Exploring the Parallel G-Quadruplex Nucleic Acid World: A Spectroscopic and Computational Investigation on the Binding of the c-myc Oncogene NHE III1 Region by the Phytochemical Polydatin
Abstract
:1. Introduction
2. Results
2.1. CD Spectroscopic Analysis of the Binding of Pu22 by tPD
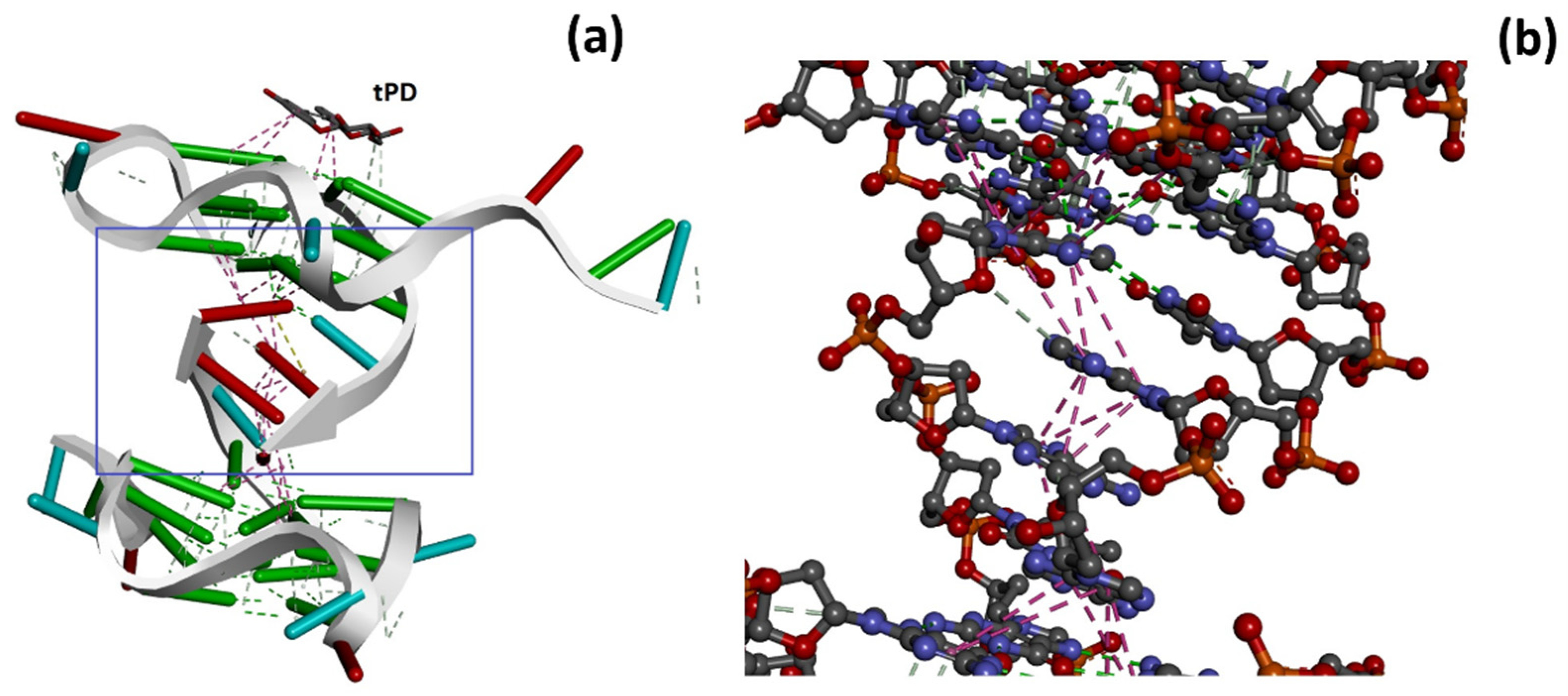
2.2. Molecular Docking Studies
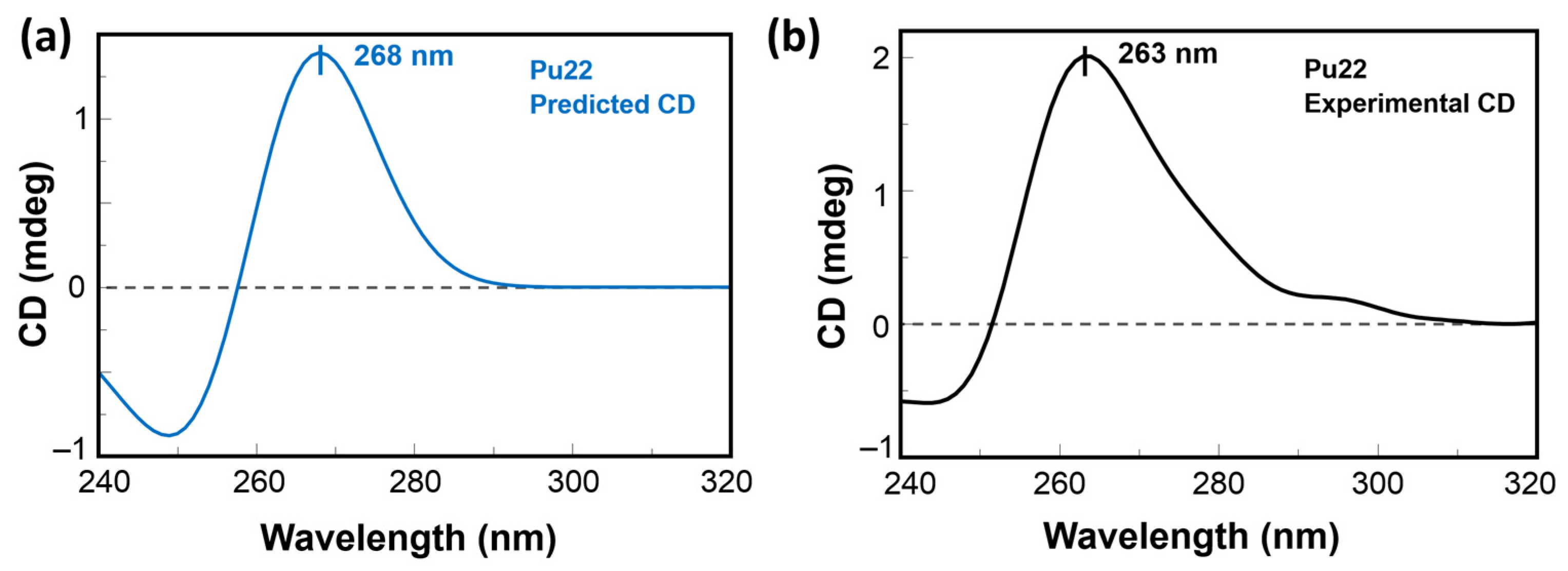
2.3. CD Predictions and Comparison with Experimental Spectroscopic Data
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. CD Studies
4.3. CD Denaturation Studies
4.4. Molecular Docking
4.5. CD Predictions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Patel, D.J.; Phan, A.T.; Kuryavyi, V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007, 35, 7429–7455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.-F.; Qin, Q.-P.; Qin, J.-L.; Liu, Y.-C.; Huang, K.-B.; Li, Y.-L.; Meng, T.; Zhang, G.-H.; Peng, Y.; Luo, X.-J. Stabilization of G-quadruplex DNA, inhibition of telomerase activity, and tumor cell apoptosis by organoplatinum (II) complexes with oxoisoaporphine. J. Med. Chem. 2015, 58, 2159–2179. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.-P.; Qin, J.-L.; Meng, T.; Lin, W.-H.; Zhang, C.-H.; Wei, Z.-Z.; Chen, J.-N.; Liu, Y.-C.; Liang, H.; Chen, Z.-F. High in vivo antitumor activity of cobalt oxoisoaporphine complexes by targeting G-quadruplex DNA, telomerase and disrupting mitochondrial functions. Eur. J. Med. Chem. 2016, 124, 380–392. [Google Scholar] [CrossRef]
- Amato, J.; Pagano, B.; Borbone, N.; Oliviero, G.; Gabelica, V.; Pauw, E.D.; D’Errico, S.; Piccialli, V.; Varra, M.; Giancola, C. Targeting G-quadruplex structure in the human c-Kit promoter with short PNA sequences. Bioconjugate Chem. 2011, 22, 654–663. [Google Scholar] [CrossRef] [Green Version]
- Esposito, V.; Galeone, A.; Mayol, L.; Oliviero, G.; Virgilio, A.; Randazzo, L. A topological classification of G-quadruplex structures. Nucleosides Nucleotides Nucleic Acids 2007, 26, 1155–1159. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009, 19, 239–250. [Google Scholar] [CrossRef]
- Onel, B.; Lin, C.; Yang, D. DNA G-quadruplex and its potential as anticancer drug target. Sci. China Chem. 2014, 57, 1605–1614. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c- MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [Green Version]
- Seenisamy, J.; Rezler, E.M.; Powell, T.J.; Tye, D.; Gokhale, V.; Joshi, C.S.; Siddiqui-Jain, A.; Hurley, L.H. The Dynamic Character of the G-Quadruplex Element in the c-MYC Promoter and Modification by TMPyP4. J. Am. Chem. Soc. 2004, 126, 8702–8709. [Google Scholar] [CrossRef]
- Rankin, S.; Reszka, A.P.; Huppert, J.; Zloh, M.; Parkinson, G.N.; Todd, A.K.; Ladame, S.; Balasubramanian, S.; Neidle, S. Putative DNA Quadruplex Formation within the Human c-kit Oncogene. J. Am. Chem. Soc. 2005, 127, 10584–10589. [Google Scholar] [CrossRef] [Green Version]
- Fernando, H.; Reszka, A.P.; Huppert, J.; Ladame, S.; Rankin, S.; Venkitaraman, A.R.; Neidle, S.; Balasubramanian, S. A Conserved Quadruplex Motif Located in a Transcription Activation Site of the Human c-kit Oncogene. Biochemistry 2006, 45, 7854–7860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef]
- Phan, A.T.; Kuryavyi, V.; Burge, S.; Neidle, S.; Patel, D.J. Structure of an Unprecedented G-Quadruplex Scaffold in the Human c-kit Promoter. J. Am. Chem. Soc. 2007, 129, 4386–4392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Agrawal, P.; Brown, R.V.; Hatzakis, E.; Hurley, L.; Yang, D. The Major G-Quadruplex Formed in the Human Platelet-Derived Growth Factor Receptor β Promoter Adopts a Novel Broken-Strand Structure in K+ Solution. J. Am. Chem. Soc. 2012, 134, 13220–13223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francisco, A.P.; Paulo, A. Oncogene expression modulation in cancer cell lines by DNA G-quadruplex-interactive small molecules. Curr. Med. Chem. 2017, 24, 4873–4904. [Google Scholar] [CrossRef]
- Ciribilli, Y.; Singh, P.; Spanel, R.; Inga, A.; Borlak, J. Decoding c-Myc networks of cell cycle and apoptosis regulated genes in a transgenic mouse model of papillary lung adenocarcinomas. Oncotarget 2015, 6, 31569. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, Q.; Wu, X.-H.; Sun, F.-Y.; Chen, L.-M.; Chen, J.-C.; Yang, S.-L.; Mei, W.-J. Ruthenium (II) complexes as apoptosis inducers by stabilizing c-myc G-quadruplex DNA. Eur. J. Med. Chem. 2014, 80, 316–324. [Google Scholar] [CrossRef]
- Platella, C.; Mazzini, S.; Napolitano, E.; Mattio, L.M.; Beretta, G.L.; Zaffaroni, N.; Pinto, A.; Montesarchio, D.; Dallavalle, S. Plant-Derived Stilbenoids as DNA-Binding Agents: From Monomers to Dimers. Chem.–A Eur. J. 2021, 27, 8832–8845. [Google Scholar] [CrossRef]
- Chaudhuri, R.; Bhattacharya, S.; Dash, J.; Bhattacharya, S. Recent Update on Targeting c-MYC G-Quadruplexes by Small Molecules for Anticancer Therapeutics. J. Med. Chem. 2020, 64, 42–70. [Google Scholar] [CrossRef]
- Sheikh-Zeineddini, N.; Safaroghli-azar, A.; Salari, S.; Bashash, D. C-Myc inhibition sensitizes pre-B ALL cells to the anti-tumor effect of vincristine by altering apoptosis and autophagy: Proposing a probable mechanism of action for 10058-F4. Eur. J. Pharmacol. 2020, 870, 172821. [Google Scholar] [CrossRef]
- Mazzini, S.; Gargallo, R.; Musso, L.; De Santis, F.; Aviñó, A.; Scaglioni, L.; Eritja, R.; Di Nicola, M.; Zunino, F.; Amatulli, A.; et al. Stabilization of c-KIT G-Quadruplex DNA Structures by the RNA Polymerase I Inhibitors BMH-21 and BA-41. Int. J. Mol. Sci. 2019, 20, 4927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chanvorachote, P.; Sriratanasak, N.; Nonpanya, N. C-myc Contributes to Malignancy of Lung Cancer: A Potential Anticancer Drug Target. Anticancer Res. 2020, 40, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Dallavalle, S.; Mattio, L.M.; Artali, R.; Musso, L.; Aviñó, A.; Fàbrega, C.; Eritja, R.; Gargallo, R.; Mazzini, S. Exploring the Interaction of Curaxin CBL0137 with G-Quadruplex DNA Oligomers. Int. J. Mol. Sci. 2021, 22, 6476. [Google Scholar] [CrossRef] [PubMed]
- Georgiades, S.N.; Abd Karim, N.H.; Suntharalingam, K.; Vilar, R. Interaction of metal complexes with G-quadruplex DNA. Angew. Chem. Int. Ed. 2010, 49, 4020–4034. [Google Scholar] [CrossRef]
- Brooks, T.A.; Hurley, L.H. Targeting MYC expression through G-quadruplexes. Genes Cancer 2010, 1, 641–649. [Google Scholar] [CrossRef]
- Yan, S.; Zheng, Z.; Cui, Y.; Mi, Y.; Liu, H.; Zhao, X.; Luo, D. C-myc g-quadruplex stabilization and cytotoxicity of an oxadiazole-bearing ruthennium (II) complex. Rev. Roum. De Chim. 2021, 66, 423–433. [Google Scholar]
- Platella, C.; Guida, S.; Bonmassar, L.; Aquino, A.; Bonmassar, E.; Ravagnan, G.; Montesarchio, D.; Roviello, G.N.; Musumeci, D.; Fuggetta, M.P. Antitumour activity of resveratrol on human melanoma cells: A possible mechanism related to its interaction with malignant cell telomerase. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2017, 1861, 2843–2851. [Google Scholar] [CrossRef]
- Quagliariello, V.; Berretta, M.; Buccolo, S.; Iovine, M.; Paccone, A.; Cavalcanti, E.; Taibi, R.; Montopoli, M.; Botti, G.; Maurea, N. Polydatin reduces cardiotoxicity and enhances the anticancer effects of sunitinib by decreasing pro-oxidative stress, pro-inflammatory cytokines, and nlrp3 inflammasome expression. Front. Oncol. 2021, 11, 680758. [Google Scholar] [CrossRef]
- Lanzilli, G.; Cottarelli, A.; Nicotera, G.; Guida, S.; Ravagnan, G.; Fuggetta, M.P. Anti-inflammatory effect of resveratrol and polydatin by in vitro IL-17 modulation. Inflammation 2012, 35, 240–248. [Google Scholar] [CrossRef] [Green Version]
- Platella, C.; Raucci, U.; Rega, N.; D’Atri, S.; Levati, L.; Roviello, G.N.; Fuggetta, M.P.; Musumeci, D.; Montesarchio, D. Shedding light on the interaction of polydatin and resveratrol with G-quadruplex and duplex DNA: A biophysical, computational and biological approach. Int. J. Biol. Macromol. 2020, 151, 1163–1172. [Google Scholar] [CrossRef]
- Bai, L.; Ma, Y.; Wang, X.; Feng, Q.; Zhang, Z.; Wang, S.; Zhang, H.; Lu, X.; Xu, Y.; Zhao, E. Polydatin Inhibits Cell Viability, Migration, and Invasion Through Suppressing the c-Myc Expression in Human Cervical Cancer. Front. Cell Dev. Biol. 2021, 9, 587218. [Google Scholar] [CrossRef] [PubMed]
- Mathad, R.I.; Hatzakis, E.; Dai, J.; Yang, D. c-MYC promoter G-quadruplex formed at the 5′-end of NHE III 1 element: Insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011, 39, 9023–9033. [Google Scholar] [CrossRef] [PubMed]
- Roviello, V.; Musumeci, D.; Mokhir, A.; Roviello, G.N. Evidence of protein binding by a nucleopeptide based on a thyminedecorated L-diaminopropanoic acid through CD and in silico studies. Curr. Med. Chem. 2021, 28, 5004–5015. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, D.; Mokhir, A.; Roviello, G.N. Synthesis and nucleic acid binding evaluation of a thyminyl l-diaminobutanoic acid-based nucleopeptide. Bioorganic Chem. 2020, 100, 103862. [Google Scholar] [CrossRef]
- Musumeci, D.; Ullah, S.; Ikram, A.; Roviello, G.N. Novel insights on nucleopeptide binding: A spectroscopic and In Silico investigation on the interaction of a thymine-bearing tetrapeptide with a homoadenine DNA. J. Mol. Liq. 2022, 347, 117975. [Google Scholar] [CrossRef]
- Amato, J.; Stellato, M.I.; Pizzo, E.; Petraccone, L.; Oliviero, G.; Borbone, N.; Piccialli, G.; Orecchia, A.; Bellei, B.; Castiglia, D. PNA as a potential modulator of COL7A1 gene expression in dominant dystrophic epidermolysis bullosa: A physico-chemical study. Mol. BioSyst. 2013, 9, 3166–3174. [Google Scholar] [CrossRef]
- Pirota, V.; Platella, C.; Musumeci, D.; Benassi, A.; Amato, J.; Pagano, B.; Colombo, G.; Freccero, M.; Doria, F.; Montesarchio, D. On the binding of naphthalene diimides to a human telomeric G-quadruplex multimer model. Int. J. Biol. Macromol. 2021, 166, 1320–1334. [Google Scholar] [CrossRef]
- Platella, C.; Capasso, D.; Riccardi, C.; Musumeci, D.; DellaGreca, M.; Montesarchio, D. Natural compounds from Juncus plants interacting with telomeric and oncogene G-quadruplex structures as potential anticancer agents. Org. Biomol. Chem. 2021, 19, 9953–9965. [Google Scholar] [CrossRef]
- Oliviero, G.; Borbone, N.; Amato, J.; D’Errico, S.; Galeone, A.; Piccialli, G.; Varra, M.; Mayol, L. Synthesis of quadruplex-forming tetra-end-linked oligonucleotides: Effects of the linker size on quadruplex topology and stability. Biopolym. Orig. Res. Biomol. 2009, 91, 466–477. [Google Scholar] [CrossRef]
- Oliviero, G.; Amato, J.; Borbone, N.; Galeone, A.; Varra, M.; Piccialli, G.; Mayol, L. Synthesis and characterization of DNA quadruplexes containing T-tetrads formed by bunch-oligonucleotides. Biopolym. Orig. Res. Biomol. 2006, 81, 194–201. [Google Scholar] [CrossRef]
- Fuggetta, M.P.; De Mico, A.; Cottarelli, A.; Morelli, F.; Zonfrillo, M.; Ulgheri, F.; Peluso, P.; Mannu, A.; Deligia, F.; Marchetti, M. Synthesis and enantiomeric separation of a novel spiroketal derivative: A potent human telomerase inhibitor with high in vitro anticancer activity. J. Med. Chem. 2016, 59, 9140–9149. [Google Scholar] [CrossRef] [PubMed]
- Arounaguiri, S.; Easwaramoorthy, D.; Ashokkumar, A.; Dattagupta, A.; Maiya, B.G. Cobalt (III), nickel (II) and ruthenium (II) complexes of 1, 10-phenanthroline family of ligands: DNA binding and photocleavage studies. J. Chem. Sci. 2000, 112, 1–17. [Google Scholar] [CrossRef]
- Ricci, C.G.; Netz, P.A. Docking studies on DNA-ligand interactions: Building and application of a protocol to identify the binding mode. J. Chem. Inf. Model. 2009, 49, 1925–1935. [Google Scholar] [CrossRef] [PubMed]
- Mulliri, S.; Laaksonen, A.; Spanu, P.; Farris, R.; Farci, M.; Mingoia, F.; Roviello, G.N.; Mocci, F. Spectroscopic and in silico studies on the interaction of substituted pyrazolo [1, 2-a] benzo [1, 2, 3, 4] tetrazine-3-one derivatives with c-myc G4-DNA. Int. J. Mol. Sci. 2021, 22, 6028. [Google Scholar] [CrossRef] [PubMed]
- Bulheller, B.M.; Hirst, J.D. DichroCalc—Circular and linear dichroism online. Bioinformatics 2009, 25, 539–540. [Google Scholar] [CrossRef]
- Kong, D.-M.; Wu, J.; Wang, N.; Yang, W.; Shen, H.-X. Peroxidase activity–structure relationship of the intermolecular four-stranded G-quadruplex–hemin complexes and their application in Hg2+ ion detection. Talanta 2009, 80, 459–465. [Google Scholar] [CrossRef]
- Egbuna, C.; Kumar, S.; Ifemeje, J.C.; Ezzat, S.M.; Kaliyaperumal, S. Phytochemicals as Lead Compounds for New Drug Discovery; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Mechchate, H.; Costa de Oliveira, R.; Es-Safi, I.; Vasconcelos Mourão, E.M.; Bouhrim, M.; Kyrylchuk, A.; Soares Pontes, G.; Bousta, D.; Grafov, A. Antileukemic Activity and Molecular Docking Study of a Polyphenolic Extract from Coriander Seeds. Pharmaceuticals 2021, 14, 770. [Google Scholar] [CrossRef]
- Artali, R.; Beretta, G.; Morazzoni, P.; Bombardelli, E.; Meneghetti, F. Green tea catechins in chemoprevention of cancer: A molecular docking investigation into their interaction with glutathione S-transferase (GST P1-1). J. Enzym. Inhib. Med. Chem. 2009, 24, 287–295. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.-Y. HDOCK: A web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Majumder, A.; Mondal, S.K.; Mukhoty, S.; Bag, S.; Mondal, A.; Begum, Y.; Sharma, K.; Banik, A. Virtual Screening and Docking Analysis of Novel Ligands for Selective Enhancement of Tea (Camellia sinensis) Flavonoids. Food Chem. X 2022, 13, 100212. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Chatterjee, O.; Banerjee, N.; Roychowdhury, T.; Dhar, G.; Mukherjee, G.; Chatterjee, S. Curcumin arrests G-quadruplex in the nuclear hyper-sensitive III1 element of c-MYC oncogene leading to apoptosis in metastatic breast cancer cells. J. Biomol. Struct. Dyn. 2021, 1–17. [Google Scholar] [CrossRef]
- Ji, D.; Juhas, M.; Tsang, C.M.; Kwok, C.K.; Li, Y.; Zhang, Y. Discovery of G-quadruplex-forming sequences in SARS-CoV-2. Brief. Bioinform. 2021, 22, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, S.V.; Wallace, F.E.; Stoddard, S.D.; Cheng, Q.; Acosta, D.; Barzani, S.; Bobay, M.; Briant, J.; Cisneros, C.; Feinstein, S. In silico design of peptide-based SARS-CoV-2 fusion inhibitors that target wt and mutant versions of SARS-CoV-2 HR1 Domains. Biophysica 2021, 1, 311–327. [Google Scholar] [CrossRef]
- Stump, S.; Mou, T.-C.; Sprang, S.R.; Natale, N.R.; Beall, H.D. Crystal structure of the major quadruplex formed in the promoter region of the human c-MYC oncogene. PLoS ONE 2018, 13, e0205584. [Google Scholar] [CrossRef] [Green Version]
- Ambrus, A.; Chen, D.; Dai, J.; Jones, R.A.; Yang, D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry 2005, 44, 2048–2058. [Google Scholar] [CrossRef]
- Jana, J.; Weisz, K. A Thermodynamic Perspective on Potential G-Quadruplex Structures as Silencer Elements in the MYC Promoter. Chem.–A Eur. J. 2020, 26, 17242–17251. [Google Scholar] [CrossRef]
- Moriyama, K.; Yoshizawa-Sugata, N.; Masai, H. Oligomer formation and G-quadruplex binding by purified murine Rif1 protein, a key organizer of higher-order chromatin architecture. J. Biol. Chem. 2018, 293, 3607–3624. [Google Scholar] [CrossRef] [Green Version]
- Marzano, M.; Falanga, A.P.; Marasco, D.; Borbone, N.; D’Errico, S.; Piccialli, G.; Roviello, G.N.; Oliviero, G. Evaluation of an Analogue of the Marine ε-PLL Peptide as a Ligand of G-quadruplex DNA Structures. Mar. Drugs 2020, 18, 49. [Google Scholar] [CrossRef] [Green Version]
- Funke, A.; Karg, B.; Dickerhoff, J.; Balke, D.; Müller, S.; Weisz, K. Ligand-Induced Dimerization of a Truncated Parallel MYC G-Quadruplex. ChemBioChem 2018, 19, 505–512. [Google Scholar] [CrossRef]
- Brinkley, R.L.; Gupta, R.B. Hydrogen bonding with aromatic rings. AIChE J. 2001, 47, 948–953. [Google Scholar] [CrossRef]
- O’Hagan, M.P.; Peñalver, P.; Gibson, R.S.L.; Morales, J.C.; Galan, M.C. Stiff-Stilbene Ligands Target G-Quadruplex DNA and Exhibit Selective Anticancer and Antiparasitic Activity. Chem.–A Eur. J. 2020, 26, 6224–6233. [Google Scholar] [CrossRef] [PubMed]
- Esaki, Y.; Islam, M.M.; Fujii, S.; Sato, S.; Takenaka, S. Design of tetraplex specific ligands: Cyclic naphthalene diimide. Chem. Commun. 2014, 50, 5967–5969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianchi, F.; Comez, L.; Biehl, R.; D’Amico, F.; Gessini, A.; Longo, M.; Masciovecchio, C.; Petrillo, C.; Radulescu, A.; Rossi, B.; et al. Structure of human telomere G-quadruplex in the presence of a model drug along the thermal unfolding pathway. Nucleic Acids Res. 2018, 46, 11927–11938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pradeep, T.P.; Tripathi, S.; Barthwal, R. Molecular recognition of parallel quadruplex [d-(TTGGGGT)] 4 by mitoxantrone: Binding with 1: 4 stoichiometry leads to telomerase inhibition. RSC Adv. 2016, 6, 71652–71661. [Google Scholar] [CrossRef]
- Pawar, S.S.; Rohane, S.H. Review on discovery studio: An important tool for molecular docking. Asian J. Res. Chem. 2021, 14, 86–88. [Google Scholar] [CrossRef]
- Menges, F. Spectragryph-Optical Spectroscopy Software, Version 1.2. 8. 2018. Available online: http://www.effemm2.de/spectragryph (accessed on 1 August 2018).
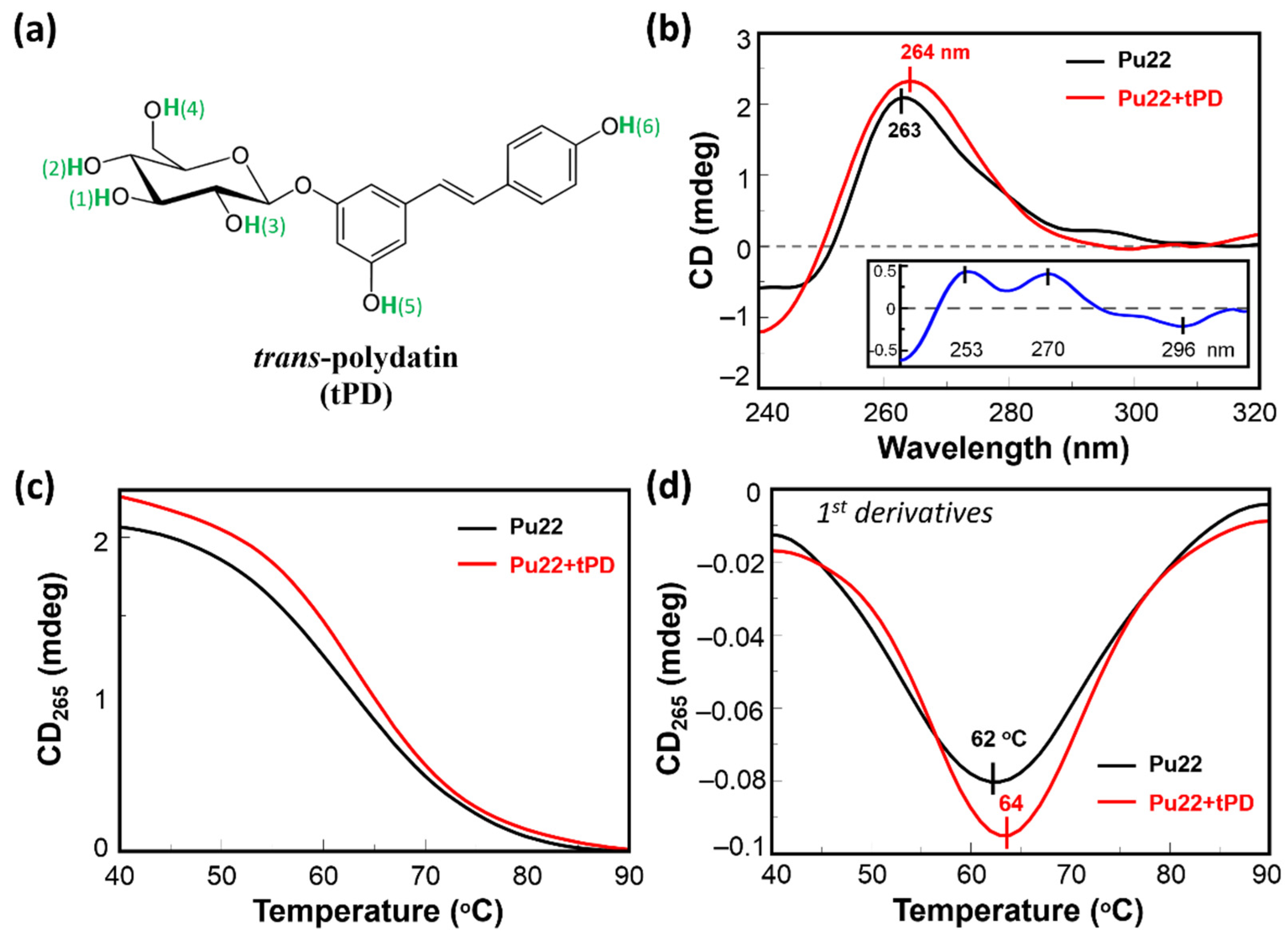
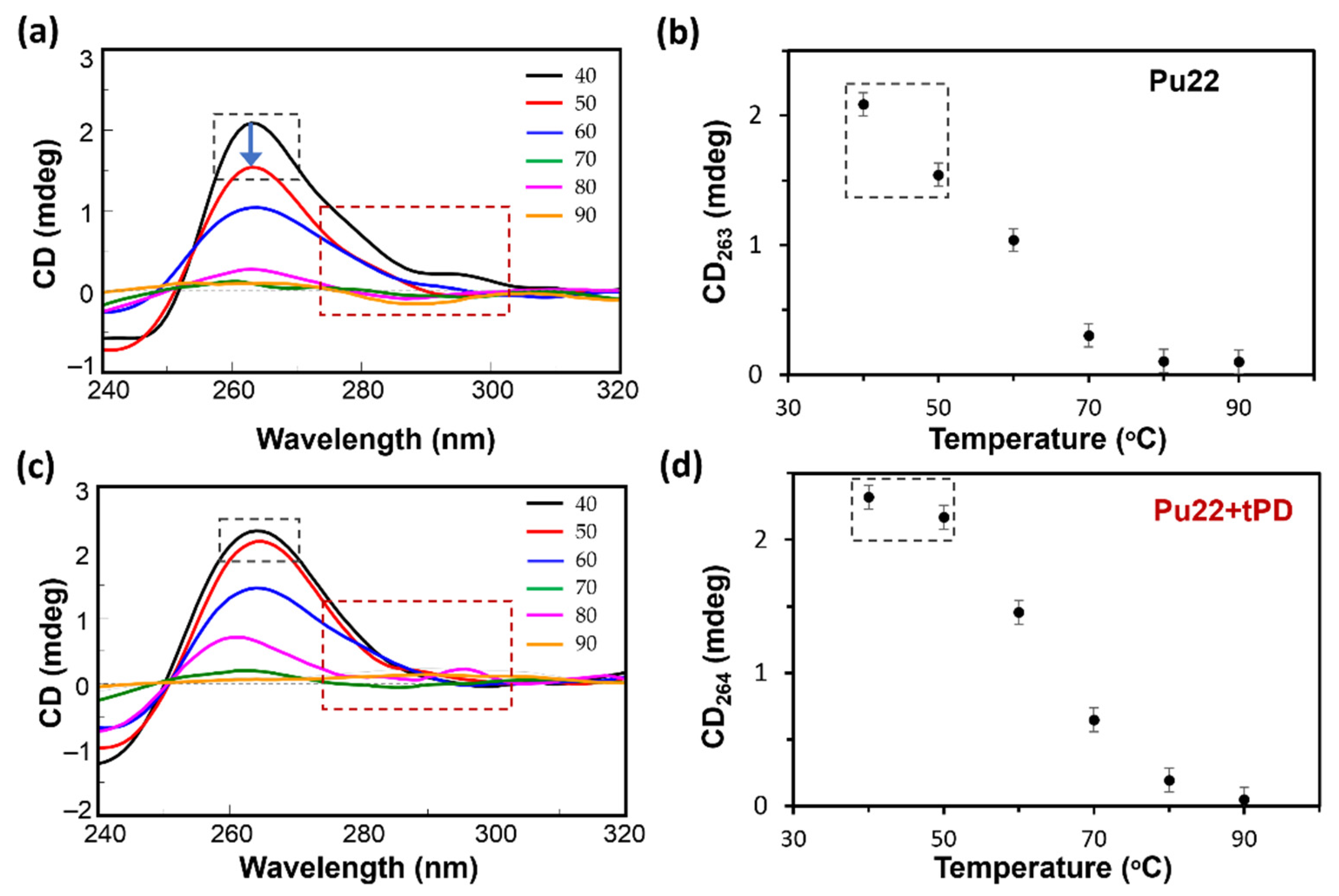
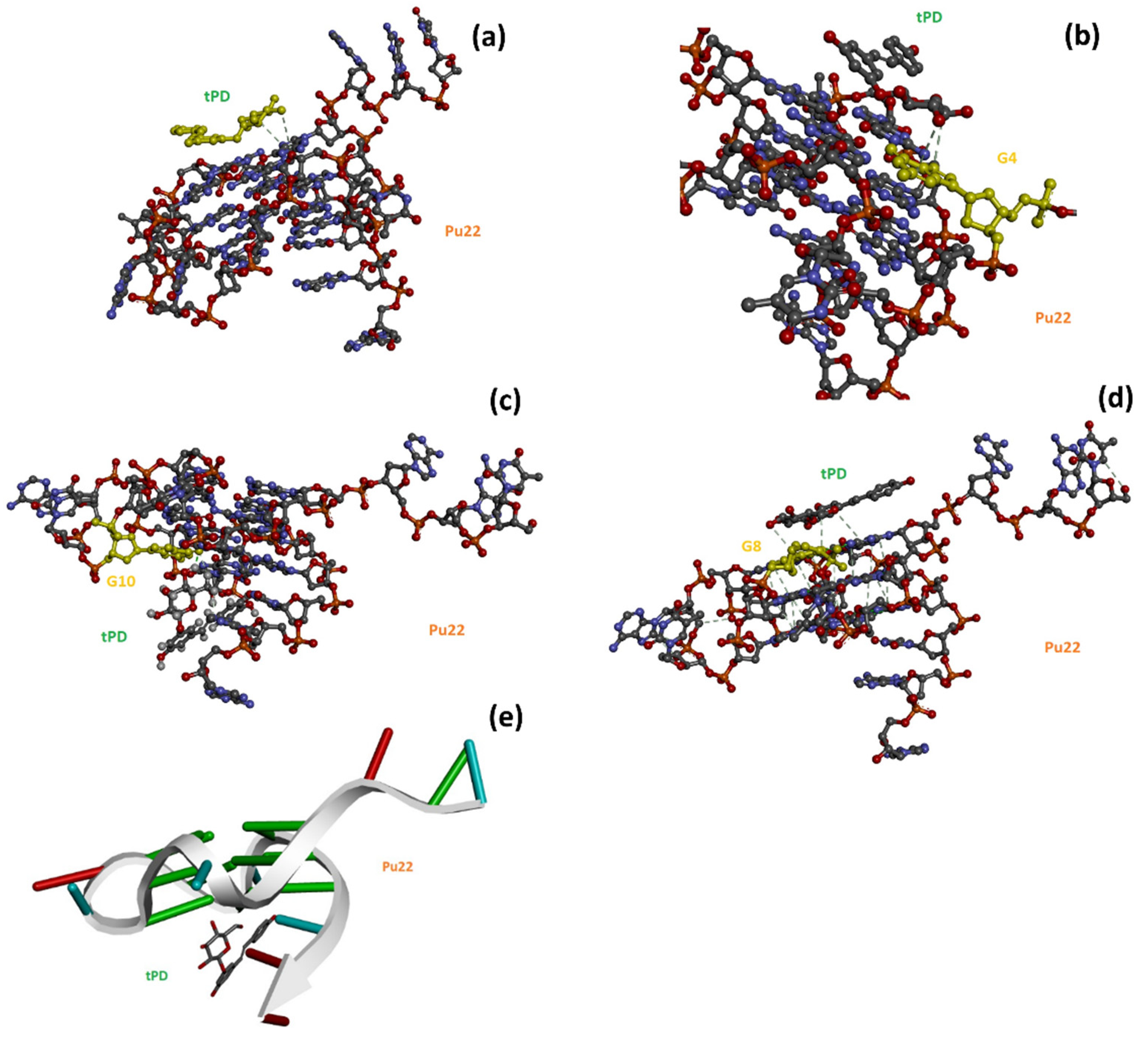
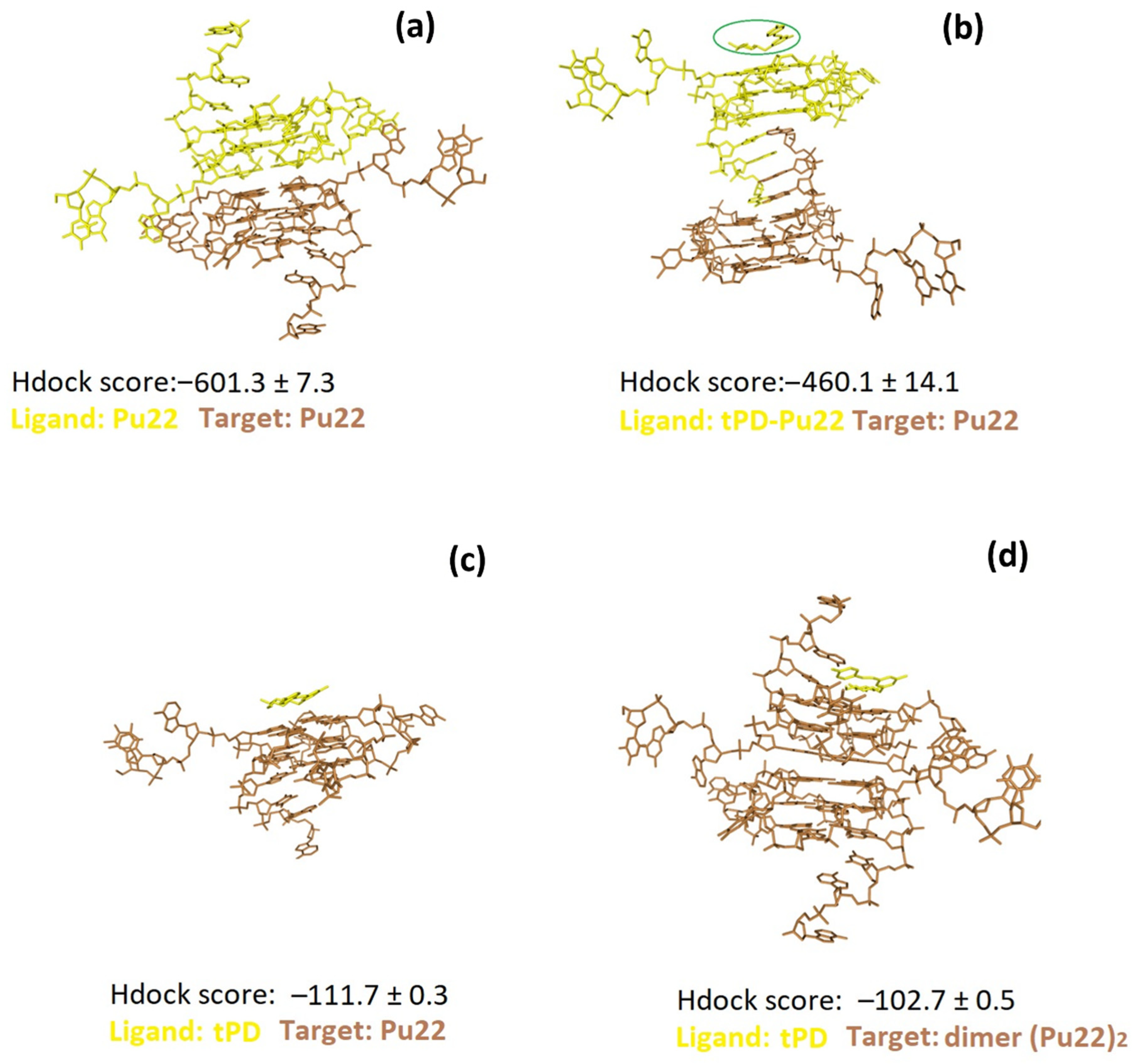
| Entry | ΔTm * (°C) | ΔCDmax 40–90 °C (mdeg) | ΔCDmax 40–50 °C (mdeg) |
|---|---|---|---|
| Pu22 | 0 | 1.99 | 0.54 |
| tPD-Pu22 | +2 | 2.23 | 0.15 |
| Complex | HDOCK Score * Top-1 Ranked Pose | HDOCK Score Mean Value (Top-1–3 Poses) ± SD | Interface Residues |
|---|---|---|---|
| tPD/Pu22 | –112.1 | –111.7 ± 0.3 | G4, G6, G8, G10, G13, G15, T16, G17, G19, T20, A21 |
| tRES/Pu22 | –120.6 | –112.9 ± 7.3 | G6, T7, G10, G15, T16, G19, T20, A21 |
| tPD/(Pu22)2 | –103.2 | –102.7 ± 0.5 | G6, G10, T11, G15, T16, G19, T20, A21, G’14, G’15, T’16, G’17, G’18, G’19, T’20, A’21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greco, F.; Musumeci, D.; Borbone, N.; Falanga, A.P.; D’Errico, S.; Terracciano, M.; Piccialli, I.; Roviello, G.N.; Oliviero, G. Exploring the Parallel G-Quadruplex Nucleic Acid World: A Spectroscopic and Computational Investigation on the Binding of the c-myc Oncogene NHE III1 Region by the Phytochemical Polydatin. Molecules 2022, 27, 2997. https://doi.org/10.3390/molecules27092997
Greco F, Musumeci D, Borbone N, Falanga AP, D’Errico S, Terracciano M, Piccialli I, Roviello GN, Oliviero G. Exploring the Parallel G-Quadruplex Nucleic Acid World: A Spectroscopic and Computational Investigation on the Binding of the c-myc Oncogene NHE III1 Region by the Phytochemical Polydatin. Molecules. 2022; 27(9):2997. https://doi.org/10.3390/molecules27092997
Chicago/Turabian StyleGreco, Francesca, Domenica Musumeci, Nicola Borbone, Andrea Patrizia Falanga, Stefano D’Errico, Monica Terracciano, Ilaria Piccialli, Giovanni Nicola Roviello, and Giorgia Oliviero. 2022. "Exploring the Parallel G-Quadruplex Nucleic Acid World: A Spectroscopic and Computational Investigation on the Binding of the c-myc Oncogene NHE III1 Region by the Phytochemical Polydatin" Molecules 27, no. 9: 2997. https://doi.org/10.3390/molecules27092997
APA StyleGreco, F., Musumeci, D., Borbone, N., Falanga, A. P., D’Errico, S., Terracciano, M., Piccialli, I., Roviello, G. N., & Oliviero, G. (2022). Exploring the Parallel G-Quadruplex Nucleic Acid World: A Spectroscopic and Computational Investigation on the Binding of the c-myc Oncogene NHE III1 Region by the Phytochemical Polydatin. Molecules, 27(9), 2997. https://doi.org/10.3390/molecules27092997














