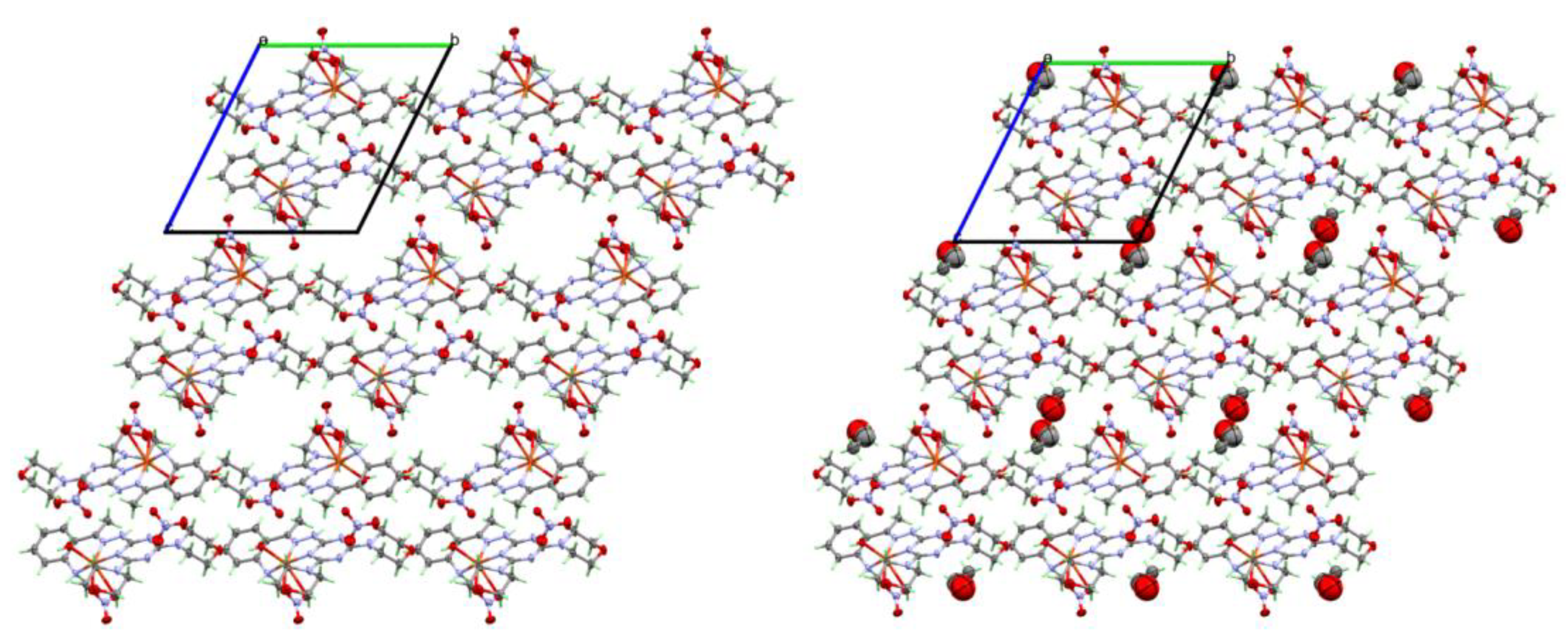
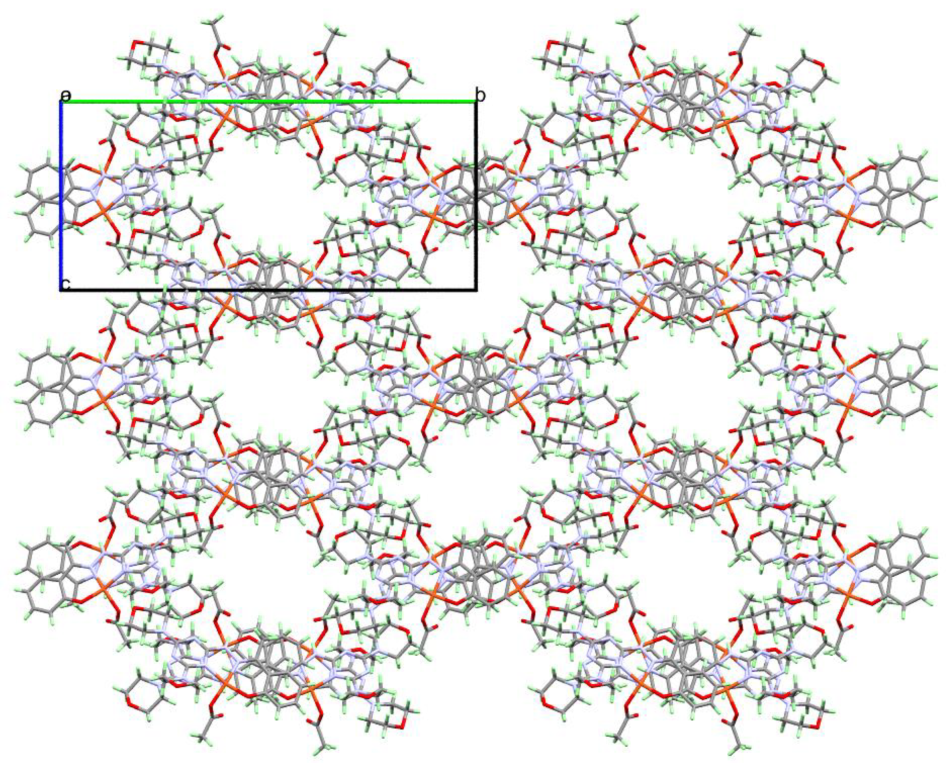
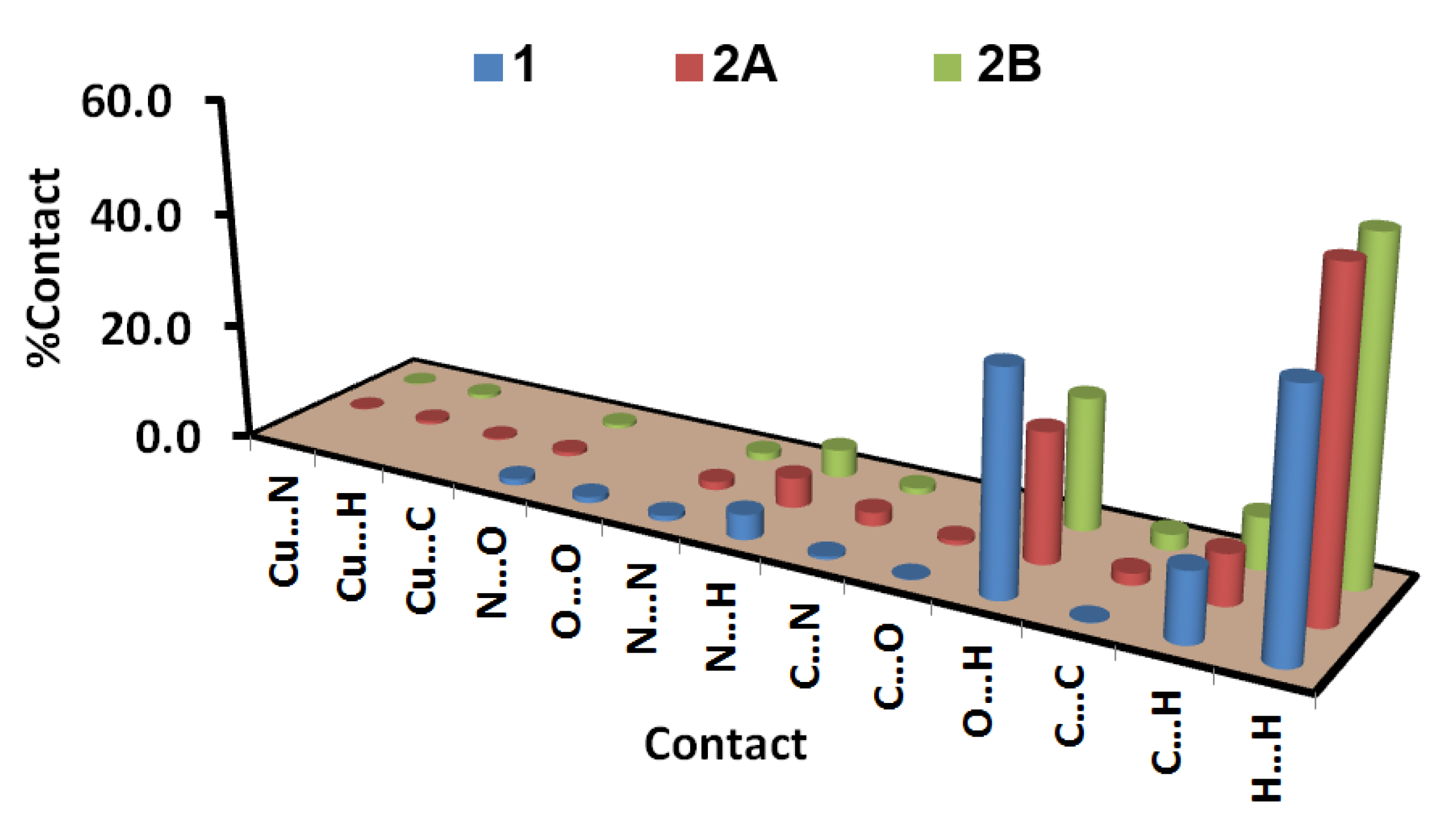
2.3. Analysis of Molecular Packing
2.4. XPS Study
2.5. FTIR Spectra
2.6. Biological Studies
2.6.1. Antimicrobial Activity
2.6.2. Antioxidant Activity
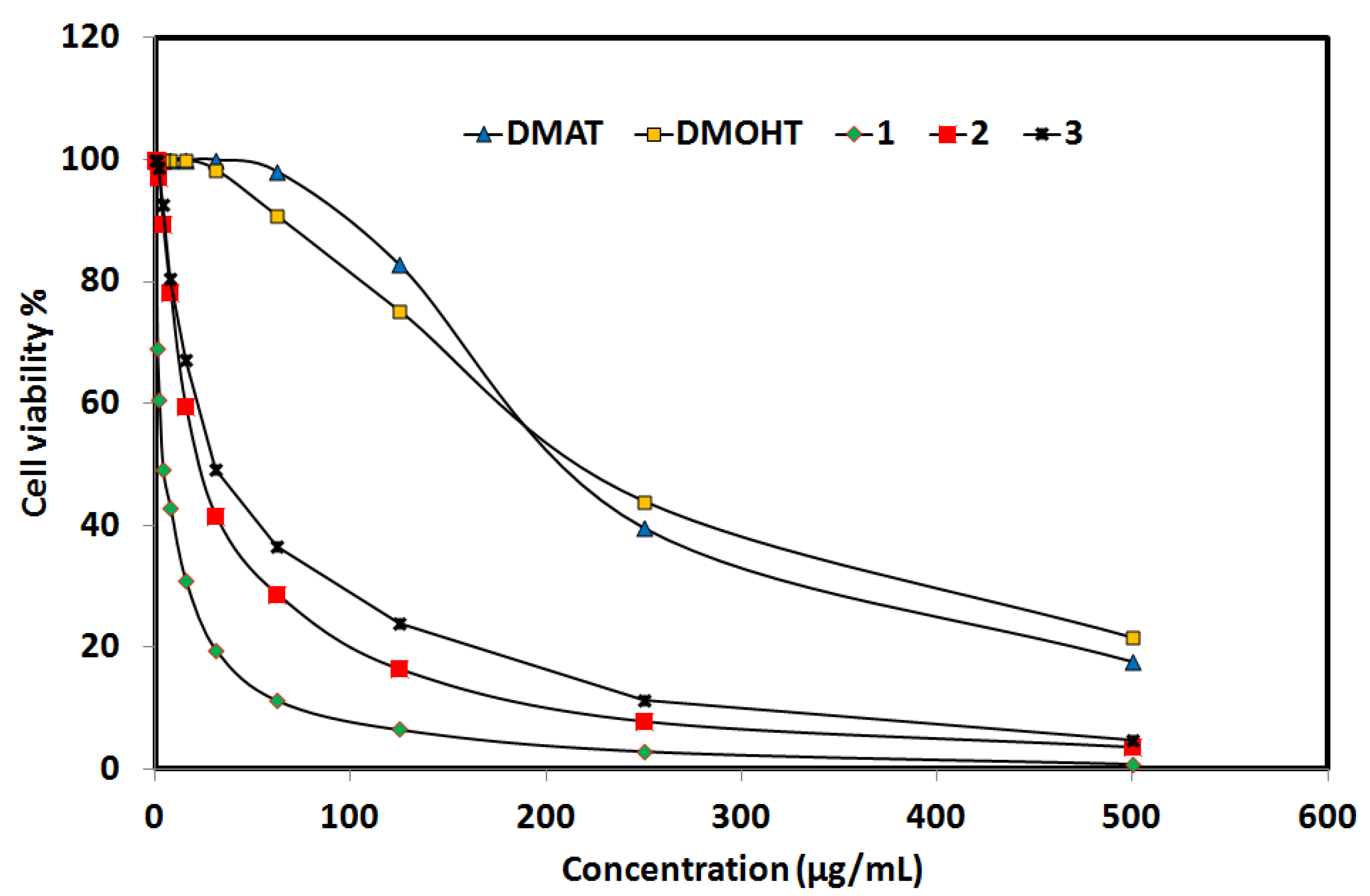
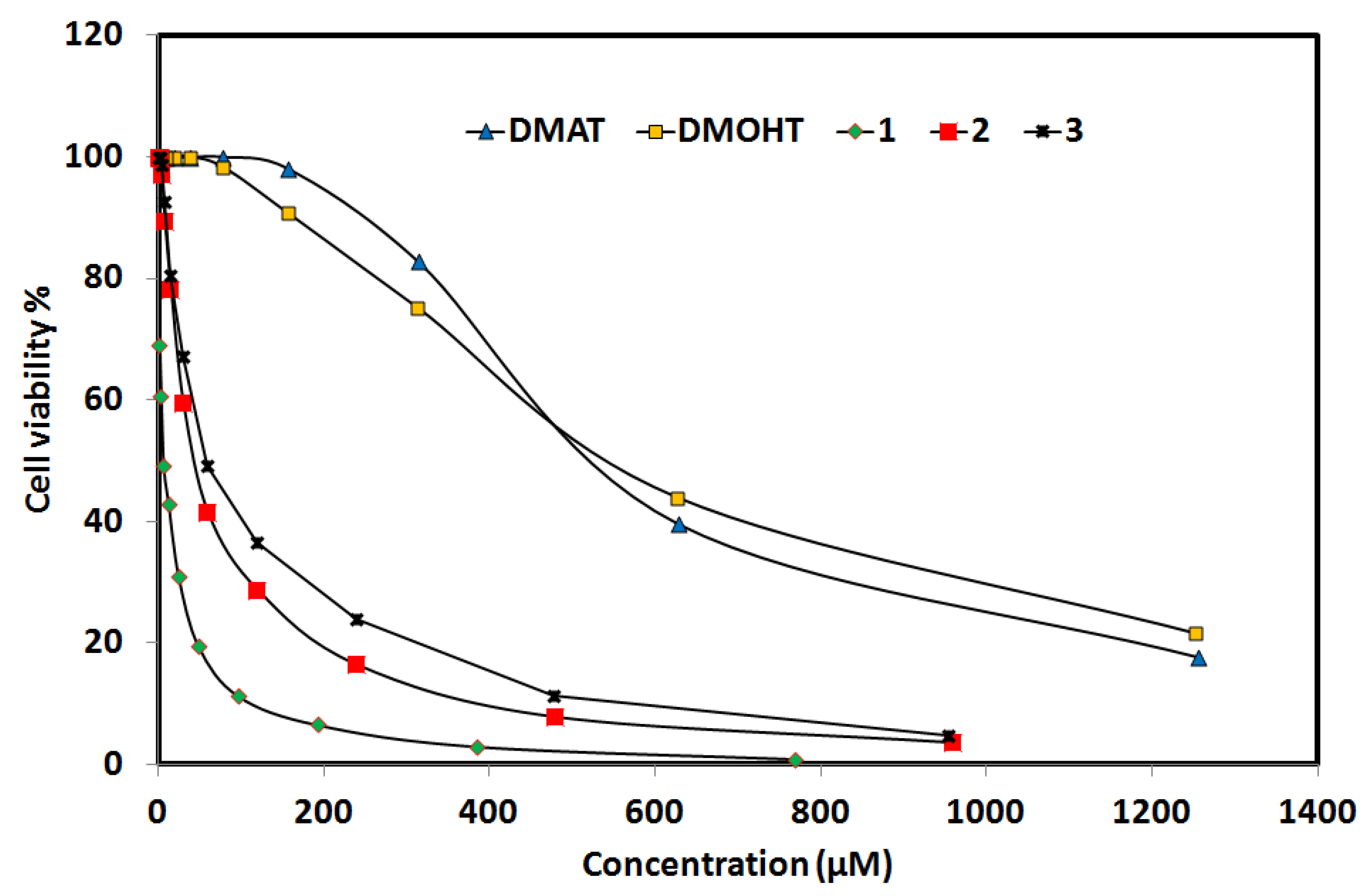
2.6.3. Anticancer Activity
3. Materials and Methods
3.1. Synthesis of Ligands
3.2. Syntheses of Cu(II) Complexes
3.2.1. Synthesis of Complex 1
3.2.2. Synthesis of Complex 2
3.2.3. Synthesis of [Cu(DMOT)(NO3)] Complex 3
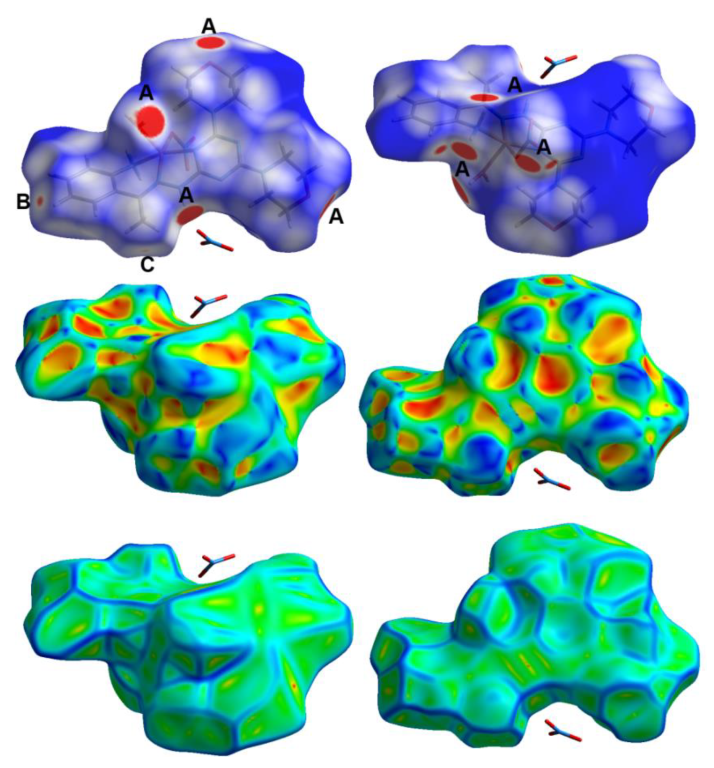
3.3. Hirshfeld Surface Analysis
3.4. Biological Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Lv, Y.; Meng, J.; Li, C.; Wang, X.; Ye, Y.; Sun, K. Update on the Synthesis of N-Heterocycles via Cyclization of Hydrazones (2017–2021). Adv. Synth. Catal. 2021, 363, 5235–5265. [Google Scholar] [CrossRef]
- Asif, M. Pharmacologically potentials of hydrazonone containing compounds: A promising scaffold. Int. J. Adv. Chem. 2014, 2, 85–103. [Google Scholar] [CrossRef] [Green Version]
- Tok, F.; Sağlık, B.N.; Özkay, Y.; Ilgın, S.; Kaplancıklı, Z.A.; Kaymakçıoğlu, B. Synthesis of new hydrazone derivatives and evaluation of their monoamine oxidase inhibitory activity. Bioorg. Chem. 2021, 114, 105038. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.C.; Sharma, D.; Sharma, A.; Saini, N.; Goyal, R.; Ola, M.; Chawla, R.; Thakur, V.K. Hydrazone comprising compounds as promising anti-infective agents: Chemistry and structure-property relationship. Mater. Today Chem. 2020, 18, 100349. [Google Scholar] [CrossRef]
- Lawrence, M.A.; Lorraine, S.C.; Wilson, K.A.; Wilson, K. Voltammetric properties and applications of hydrazones and azo moieties. Polyhedron 2019, 173, 114111. [Google Scholar] [CrossRef]
- Mali, S.N.; Thorat, B.R.; Gupta, D.R.; Pandey, A. Mini-Review of the Importance of Hydrazides and Their Derivatives-Synthesis and Biological Activity. Eng. Proc. 2021, 11, 21. [Google Scholar]
- Zhai, P.; Li, W.; Lin, J.; Li, X.; Wei, W.L.; Chen, W. Hydrazones as Substrates in the Synthesis of Isoxazolidines via a KOH-Promoted One-Pot Three-Component Cycloaddition with Nitroso Compounds and Olefins. J. Org. Chem. 2021, 86, 17710–17721. [Google Scholar] [CrossRef]
- Suvarapu, L.N.; Seo, Y.K.; Baek, S.O.; Ammireddy, V.R. Review on analytical and biological applications of hydrazones and their metal complexes. E-J. Chem. 2012, 9, 1288–1304. [Google Scholar] [CrossRef]
- Cao, W.; Liu, Y.; Zhang, T.; Jia, J. Synthesis, characterization, theoretical and antimicrobial studies of tridentate hydrazone metal complexes of Zn (II), Cd (II), Cu (II) and Co (III). Polyhedron 2018, 147, 62–68. [Google Scholar] [CrossRef]
- Bedia, K.K.; Elçin, O.; Seda, U.; Fatma, K.; Nathaly, S.; Sevim, R.; Dimoglo, A. Synthesis and characterization of novel hydrazide–hydrazones and the study of their structure–antituberculosis activity. Eur. J. Med. Chem. 2006, 41, 1253–1261. [Google Scholar] [CrossRef]
- Hussain, I.; Ullah, A.; Khan, A.U.; Khan, W.U.; Ullah, R. Synthesis, characterization and biological activities of hydrazone schiff base and its novel metals complexes. Sains Malays. 2019, 48, 1439–1446. [Google Scholar] [CrossRef]
- Iskander, M.F.; Khalil, T.E.; Foro, S. Synthesis, characterization and X-ray crystal structures of nickel (II) and copper (II) complexes derived from salicylaldehyde cyanoacetylhydrazone. An unexpected ligand transformation. Inorg. Chim. Acta 2016, 453, 633–646. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, Y.F.; Ding, H.; Li, X.; Lang, X. Hydrazone-linked 2D porphyrinic covalent organic framework photocatalysis for visible light-driven aerobic oxidation of amines to imines. J. Colloid Interface Sci. 2022, 610, 44654. [Google Scholar] [CrossRef] [PubMed]
- Khattab, T.A. From chromic switchable hydrazones to smart materials. Mater. Chem. Phys. 2020, 254, 123456. [Google Scholar] [CrossRef]
- Singh, R.B.; Jain, P.; Singh, R.P. Hydrazones as analytical reagents: A review. Talanta 1982, 29, 77–84. [Google Scholar] [CrossRef]
- Wahbeh, J.; Milkowski, S. The use of hydrazones for biomedical applications. Slas Technol. Transl. Life Sci. Innov. 2019, 24, 161–168. [Google Scholar] [CrossRef]
- Rollas, S.; Küçükgüzel, S.G. Biological activities of hydrazone derivatives. Molecules 2007, 12, 1910–1939. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.R.; Marella, A.; Alam, M.T.; Naz, R.; Akhter, M.; Shaquiquzzaman, M.; Saha, R.; Tanwar, O.; Alam, M.M.; Hooda, J. Review of biological activities of hydrazones. Indones. J. Pharm. 2012, 23, 193–202. [Google Scholar]
- Oliveira, C.B.J.; França, T.C.; LaPlante, S.R.; Villar, J.D. Synthesis and biological activity of hydrazones and derivatives: A review. Mini. Rev. Med. Chem. 2020, 20, 342–368. [Google Scholar]
- Gemma, S.; Kukreja, G.; Fattorusso, C.; Persico, M.; Romano, M.P.; Altarelli, M.; Savini, L.; Campiani, G.; Fattorusso, E.; Basilico, N.; et al. Synthesis of N1-arylidene-N2-quinolyl-and N2-acrydinylhydrazones as potent antimalarial agents active against CQ-resistant P. falciparum strains. Bioorg. Med. Chem. Let. 2006, 16, 5384–5388. [Google Scholar] [CrossRef]
- Lasri, J.; Haukka, M.; Al-Rasheed, H.H.; Abutaha, N.; El-Faham, A.; Soliman, S.M. Synthesis, structure and in vitro anticancer activity of Pd(II) complex of pyrazolyl-s-triazine ligand; A new example of metal-mediated hydrolysis of s-Triazine pincer ligand. Crystals 2021, 11, 119. [Google Scholar] [CrossRef]
- Yu, M.H.; Liu, X.T.; Space, B.; Chang, Z.; Bu, X.H. Metal-organic materials with triazine-based ligands: From structures to properties and applications. Coord. Chem. Rev. 2021, 427, 213518. [Google Scholar] [CrossRef]
- Hoog, P.; Gamez, P.; Driessen, W.L.; Reedijk, J. New polydentate and polynucleating N-donor ligands from amines and 2,4,6-trichloro-1,3,5-triazine. Tetrahedron Lett. 2002, 43, 6783–6786. [Google Scholar] [CrossRef]
- Patel, R.V.; Kumari, P.; Rajani, D.P.; Chikhalia, K.H. Synthesis, characterization and pharmacological activities of 2-[4-cyano-(3-trifluoromethyl) phenyl amino)]-4-(4-quinoline/coumarin-4-yloxy)-6-(fluoropiperazinyl)-s-triazines. J. Fluor. Chem. 2011, 132, 617–627. [Google Scholar] [CrossRef]
- Gamez, P.; Reedijk, J. 1,3,5-Triazine-based synthons in supramolecular chemistry. Eur. J. Inorg. Chem. 2006, 2006, 29–42. [Google Scholar] [CrossRef]
- Yang, C.; Chen, X.M.; Zhang, W.H.; Chen, J.; Yang, Y.S.; Gong, M.L. Synthesis, crystal structure and luminescence properties of a europium(III) complex with a new planar aromatic tridentate N3 ligand. J. Chem. Soc. Dalton Trans. 1996, 8, 1767–1768. [Google Scholar] [CrossRef]
- Barakat, A.; El-Faham, A.; Haukka, M.; Al-Majid, A.M.; Soliman, S.M. s-Triazine pincer ligands: Synthesis of their metal complexes, coordination behavior, and applications. Appl. Organomet. Chem. 2021, 35, e6317. [Google Scholar] [CrossRef]
- Mooibroek, T.J.; Gamez, P. The s-Triazine ring, a remarkable unit to generate supramolecular interactions. Inorg. Chim. Acta 2007, 360, 381–404. [Google Scholar] [CrossRef]
- Ghosh, C.; Nandi, A.; Basu, S. Supramolecular self-assembly of triazine-based small molecules: Targeting the endoplasmic reticulum in cancer cells. Nanoscale 2019, 11, 3326–3335. [Google Scholar] [CrossRef]
- Feringán, B.; Termine, R.; Golemme, A.; Granadino, J.M.; Navarro, A.; Giménez, R.; Sierra, T. Triphenylamine-and triazine-containing hydrogen bonded complexes: Liquid crystalline supramolecular semiconductors. J. Mater. Chem. C 2021, 9, 1972–1982. [Google Scholar] [CrossRef]
- Blotny, G. Recent applications of 2,4,6-trichloro-1,3,5-triazine and its derivatives in organic synthesis. Tetrahedron 2006, 62, 9507–9522. [Google Scholar] [CrossRef]
- Kolesinska, B.; Kaminski, Z.J. The umpolung of substituent effect in nucleophilic aromatic substitution. A new approach to the synthesis of N, N-disubstituted melamines (triazine triskelions) under mild reaction conditions. Tetrahedron 2009, 65, 3573–3576. [Google Scholar] [CrossRef]
- Das, A.; Demeshko, S.; Dechert, S.; Meyer, F. A New Triazine-Based Tricompartmental Ligand for Stepwise Assembly of Mononuclear, Dinuclear, and 1D-Polymeric Heptacoordinate Manganese(II)/Azido Complexes. Eur. J. Inorg. Chem. 2011, 2011, 1240–1248. [Google Scholar] [CrossRef]
- Beilstein, P.; Cook, A.M.; Huetter, R. Determination of seventeen s-Triazine herbicides and derivatives by high-pressure liquid chromatography. J. Agric. Food Chem. 1981, 29, 1132–1135. [Google Scholar] [CrossRef] [Green Version]
- Giacomelli, G.; Porcheddu, A.; Luca, L.D. [1,3,5]-Triazine: A versatile heterocycle in current applications of organic chemistry. Curr. Org. Chem. 2004, 8, 1497–1519. [Google Scholar] [CrossRef]
- Singh, S.; Mandal, M.K.; Masih, A.; Saha, A.; Ghosh, S.K.; Bhat, H.R.; Singh, U.P. 1,3,5-Triazine: A versatile pharmacophore with diverse biological activities. Arch. Pharm. 2021, 354, 2000363. [Google Scholar] [CrossRef]
- Solankee, A.; Kapadia, K.; Ćirić, A.; Soković, M.; Doytchinova, I.; Geronikaki, A. Synthesis of some new s-Triazine based chalcones and their derivatives as potent antimicrobial agents. Eur. J. Med. Chem. 2010, 45, 510–518. [Google Scholar] [CrossRef]
- Barakat, A.; El-Senduny, F.F.; Almarhoon, Z.; Al-Rasheed, H.H.; Badria, F.A.; Al-Majid, A.M.; Ghabbour, H.A.; El-Faham, A. Synthesis, X-ray crystal structures, and preliminary antiproliferative activities of new s-Triazine-hydroxybenzylidene hydrazone derivatives. J. Chem. 2019, 2019, 9403908. [Google Scholar] [CrossRef] [Green Version]
- Al-Rasheed, H.; Dahlous, K.; Sharma, A.; Sholkamy, E.; El-Faham, A.; Torre, B.G.; Albericio, F. Barbiturate-and Thiobarbituarte-based s-Triazine hydrazone derivatives with promising antiproliferative activities. ACS Omega 2020, 5, 15805–15811. [Google Scholar] [CrossRef]
- Menear, K.A.; Gomez, S.; Malagu, K.; Bailey, C.; Blackburn, K.; Cockcroft, X.L.; Ewen, S.; Fundo, A.; Gall, A.; Hermann, G.; et al. Identification and optimisation of novel and selective small molecular weight kinase inhibitors of mTOR. Bioorganic Med. Chem. Lett. 2009, 19, 5898–5901. [Google Scholar] [CrossRef]
- Bai, F.; Liu, H.; Tong, L.; Zhou, W.; Liu, L.; Zhao, Z.; Liu, X.; Jiang, H.; Wang, X.; Xie, H.; et al. Discovery of novel selective inhibitors for EGFR-T790M/L858R. Bioorganic Med. Chem. Lett. 2012, 22, 1365–1370. [Google Scholar] [CrossRef] [PubMed]
- Shanmugakala, R.; Tharmaraj, P.; Sheela, C.D.; Anitha, C. Synthesis and studies on s-Triazine-based ligand and its metal complexes. Int. J. Inorg. Chem. 2012, 2012, 301086. [Google Scholar] [CrossRef]
- Samy, F.; Taha, A. Synthesis, Spectroscopic, Biological and Theoretical Studies of Nano Complexes Derived from Triazine Hydrazone. Egypt. J. Chem. 2018, 61, 731–746. [Google Scholar]
- Fouad, R.; Adly, O.M. Novel Cu2+ and Zn2+ nanocomplexes drug based on hydrazone ligand bearings chromone and triazine moieties: Structural, spectral, DFT, molecular docking and cytotoxic studies. J. Mol. Struct. 2021, 1225, 129158. [Google Scholar] [CrossRef]
- Samy, F.; Shebl, M. Synthesis, spectroscopic, biological, and theoretical studies of new complexes from (E)-3-(2-(5, 6-diphenyl-1, 2, 4-triazin-3-yl) hydrazono) butan-2-one oxime. Appl. Organomet. Chem. 2020, 34, e5502. [Google Scholar] [CrossRef]
- Wu, P.; He, Y.; Wang, H.; Zhou, Y.G.; Yu, Z. Copper (II)-Catalyzed C–H Nitrogenation/Annulation Cascade of Ketene N, S-Acetals with Aryldiazonium Salts: A Direct Access to N2-Substituted Triazole and Triazine Derivatives. Org. Lett. 2019, 22, 310–315. [Google Scholar] [CrossRef]
- Crisponi, G.; Nurchi, V.M.; Fanni, D.; Gerosa, C.; Nemolato, S.; Faa, G. Copper-related diseases: From chemistry to molecular pathology. Coord. Chem. Rev. 2010, 254, 876–889. [Google Scholar] [CrossRef]
- Iakovidis, I.; Delimaris, I.; Piperakis, S.M. Copper and its complexes in medicine: A biochemical approach. Mol. Biol. Int. 2011, 2011, 594529. [Google Scholar] [CrossRef] [Green Version]
- Daniel, K.G.; Gupta, P.; Harbach, R.H.; Guida, W.C.; Dou, Q.P. Organic copper complexes as a new class of proteasome inhibitors and apoptosis inducers in human cancer cells. Biochem. Pharmacol. 2004, 67, 1139–1151. [Google Scholar] [CrossRef]
- Balamurugan, K.; Schaffner, W. Copper homeostasis in eukaryotes: Teetering on a tightrope. Biochim. Biophys. Acta Mol. Cell Res. 2006, 1763, 737–746. [Google Scholar] [CrossRef] [Green Version]
- Adams, J.Y.; Keen, C.L. Copper, oxidative stress, and human health. Mol. Asp. Med. 2005, 26, 268–298. [Google Scholar] [CrossRef] [PubMed]
- Arredondo, M.; Núñez, M.T. Iron and copper metabolism. Mol. Asp. Med. 2005, 26, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Noyce, J.O.; Michels, H.; Keevil, C.W. Inactivation of influenza A virus on copper versus stainless steel surfaces. Appl. Environ. Microbiol. 2007, 73, 2748–2750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowndes, S.A.; Harris, A.L. The role of copper in tumour angiogenesis. J. Mammary Gland Biol. Neoplasia 2005, 10, 299–310. [Google Scholar] [CrossRef] [PubMed]
- El-Faham, A.; Soliman, S.M.; Ghabbour, H.A.; Elnakady, Y.A.; Mohaya, T.A.; Siddiqui, M.R.H.; Albericio, F. Ultrasonic promoted synthesis of novel s-Triazine-Schiff base derivatives; molecular structure, spectroscopic studies, and their preliminary anti-proliferative activities. J. Mol. Str. 2016, 1125, 121–135. [Google Scholar] [CrossRef]
- Heide, P.V. X-ray Photoelectron Spectroscopy: An Introduction to Principles and Practices; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; pp. 3–6. [Google Scholar]
- Brown, D.G.; Weser, U. XPS Spectra of Copper and Nickel Biuret Complexes—Observations of Intense Satellite Structure in the 2P Spectrum of a Copper(III) System. J. Inorg. Biochem. 1979, 34, 989–994. [Google Scholar] [CrossRef]
- Biesinger, M.C.; Payne, B.P.; Grosvenor, A.P.; Lau, L.W.M.; Gerson, A.R.; Smart, R.S.C. Resolving surface chemical states in XPS analysis of first row transition metals, oxides and hydroxides: Cr, Mn, Fe, Co and Ni. Appl. Surf. Sci. 2011, 257, 2717–2730. [Google Scholar] [CrossRef]
- Haber, V.; Ptáček, P. XPS study of metal complexes with an unsymmetrical tetradentate Schiff base. Inorg. Chim. Acta 1991, 179, 267–270. [Google Scholar] [CrossRef]
- Nakamoto, K. Infrared and Raman Spectra of Inorganic and Coordination Compounds Part B: Applications in Coordination, Organometallic, and Bioinorganic Chemistry, 6th ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009. [Google Scholar]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. Crystal Explorer17; University of Western Australia: Perth, Australia, 2017; Available online: https://crystalexplorer.scb.uwa.edu.au/ (accessed on 20 July 2020).



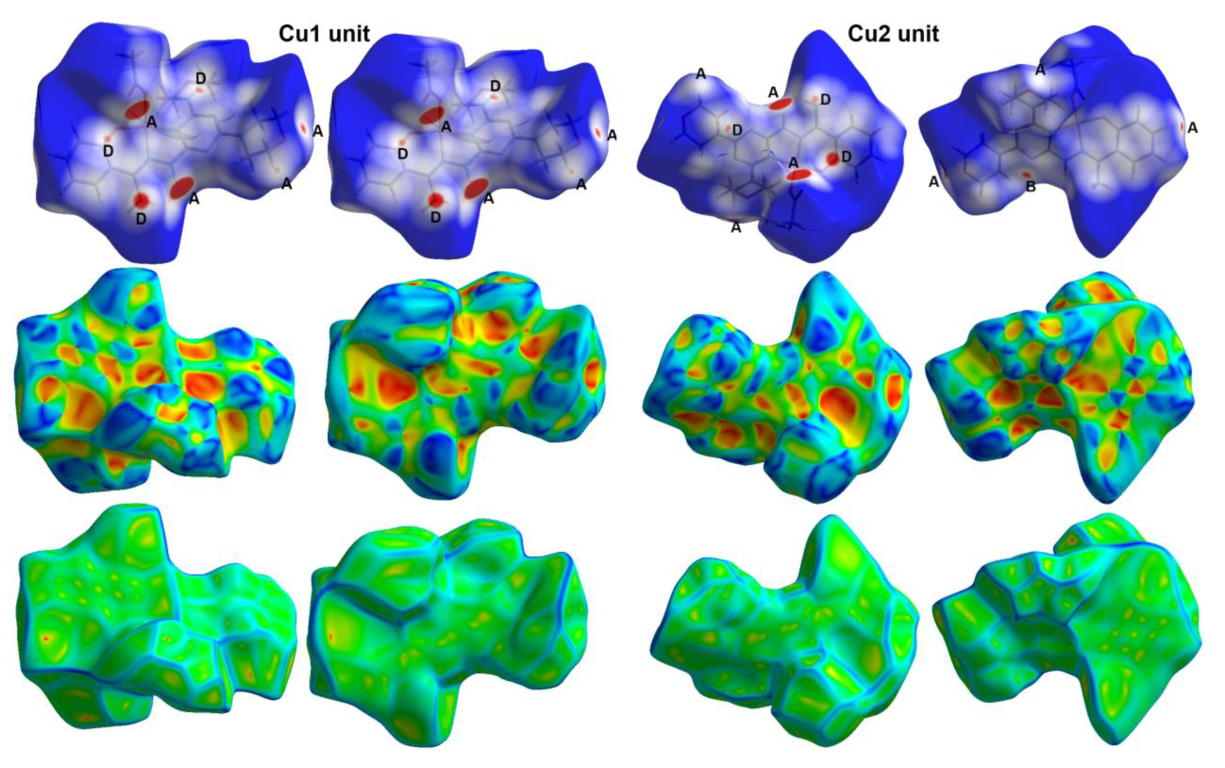

| Bond | Distance | Bond | Distance |
|---|---|---|---|
| Cu1-N1 | 1.989 (2) | Cu1-O4 | 2.006 (2) |
| Cu1-N2 | 1.990 (2) | Cu1-O1 | 2.185 (2) |
| Cu1-N4 | 2.022 (2) | Cu1-O5 | 2.651 (2) |
| Bonds | Angle | Bonds | Angle |
| N1-Cu1-N4 | 163.73 (9) | O4-Cu1-N4 | 95.83 (8) |
| N1-Cu1-O1 | 92.13 (9) | O4-Cu1-O1 | 103.61 (9) |
| N1-Cu1-O4 | 88.16 (9) | O5-Cu1-O4 | 53.28 (8) |
| N2-Cu1-N4 | 82.26 (8) | O5-Cu1-N1 | 83.17 (9) |
| N2-Cu1-O1 | 98.06 (10) | O5-Cu1-N2 | 104.87 (9) |
| N2-Cu1-O4 | 158.14 (9) | O5-Cu1-N4 | 86.58 (9) |
| N4-Cu1-O1 | 102.19 (9) | O5-Cu1-O1 | 156.38 (9) |
| N1-Cu1-N2 | 88.17 (9) |
| Atoms | D-H (Å) | H…A (Å) | D…A (Å) | D-H…A (º) |
|---|---|---|---|---|
| N1-H1A...O6 i | 0.84 (3) | 2.17 (4) | 2.975 (4) | 161 (3) |
| N1-H1B...O2 ii | 0.85 (3) | 2.14 (3) | 2.932 (3) | 155 (2) |
| O1-H1C...O7 iii | 0.81 (3) | 2.01 (3) | 2.787 (4) | 162 (4) |
| O1-H1D...O3 iv | 0.79 (4) | 2.00 (4) | 2.767 (4) | 163 (4) |
| N3-H3A...O7 | 0.84 (3) | 2.12 (3) | 2.936 (4) | 165 (3) |
| Symm. Code: (i) 2-x,1-y,1-z; (ii) -1 + x,y,z; (iii) 2-x,1-y,1-z; (iv) x,1 + y,z | ||||
| Bond | Distance | Bond | Distance |
|---|---|---|---|
| Cu1-O3 | 1.905(7) | Cu2-O10 | 1.902(8) |
| Cu1-O1 | 1.873(6) | Cu2-O9 | 1.852(7) |
| Cu1-N2 | 1.945(8) | Cu2-N10 | 1.938(8) |
| Cu1-N4 | 1.976(7) | Cu2-N12 | 1.984(8) |
| Bonds | Angle | Bonds | Angle |
| O3-Cu1-N2 | 160.9(3) | O10-Cu2-N10 | 164.1(3) |
| O3-Cu1-N4 | 97.4(3) | O10-Cu2-N12 | 96.2(3) |
| O1-Cu1-O3 | 93.0(3) | O9-Cu2-O10 | 92.5(3) |
| O1-Cu1-N2 | 93.2(3) | O9-Cu2-N10 | 92.5(4) |
| O1-Cu1-N4 | 160.2(3) | O9-Cu2-N12 | 163.9(3) |
| N2-Cu1-N4 | 82.4(3) | N10-Cu2-N12 | 82.1(4) |
| Contact | Distance | Contact | Distance |
|---|---|---|---|
| 1 | 2 | ||
| O5…H14A | 2.601 | O8…H3A | 2.397 |
| O2…H1B | 1.996 | O4…H11 | 1.857 |
| O7…H1C | 1.841 | O2…H3 | 1.802 |
| O8…H1C | 2.543 | O5…H24 | 2.483 |
| O3…H1D | 1.820 | O7…H40A | 2.584 |
| O8…H19A | 2.577 | O6…H16B | 2.551 |
| O6…H2A | 2.419 | O6…H35B | 2.361 |
| O6…H1A | 2.008 | O7…H21B | 2.602 |
| O7…H3A | 1.955 | C22…H11B | 2.547 |
| O7…H11C | 2.593 | C1…H32B | 2.713 |
| O9…H11C | 2.509 | C10…H39B | 2.762 |
| O9…H3A | 2.483 | C21…H39B | 2.710 |
| C11…C11 | 3.350 | N14…H14B | 2.490 |
| H4…H4 | 2.074 | C1…C7 | 3.332 |
| C6…C6 | 3.399 | ||
| Microbe | Type | DMAT | DMOHT | 1 | 2 | 3 | Control |
|---|---|---|---|---|---|---|---|
| A. fumigatus | Fungi | NA * | NA * | 14 | 15 | NA | 17 a |
| C. albicans | NA * | NA * | NA | 10 | NA | 20 a | |
| S. aureus | Gram positive bacteria | NA * | NA * | 10 | 11 | 12 | 24 b |
| B. subtilis | NA * | NA * | 25 | 25 | 24 | 26 b | |
| E. coli | Gram negative bacteria | NA * | NA * | 11 | 14 | 15 | 30 b |
| P. vulgaris | NA * | NA * | 15 | 15 | 13 | 25 b |
| Microbe | 1 | 2 | 3 | Control |
|---|---|---|---|---|
| A. fumigatus | 1250 | 625 | ND * | 156.25 a |
| C. albicans | ND* | 5000 | ND * | 312.5 a |
| S. aureus | 2500 | 1250 | 1250 | 9.7 b |
| B. subtilis | 39.06 | 39.06 | 78.125 | 4.8 b |
| E. coli | 2500 | 1250 | 312.5 | 4.8 b |
| P. vulgaris | 625 | 1250 | 1250 | 4.8 b |
| Conc. (μg/mL) | DPPH Scavenging % | |||||
|---|---|---|---|---|---|---|
| Ascorbic Acid | DMAT | DMOHT | 1 | 2 | 3 | |
| 1280 | 98.91 | 95.31 | 93.87 | 96.84 | 94.54 | 95.76 |
| 640 | 97.83 | 94.08 | 88.54 | 95.07 | 92.15 | 94.29 |
| 320 | 95.64 | 91.37 | 84.93 | 92.48 | 88.03 | 90.81 |
| 160 | 92.31 | 86.59 | 73.64 | 87.65 | 83.14 | 85.92 |
| 80 | 90.25 | 79.28 | 61.32 | 82.31 | 78.62 | 81.47 |
| 40 | 83.09 | 70.89 | 47.09 | 77.49 | 67.84 | 68.9 |
| 20 | 71.38 | 54.23 | 31.95 | 68.61 | 49.08 | 51.32 |
| 10 | 48.52 | 35.17 | 19.87 | 40.93 | 36.52 | 32.95 |
| 5 | 40.36 | 28.65 | 10.48 | 27.86 | 17.66 | 18.73 |
| 2.5 | 34.57 | 20.79 | 5.73 | 21.32 | 9.39 | 11.46 |
| IC50 | 10.62 ± 0.84 | 17.83 ± 0.65 | 48.2 ± 1.08 | 13.34 ± 0.58 | 20.93 ± 0.91 | 19.31 ± 0.73 |
| DMAT | DMOHT | Complex 1 | Complex 2 | Complex 3 | cis-Platin | |
|---|---|---|---|---|---|---|
| IC50 | 551.48 | 564.51 | 5.94 | 141.47 | 58.34 | 25.01 |
| CC50 | 716.04 | 999.90 | 37.67 | 101.47 | 172.96 | 242.25 |
| SI = CC50/IC50 | 1.30 | 1.77 | 6.34 | 0.72 | 2.96 | 9.69 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalil, T.E.; Dahlous, K.A.; Soliman, S.M.; Khalil, N.A.; El-Faham, A.; El-Dissouky, A. Synthesis, X-ray Structure and Biological Studies of New Self-Assembled Cu(II) Complexes Derived from s-Triazine Schiff Base Ligand. Molecules 2022, 27, 2989. https://doi.org/10.3390/molecules27092989
Khalil TE, Dahlous KA, Soliman SM, Khalil NA, El-Faham A, El-Dissouky A. Synthesis, X-ray Structure and Biological Studies of New Self-Assembled Cu(II) Complexes Derived from s-Triazine Schiff Base Ligand. Molecules. 2022; 27(9):2989. https://doi.org/10.3390/molecules27092989
Chicago/Turabian StyleKhalil, Tarek E., Kholood A. Dahlous, Saied M. Soliman, Nessma A. Khalil, Ayman El-Faham, and Ali El-Dissouky. 2022. "Synthesis, X-ray Structure and Biological Studies of New Self-Assembled Cu(II) Complexes Derived from s-Triazine Schiff Base Ligand" Molecules 27, no. 9: 2989. https://doi.org/10.3390/molecules27092989






