Collagen Type II—Chitosan Interactions as Dependent on Hydroxylation and Acetylation Inferred from Molecular Dynamics Simulations
Abstract
:1. Introduction
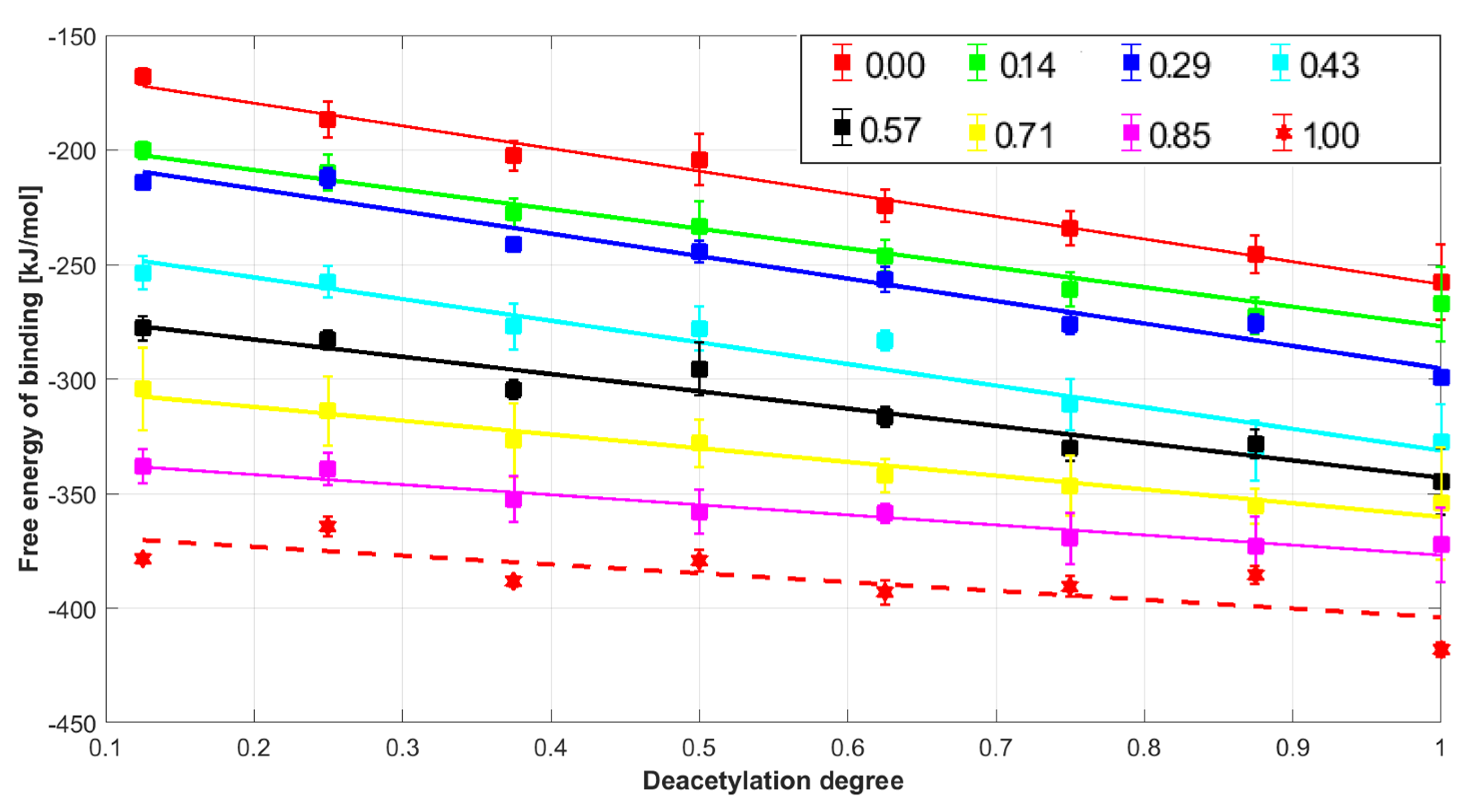
2. Results and Discussion
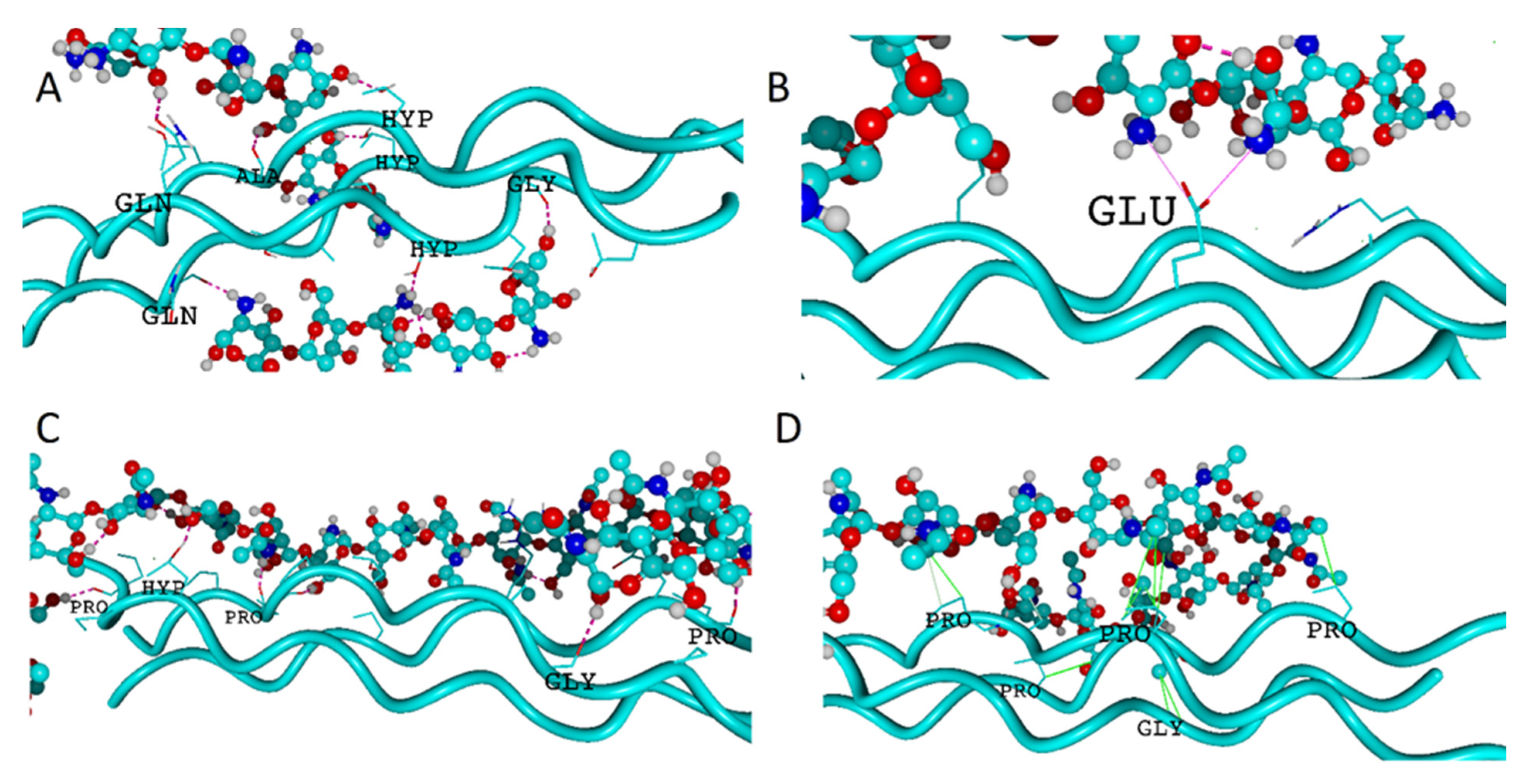
2.1. Hydrogen Bonds
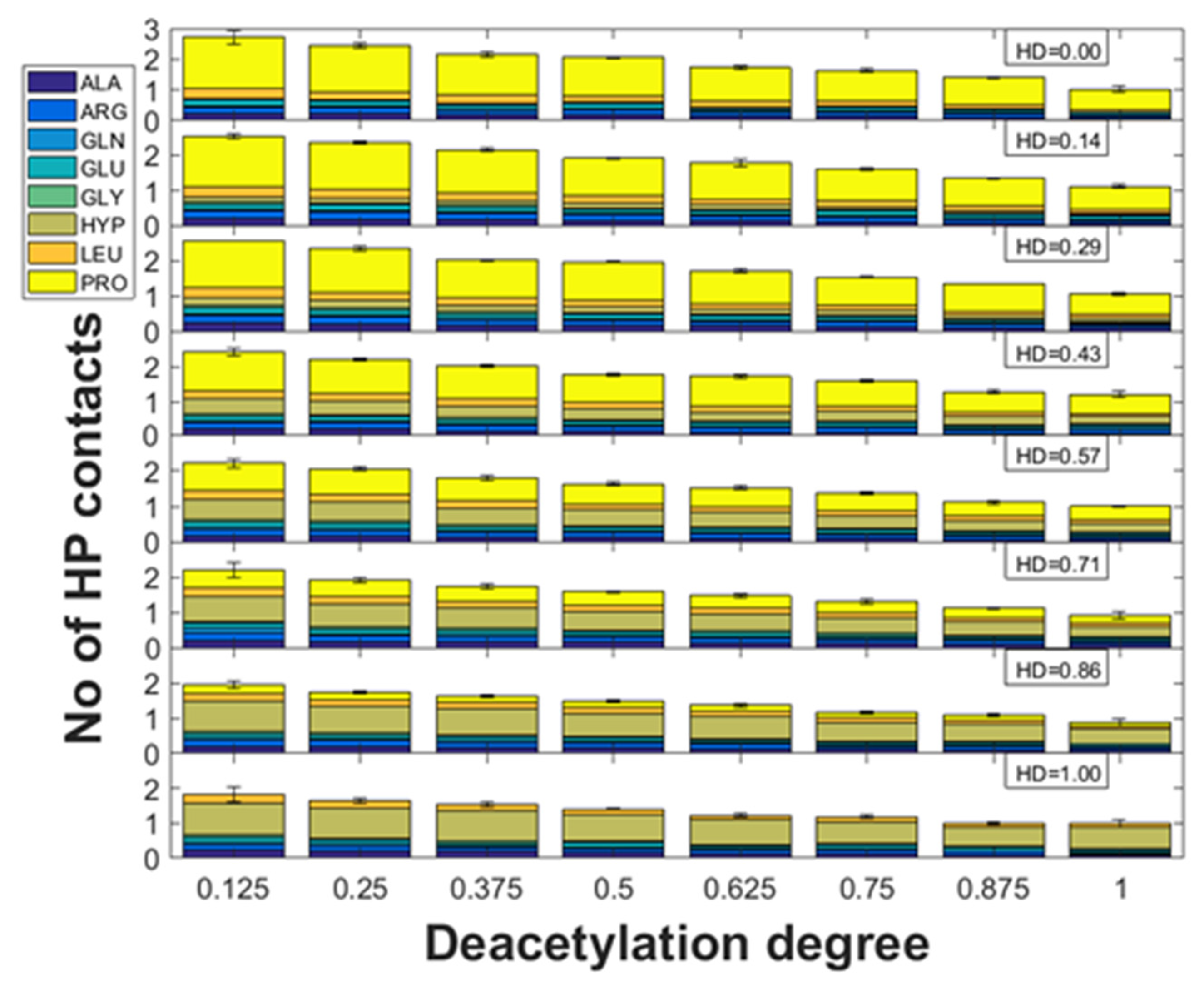
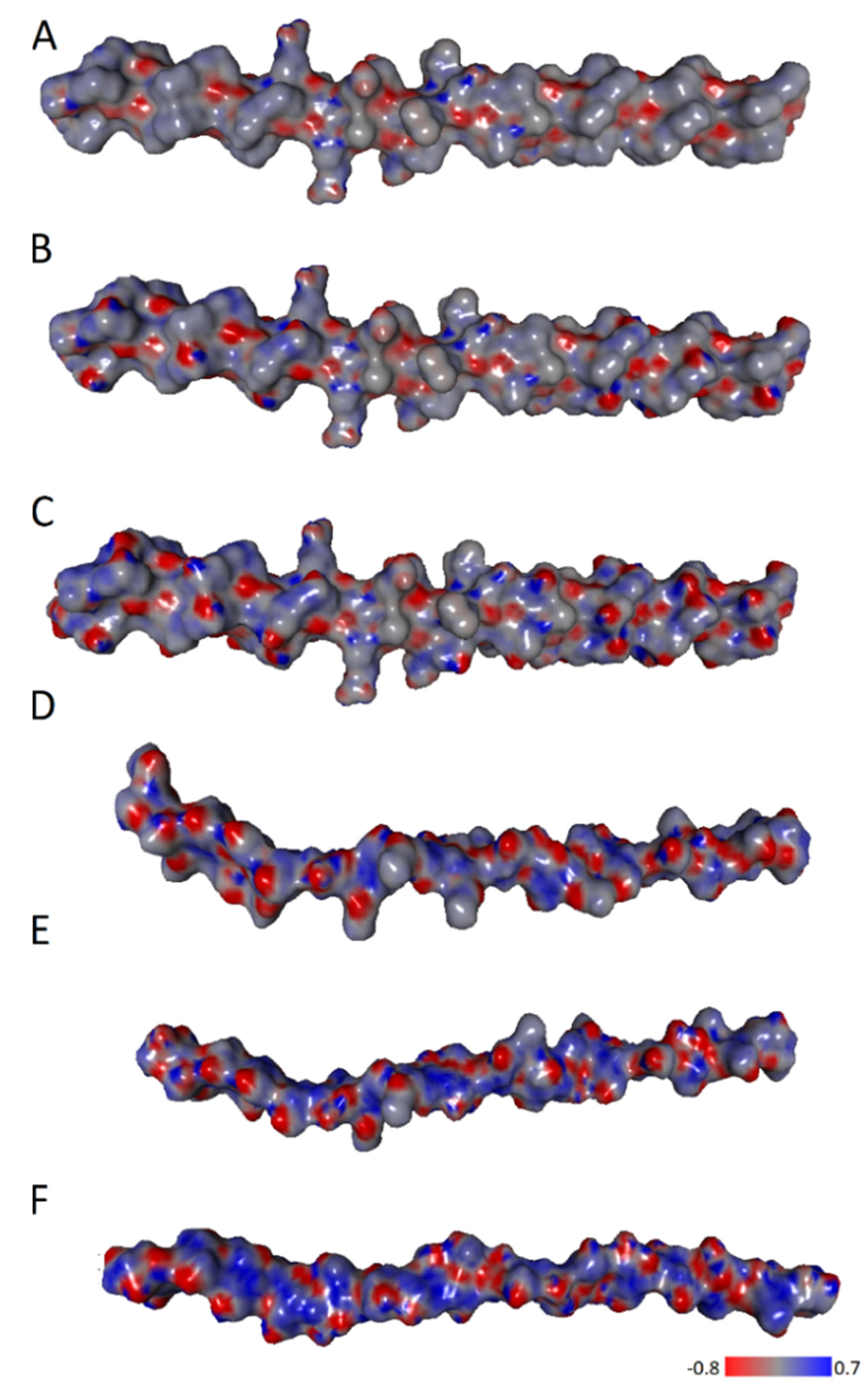
2.2. Hydrophobic Interactions
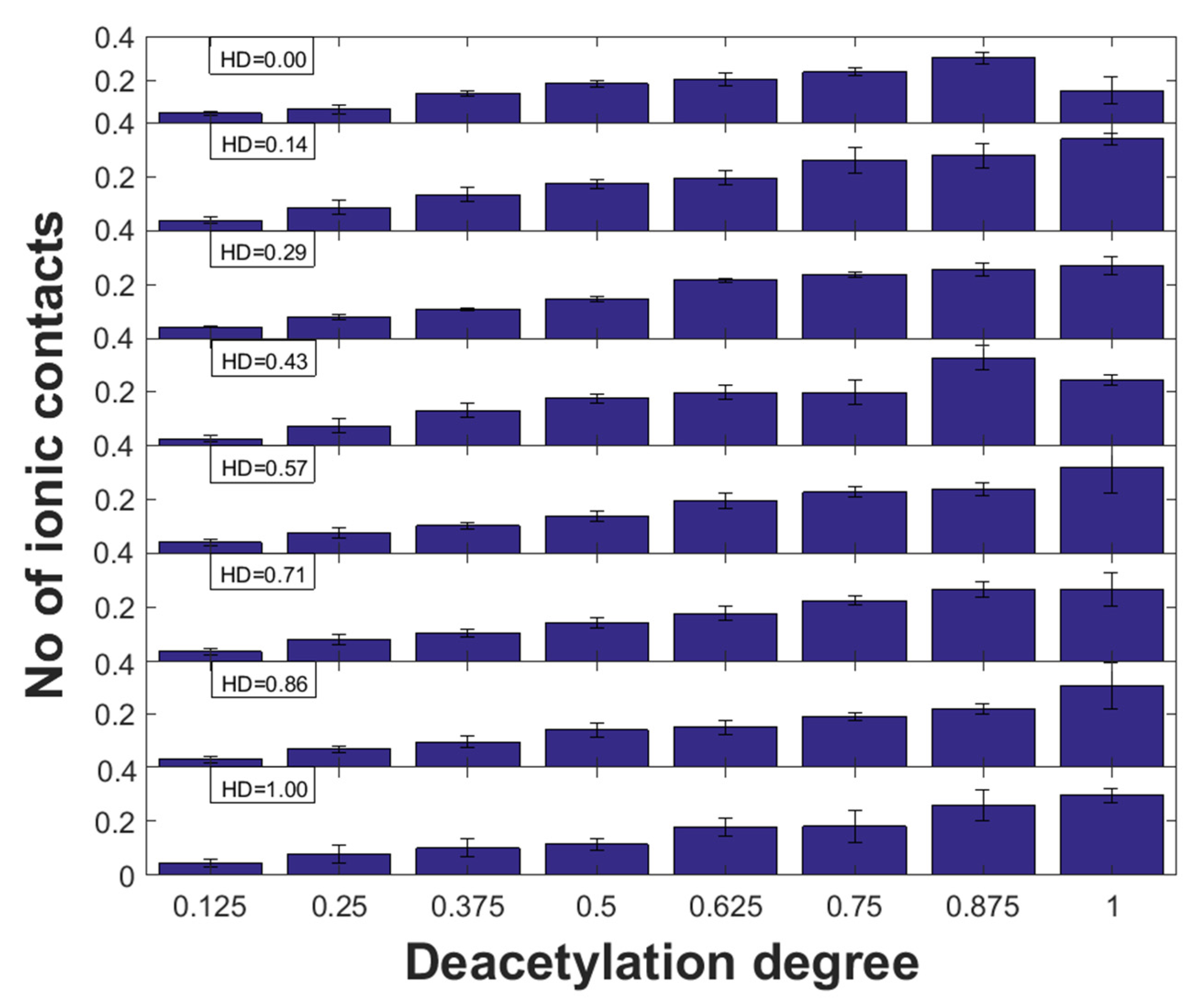
2.3. Ionic Interactions
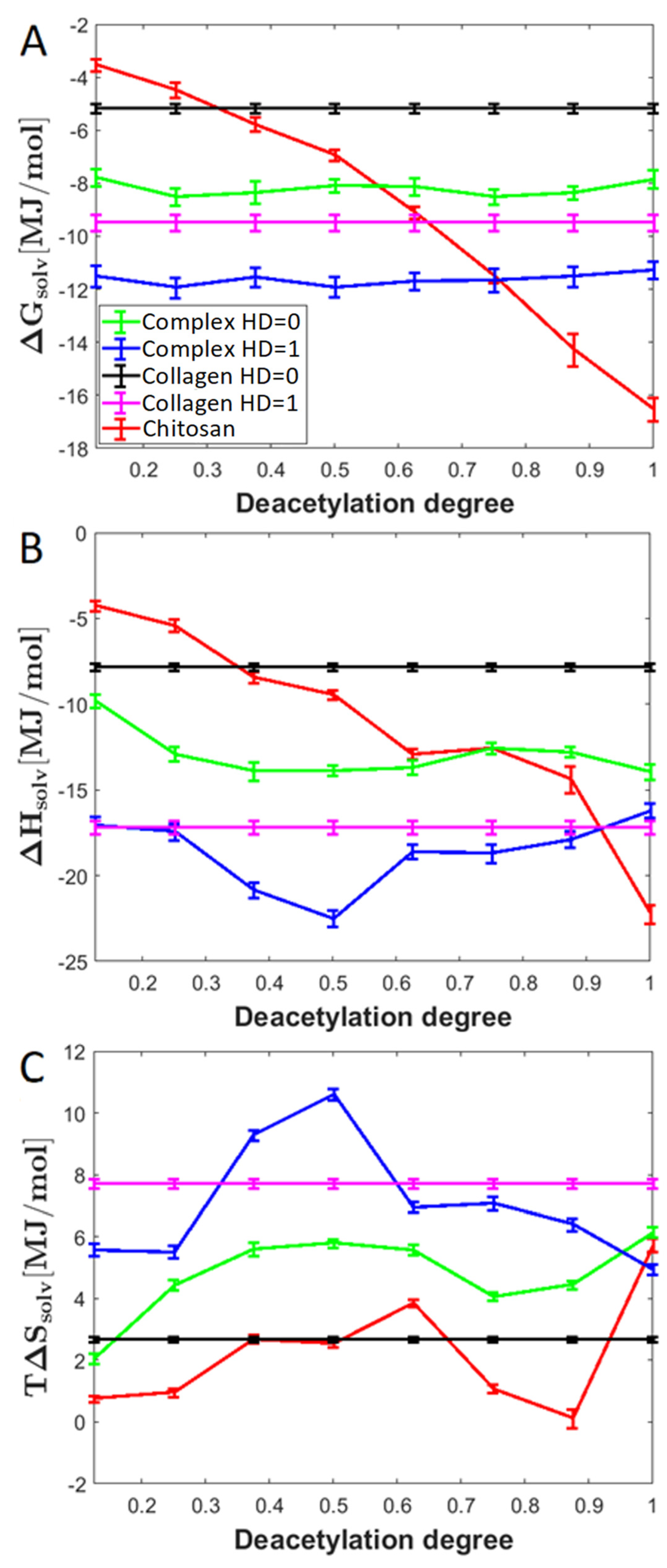
2.4. Entropic and Enthalpic Contributions to Solvation
3. Methods
3.1. Docking and Molecular Dynamics Simulation Setup
3.2. Binding Free Energy Determination
3.3. Determination of Free Energy and Entropy of Solvation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Han, L.; Grodzinsky, A.J.; Ortiz, C. Nanomechanics of the cartilage extracellular matrix. Annu. Rev. Mater. Res. 2011, 41, 133–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.; Torres, J.E.; Hakim, M.; Babiak, P.M.; Pal, P.; Battistoni, C.M.; Nguyen, M.; Panitch, A.; Solorio, L.; Liu, J.C. Collagen- and hyaluronic acid-based hydrogels and their biomedical applications. Mater. Sci. Eng. R Reports 2021, 146, 100641. [Google Scholar] [CrossRef] [PubMed]
- Mitura, S.; Sionkowska, A.; Jaiswal, A. Biopolymers for hydrogels in cosmetics: Review. J. Mater. Sci. Mater. Med. 2020, 31, 50. [Google Scholar] [CrossRef] [PubMed]
- Sionkowska, A.; Gadomska, M.; Musiał, K.; Piatek, J. Hyaluronic Acid as a Component of Natural Polymer Blends for Biomedical Applications: A Review. Molecules 2020, 25, 4035. [Google Scholar] [CrossRef] [PubMed]
- Mazeau, K.; Rinaudo, M. Comparative properties of hyaluronan and chitosan in aqueous environment. Polym. Sci.-Ser. C 2012, 54, 96–107. [Google Scholar] [CrossRef]
- Wu, X.; Black, L.; Santacana-Laffitte, G.; Patrick, C.W. Preparation and assessment of glutaraldehyde-crosslinked collagen-chitosan hydrogels for adipose tissue engineering. J. Biomed. Mater. Res.-Part A 2007, 81, 59–65. [Google Scholar] [CrossRef]
- Yan, L.P.; Wang, Y.J.; Ren, L.; Wu, G.; Caridade, S.G.; Fan, J.B.; Wang, L.Y.; Ji, P.H.; Oliveira, J.M.; Oliveira, J.T.; et al. Genipin-cross-linked collagen/chitosan biomimetic scaffolds for articular cartilage tissue engineering applications. J. Biomed. Mater. Res.-Part A 2010, 95 Pt A, 465–475. [Google Scholar] [CrossRef] [Green Version]
- Kim, I.Y.; Seo, S.J.; Moon, H.S.; Yoo, M.K.; Park, I.Y.; Kim, B.C.; Cho, C.S. Chitosan and its derivatives for tissue engineering applications. Biotechnol. Adv. 2008, 26, 1–21. [Google Scholar] [CrossRef]
- Tangsadthakun, C.; Kanokpanont, S.; Sanchavanakit, N.; Banaprasert, T.; Damrongsakkul, S. Properties of Collagen/Chitosan Scaffolds for Skin Tissue Engineering. J. Met. Mater. Miner. 2017, 16, 37–44. [Google Scholar]
- Zhu, C.; Fan, D.; Duan, Z.; Xue, W.; Shang, L.; Chen, F.; Luo, Y. Initial investigation of novel human-like collagen/chitosan scaffold for vascular tissue engineering. J. Biomed. Mater. Res.-Part A 2009, 89, 829–840. [Google Scholar] [CrossRef]
- Jithendra, P.; Rajam, A.M.; Kalaivani, T.; Mandal, A.B.; Rose, C. Preparation and characterization of aloe vera blended Collagen-Chitosan composite scaffold for tissue engineering applications. ACS Appl. Mater. Interfaces 2013, 5, 7291–7298. [Google Scholar] [CrossRef] [PubMed]
- Rafat, M.; Li, F.; Fagerholm, P.; Lagali, N.S.; Watsky, M.A.; Munger, R.; Matsuura, T.; Griffith, M. PEG-stabilized carbodiimide crosslinked collagen-chitosan hydrogels for corneal tissue engineering. Biomaterials 2008, 29, 3960–3972. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Krishnaraj, R.; Desai, T.A. Evaluation of nanostructured composite collagen–chitosan matrices for tissue engineering. Tissue Eng. 2001, 7, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, L.L.; Resende, C.X.; Tavares, D.S.; Soares, G.A.; Castro, L.O.; Granjeiro, J.M. Cytocompatibility of chitosan and collagen-chitosan scaffolds for tissue engineering. Polimeros 2011, 21, 1–6. [Google Scholar] [CrossRef]
- Ma, L.; Gao, C.; Mao, Z.; Zhou, J.; Shen, J.; Hu, X.; Han, C. Collagen/chitosan porous scaffolds with improved biostability for skin tissue engineering. Biomaterials 2003, 24, 4833–4841. [Google Scholar] [CrossRef]
- Chicatun, F.; Griffanti, G.; McKee, M.D.; Nazhat, S.N. Collagen/chitosan composite scaffolds for bone and cartilage tissue engineering. In Biomedical Composites; Woodhead Publishing: Sawston, UK, 2017; pp. 163–198. [Google Scholar]
- Jiang, Y.; Fu, C.; Wu, S.; Liu, G.; Guo, J.; Su, Z. Determination of the deacetylation degree of chitooligosaccharides. Mar. Drugs 2017, 15, 332. [Google Scholar] [CrossRef]
- Jones, M.; Kujundzic, M.; John, S.; Bismarck, A. Crab vs. Mushroom: A review of crustacean and fungal chitin in wound treatment. Mar. Drugs 2020, 18, 64. [Google Scholar] [CrossRef] [Green Version]
- Pellis, A.; Guebitz, G.M.; Nyanhongo, G.S. Chitosan: Sources, Processing and Modification Techniques. Gels 2022, 8, 393. [Google Scholar] [CrossRef]
- El Knidri, H.; El Khalfaouy, R.; Laajeb, A.; Addaou, A.; Lahsini, A. Eco-friendly extraction and characterization of chitin and chitosan from the shrimp shell waste via microwave irradiation. Process Saf. Environ. Prot. 2016, 104, 395–405. [Google Scholar] [CrossRef]
- Oladzadabbasabadi, N.; Mohammadi Nafchi, A.; Ariffin, F.; Wijekoon, M.M.J.O.; Al-Hassan, A.A.; Dheyab, M.A.; Ghasemlou, M. Recent advances in extraction, modification, and application of chitosan in packaging industry. Carbohydr. Polym. 2022, 277, 118876. [Google Scholar] [CrossRef]
- Rodríguez-Vázquez, M.; Vega-Ruiz, B.; Ramos-Zúñiga, R.; Saldaña-Koppel, D.A.; Quiñones-Olvera, L.F. Chitosan and Its Potential Use as a Scaffold for Tissue Engineering in Regenerative Medicine. Biomed Res. Int. 2015, 2015, 821279. [Google Scholar] [CrossRef] [PubMed]
- Domalik-Pyzik, P.; Chłopek, J.; Pielichowska, K. Chitosan-Based Hydrogels: Preparation, Properties, and Applications. In Cellulose-Based Superabsorbent Hydrogels. Polymers and Polymeric Composites: A Reference Series; Mondal, M., Ed.; Springer: Cham, Switzerland, 2019; pp. 1665–1693. [Google Scholar]
- Mathaba, M.; Daramola, M.O. Effect of chitosan’s degree of deacetylation on the performance of pes membrane infused with chitosan during amd treatment. Membranes 2020, 10, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasweefali, M.K.; Sabu, S.; Sunooj, K.V.; Sasidharan, A.; Xavier, K.A.M. Consequences of chemical deacetylation on physicochemical, structural and functional characteristics of chitosan extracted from deep-sea mud shrimp. Carbohydr. Polym. Technol. Appl. 2021, 2, 100032. [Google Scholar] [CrossRef]
- Yuan, Y.; Chesnutt, B.M.; Haggard, W.O.; Bumgardner, J.D. Deacetylation of Chitosan: Material Characterization and in vitro Evaluation via Albumin Adsorption and Pre-Osteoblastic Cell Cultures. Materials 2011, 4, 1399–1416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sionkowska, A.; Wisniewski, M.; Skopinska, J.; Kennedy, C.J.; Wess, T.J. Molecular interactions in collagen and chitosan blends. Biomaterials 2004, 25, 795–801. [Google Scholar] [CrossRef]
- Hou, C.; Gao, L.; Wang, Z.; Rao, W.; Du, M.; Zhang, D. Mechanical properties, thermal stability, and solubility of sheep bone collagen–chitosan films. J. Food Process Eng. 2020, 43, e13086. [Google Scholar] [CrossRef]
- Machado, A.A.S.; Martins, V.C.A.; Plepis, A.M.G. Thermal and rheological behavior of collagen: Chitosan blends. J. Therm. Anal. Calorim. 2002, 67, 491–498. [Google Scholar] [CrossRef]
- Hsu, S.H.; Whu, S.W.; Tsai, C.L.; Wu, Y.H.; Chen, H.W.; Hsieh, K.H. Chitosan as scaffold materials: Effects of molecular weight and degree of deacetylation. J. Polym. Res. 2004, 11, 141–147. [Google Scholar] [CrossRef]
- Varma, S.; Orgel, J.P.R.O.; Schieber, J.D. Contrasting local and macroscopic effects of collagen hydroxylation. Int. J. Mol. Sci. 2021, 22, 9068. [Google Scholar] [CrossRef]
- Rosenbloom, J.; Harsch, M.; Jimenez, S. Hydroxyproline content determines the denaturation temperature of chick tendon collagen. Arch. Biochem. Biophys. 1973, 158, 478–484. [Google Scholar] [CrossRef]
- Sipila, K.H.; Drushinin, K.; Rappu, P.; Jokinen, J.; Salminen, T.A.; Salo, A.M.; Käpyla, J.; Myllyharju, J.; Heino, J. Proline hydroxylation in collagen supports integrin binding by two distinct mechanisms. J. Biol. Chem. 2018, 293, 7645–7658. [Google Scholar] [CrossRef] [PubMed]
- Martinou, A.; Kafetzopoulos, D.; Bouriotis, V. Chitin deacetylation by enzymatic means: Monitoring of deacetylation processes. Carbohydr. Res. 1995, 273, 235–242. [Google Scholar] [CrossRef]
- Jaworska, M.M. Kinetics of enzymatic deacetylation of chitosan. Cellulose 2012, 19, 363–369. [Google Scholar] [CrossRef] [Green Version]
- Harmsen, R.A.G.; Tuveng, T.R.; Antonsen, S.G.; Eijsink, V.G.H.; Sørlie, M. Can we make Chitosan by Enzymatic Deacetylation of Chitin? Molecules 2019, 24, 3862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- No, H.K.; Cho, Y.I.; Kim, H.R.; Meyers, S.P. Effective Deacetylation of Chitin under Conditions of 15 psi/121 °C. J. Agric. Food Chem. 2000, 48, 2625–2627. [Google Scholar] [CrossRef]
- He, X.; Li, K.; Xing, R.; Liu, S.; Hu, L.; Li, P. The production of fully deacetylated chitosan by compression method. Egypt. J. Aquat. Res. 2016, 42, 75–81. [Google Scholar] [CrossRef] [Green Version]
- Tsaih, M.L.; Chen, R.H. The effect of reaction time and temperature during heterogenous alkali deacetylation on degree of deacetylation and molecular weight of resulting chitosan. J. Appl. Polym. Sci. 2003, 88, 2917–2923. [Google Scholar] [CrossRef]
- An, B.; Kaplan, D.L.; Brodsky, B. Engineered recombinant bacterial collagen as an alternative collagen-based biomaterial for tissue engineering. Front. Chem. 2014, 2, 40. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Gan, Q.; Clough, R.C.; Pappu, K.M.; Howard, J.A.; Baez, J.A.; Wang, K. Hydroxylation of recombinant human collagen type I alpha 1 in transgenic maize co-expressed with a recombinant human prolyl 4-hydroxylase. BMC Biotechnol. 2011, 11, 69. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Lew, J.; Premkumar, J.; Poh, C.L.; Naing, M.W. Production of recombinant collagen: State of the art and challenges. Eng. Biol. 2017, 1, 18–23. [Google Scholar] [CrossRef] [Green Version]
- Acevedo, C.A.; Diáz-Calderón, P.; López, D.; Enrione, J. Assessment of gelatin-chitosan interactions in films by a chemometrics approach. CYTA-J. Food 2015, 13, 227–234. [Google Scholar] [CrossRef]
- Ledward, D.A. Gelation of gelatin. In Functional Properties of Food Macromolecules; Mitchell, J.R., Ledward, D.A., Eds.; Elsevier Applied Science: London, UK, 1986; pp. 171–201. [Google Scholar]
- Gómez-Estaca, J.; Gómez-Guillén, M.C.; Fernández-Martín, F.; Montero, P. Effects of gelatin origin, bovine-hide and tuna-skin, on the properties of compound gelatin-chitosan films. Food Hydrocoll. 2011, 25, 1461–1469. [Google Scholar] [CrossRef] [Green Version]
- Pietrucha, K. Changes in denaturation and rheological properties of collagen-hyaluronic acid scaffolds as a result of temperature dependencies. Int. J. Biol. Macromol. 2005, 36, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Taravel, M.N.; Domard, A. Collagen and its interaction with chitosan. II. Influence of the physicochemical characteristics of collagen. Biomaterials 1995, 16, 865–871. [Google Scholar] [CrossRef]
- Martínez, A.; Blanco, M.D.; Davidenko, N.; Cameron, R.E. Tailoring chitosan/collagen scaffolds for tissue engineering: Effect of composition and different crosslinking agents on scaffold properties. Carbohydr. Polym. 2015, 132, 606–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakhem, E.; Bitar, K. Development of Chitosan Scaffolds with Enhanced Mechanical Properties for Intestinal Tissue Engineering Applications. J. Funct. Biomater. 2015, 6, 999–1011. [Google Scholar] [CrossRef] [Green Version]
- Han, C.M.; Zhang, L.P.; Sun, J.Z.; Shi, H.F.; Zhou, J.; Gao, C.Y. Application of collagen-chitosan/fibrin glue asymmetric scaffolds in skin tissue engineering. J. Zhejiang Univ. Sci. B 2010, 11, 524–530. [Google Scholar] [CrossRef]
- Carvalho, D.N.; Lopez-Cebral, R.; Sousa, R.O.; Alves, A.L.; Reys, L.L.; Silva, S.S.; Oliveira, J.M.; Reis, R.L.; Silva, T.H. Marine collagen-chitosan-fucoidan cryogels as cell-laden biocomposites envisaging tissue engineering. Biomed. Mater. 2020, 15, 055030. [Google Scholar] [CrossRef]
- Moon, H.; Choy, S.; Park, Y.; Jung, Y.M.; Koo, J.M.; Hwang, D.S. Different Molecular Interaction between Collagen and α- or β-Chitin in Mechanically Improved Electrospun Composite. Mar. Drugs 2019, 17, 318. [Google Scholar] [CrossRef] [Green Version]
- Krieger, E.; Dunbrack, R.L.; Hooft, R.W.W.; Krieger, B. Assignment of protonation states in proteins and ligands: Combining pK a prediction with hydrogen bonding network optimization. Methods Mol. Biol. 2012, 819, 405–421. [Google Scholar] [CrossRef]
- Indrani, D.J.; Lukitowati, F.; Yulizar, Y. Preparation of Chitosan/Collagen Blend Membranes for Wound Dressing: A Study on FTIR Spectroscopy and Mechanical Properties. IOP Conf. Ser. Mater. Sci. Eng. 2017, 202, 012020. [Google Scholar] [CrossRef]
- Staroszczyk, H.; Sztuka, K.; Wolska, J.; Wojtasz-Paja̧k, A.; Kołodziejska, I. Interactions of fish gelatin and chitosan in uncrosslinked and crosslinked with EDC films: FT-IR study. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2014, 117, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Langrock, T.; Hoffmann, R. Analysis of Hydroxyproline in Collagen Hydrolysates. In Amino Acid Analysis. Methods in Molecular Biology; Alterman, M., Ed.; Humana: New York, NY, USA, 2019; Volume 2030, pp. 47–56. [Google Scholar]
- The MathWorks Inc. MATLAB; The MathWorks Inc.: Natick, MA, USA, 2022. [Google Scholar]
- Pokidysheva, E.; Boudko, S.; Vranka, J.; Zientek, K.; Maddox, K.; Moser, M.; Fässler, R.; Ware, J.; Bächinger, H.P. Biological role of prolyl 3-hydroxylation in type IV collagen. Proc. Natl. Acad. Sci. USA 2014, 111, 161–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rappu, P.; Salo, A.M.; Myllyharju, J.; Heino, J. Role of prolyl hydroxylation in the molecular interactions of collagens. Essays Biochem. 2019, 63, 325–335. [Google Scholar] [CrossRef] [Green Version]
- Kivirikko, K.I.; Myllylä, R.; Pihlajaniemi, T. Hydroxylation of proline and lysine residues in collagens and other animal and plant proteins. In Post-Translational Modifications of Proteins; Harding, J.J., Crabbe, M.J.C., Eds.; CRC Press: Boca Raton, FL, USA, 1991; pp. 1–51. ISBN 9780849341717. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A Point-Charge Force Field for Molecular Mechanics Simulations of Proteins Based on Condensed-Phase Quantum Mechanical Calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; González-Outeiriño, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: A generalizable biomolecular force field. carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef] [Green Version]
- Kulke, M.; Geist, N.; Friedrichs, W.; Langel, W. Molecular dynamics simulations on networks of heparin and collagen. Proteins Struct. Funct. Bioinform. 2017, 85, 1119–1130. [Google Scholar] [CrossRef]
- Roy, A.; Gauld, J.W. Molecular Dynamics Investigation on the Effects of Protonation and Lysyl Hydroxylation on Sulfilimine Cross-links in Collagen IV. ACS Omega 2022, 7, 39680–39689. [Google Scholar] [CrossRef]
- Cole, C.C.; Misiura, M.; Hulgan, S.A.H.; Peterson, C.M.; Williams, J.W.; Kolomeisky, A.B.; Hartgerink, J.D. Cation−π Interactions and Their Role in Assembling Collagen Triple Helices. Biomacromolecules 2022, 23, 4645–4654. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. YASARA View—Molecular graphics for all devices—From smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef] [PubMed]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Holst, M.; Baker, N.; Wang, F. Adaptive multilevel finite element solution of the Poisson-Boltzmann equation I. Algorithms and examples. J. Comput. Chem. 2000, 21, 1319–1342. [Google Scholar] [CrossRef]
- Baker, N.; Holst, M.; Wang, F. Adaptive multilevel finite element solution of the Poisson-Boltzmann equation II. Refinement at solvent-accessible surfaces in biomolecular systems. J. Comput. Chem. 2000, 21, 1343–1352. [Google Scholar] [CrossRef]
- Syme, N.R.; Dennis, C.; Bronowska, A.; Paesen, G.C.; Homans, S.W. Comparison of entropic contributions to binding in a “hydrophilic” versus “hydrophobic” ligand-protein interaction. J. Am. Chem. Soc. 2010, 132, 8682–8689. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Przybyłek, M.; Bełdowski, P.; Wieland, F.; Cysewski, P.; Sionkowska, A. Collagen Type II—Chitosan Interactions as Dependent on Hydroxylation and Acetylation Inferred from Molecular Dynamics Simulations. Molecules 2023, 28, 154. https://doi.org/10.3390/molecules28010154
Przybyłek M, Bełdowski P, Wieland F, Cysewski P, Sionkowska A. Collagen Type II—Chitosan Interactions as Dependent on Hydroxylation and Acetylation Inferred from Molecular Dynamics Simulations. Molecules. 2023; 28(1):154. https://doi.org/10.3390/molecules28010154
Chicago/Turabian StylePrzybyłek, Maciej, Piotr Bełdowski, Florian Wieland, Piotr Cysewski, and Alina Sionkowska. 2023. "Collagen Type II—Chitosan Interactions as Dependent on Hydroxylation and Acetylation Inferred from Molecular Dynamics Simulations" Molecules 28, no. 1: 154. https://doi.org/10.3390/molecules28010154
APA StylePrzybyłek, M., Bełdowski, P., Wieland, F., Cysewski, P., & Sionkowska, A. (2023). Collagen Type II—Chitosan Interactions as Dependent on Hydroxylation and Acetylation Inferred from Molecular Dynamics Simulations. Molecules, 28(1), 154. https://doi.org/10.3390/molecules28010154









