Structural Modeling of Nanobodies: A Benchmark of State-of-the-Art Artificial Intelligence Programs
Abstract
1. Introduction
2. Results and Discussion
2.1. Dataset Selection and Validation
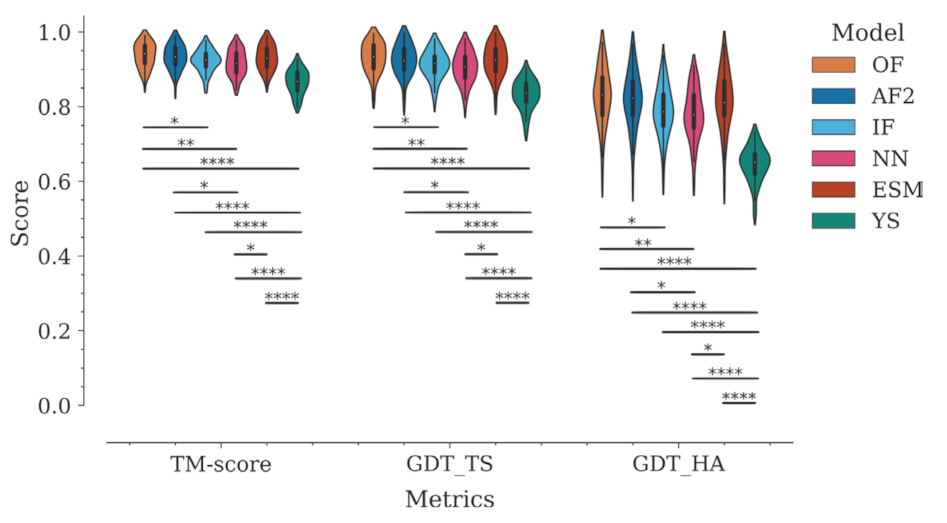
2.2. Structure Prediction Accuracy
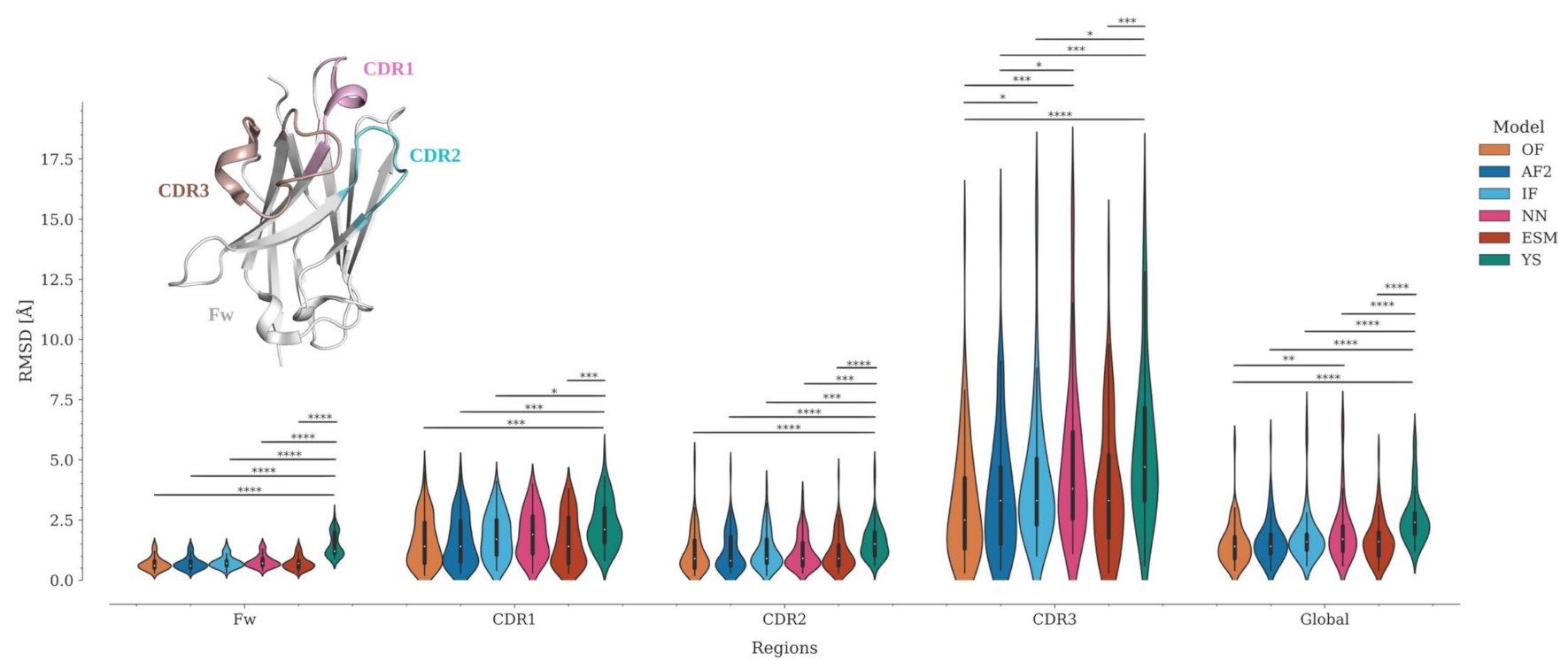
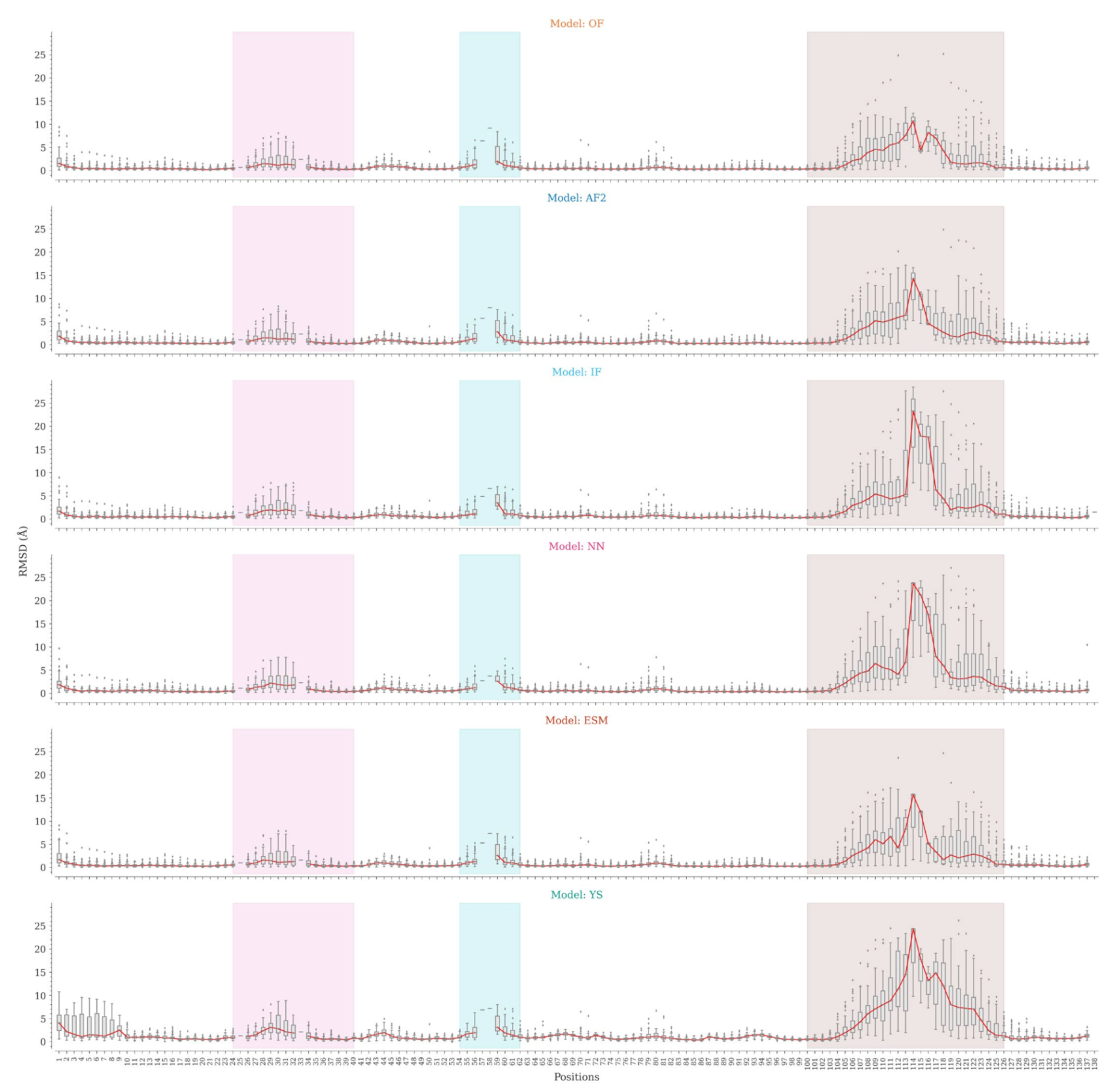
2.3. Structure Prediction Accuracy by Sequence Position
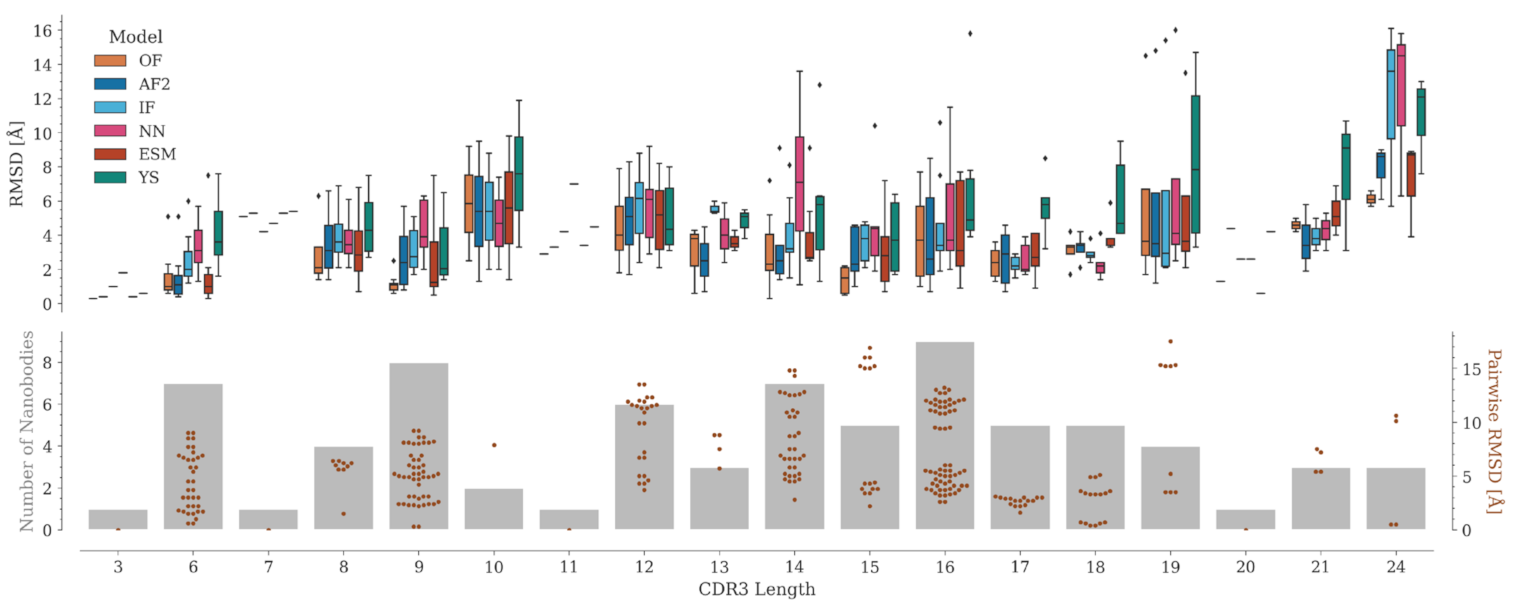
2.4. CDR3 Structure Prediction Accuracy
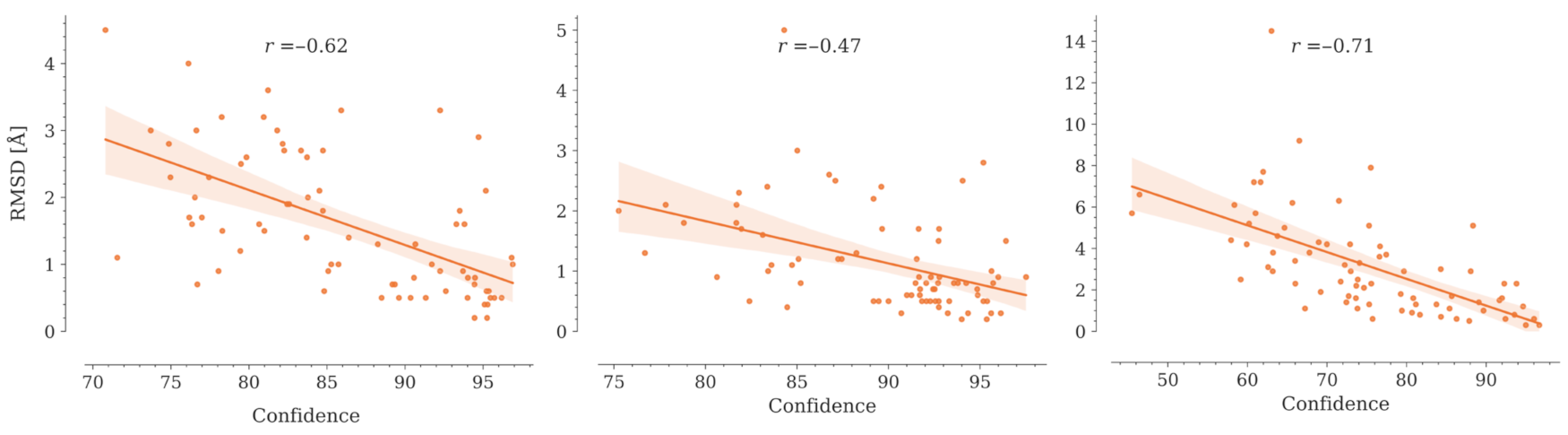
2.5. Nanobody Modeling Confidence
2.6. Structure Prediction Accuracy Varying Modeling Parameters
2.6.1. Number of Recycles
2.6.2. Modeling Nanobodies in Complex with Their Antigens with AlphaFold-Multimer
2.6.3. Energy Minimization
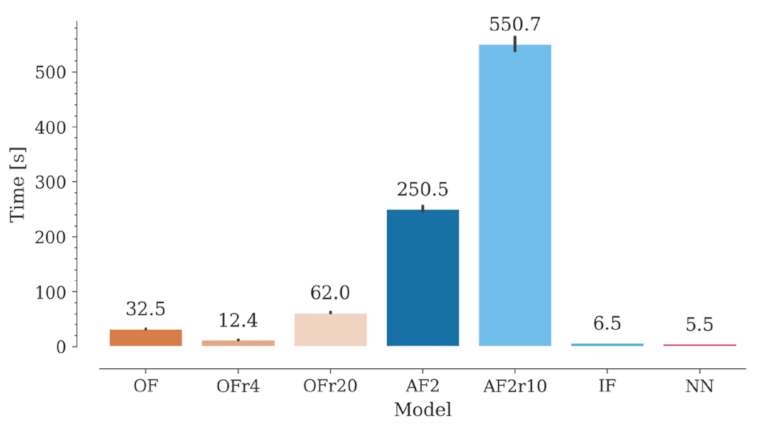
2.7. Computation Time
3. Materials and Methods
3.1. Benchmark Dataset
3.2. Artificial Intelligence Models
3.2.1. AlphaFold2
3.2.2. OmegaFold
3.2.3. ESMFold
3.2.4. Yang-Server
3.2.5. IgFold
3.2.6. Nanonet
3.3. Performance Evaluation Metrics
3.3.1. Structural Similarity Metrics
3.3.2. Statistics
3.3.3. Execution Environment
3.3.4. Energy Minimization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hammers, C.; Songa, E.B.; Bendahman, N.; Hammers, R. Naturally Occurring Antibodies Devoid of Light Chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef] [PubMed]
- Steeland, S.; Vandenbroucke, R.E.; Libert, C. Nanobodies as Therapeutics: Big Opportunities for Small Antibodies. Drug Discov. Today 2016, 21, 1076–1113. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Sapienza, G.; Rossotti, M.A.; Tabares-da Rosa, S. Single-Domain Antibodies as Versatile Affinity Reagents for Analytical and Diagnostic Applications. Front. Immunol. 2017, 8, 977. [Google Scholar] [CrossRef]
- Zare, H.; Aghamollaei, H.; Hosseindokht, M.; Heiat, M.; Razei, A.; Bakherad, H. Nanobodies, the Potent Agents to Detect and Treat the Coronavirus Infections: A Systematic Review. Mol. Cell. Probes 2021, 55, 101692. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S. Applications of Nanobodies. Annu. Rev. Anim. Biosci. 2021, 9, 401–421. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.Y.; Shah, K. Nanobodies: Next Generation of Cancer Diagnostics and Therapeutics. Front. Oncol. 2020, 10, 1182. [Google Scholar] [CrossRef]
- Wang, J.; Kang, G.; Yuan, H.; Cao, X.; Huang, H.; de Marco, A. Research Progress and Applications of Multivalent, Multispecific and Modified Nanobodies for Disease Treatment. Front. Immunol. 2022, 12, 6013. [Google Scholar] [CrossRef]
- Njeru, F.N.; Kusolwa, P.M. Nanobodies: Their Potential for Applications in Biotechnology, Diagnosis and Antiviral Properties in Africa; Focus on Application in Agriculture. Biotechnol. Biotechnol. Equip. 2021, 35, 1331–1342. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Q.; Sun, Z.; Wang, Y.; Su, B.; Zhang, C.; Cao, H.; Liu, X. Nanobody Affinity Improvement: Directed Evolution of the Anti-Ochratoxin a Single Domain Antibody. Int. J. Biol. Macromol 2020, 151, 312–321. [Google Scholar] [CrossRef]
- Soler, M.A.; Fortuna, S.; de Marco, A.; Laio, A. Binding Affinity Prediction of Nanobody-Protein Complexes by Scoring of Molecular Dynamics Trajectories. Phys. Chem. Chem. Phys. 2018, 20, 3438–3444. [Google Scholar] [CrossRef]
- Hacisuleyman, A.; Erman, B. ModiBodies: A Computational Method for Modifying Nanobodies to Improve Their Antigen Binding Affinity and Specificity. bioRxiv 2019. [Google Scholar] [CrossRef]
- Cohen, T.; Halfon, M.; Schneidman-Duhovny, D. NanoNet: Rapid and Accurate End-to-End Nanobody Modeling by Deep Learning. Front. Immunol. 2022, 13, 4319. [Google Scholar] [CrossRef] [PubMed]
- Ruffolo, J.A.; Chu, L.-S.; Mahajan, S.P.; Gray, J.J. Fast, Accurate Antibody Structure Prediction from Deep Learning on Massive Set of Natural Antibodies. bioRxiv 2022. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; di Costanzo, L.; Christie, C.; Dalenberg, K.; Duarte, J.M.; Dutta, S.; et al. RCSB Protein Data Bank: Biological Macromolecular Structures Enabling Research and Education in Fundamental Biology, Biomedicine, Biotechnology and Energy. Nucleic Acids Res. 2019, 47, D464–D474. [Google Scholar] [CrossRef]
- Mitchell, L.S.; Colwell, L.J. Analysis of Nanobody Paratopes Reveals Greater Diversity than Classical Antibodies. Protein Eng. Des. Sel. 2018, 31, 267–275. [Google Scholar] [CrossRef]
- Xi, X.; Sun, W.; Su, H.; Zhang, X.; Sun, F. Identification of a Novel Anti-EGFR Nanobody by Phage Display and Its Distinct Paratope and Epitope via Homology Modeling and Molecular Docking. Mol. Immunol. 2020, 128, 165–174. [Google Scholar] [CrossRef]
- Cheng, X.; Wang, J.; Kang, G.; Hu, M.; Yuan, B.; Zhang, Y.; Huang, H. Homology Modeling-Based in Silico Affinity Maturation Improves the Affinity of a Nanobody. Int. J. Mol. Sci. 2019, 20, 4187. [Google Scholar] [CrossRef]
- Zou, J. Artificial Intelligence Revolution in Structure Prediction for Entire Proteomes. MedComm—Future Med. 2022, 1, e19. [Google Scholar] [CrossRef]
- Bertoline, L.M.F.; Lima, A.N.; Krieger, J.E.; Teixeira, S.K. Before and after AlphaFold2: An Overview of Protein Structure Prediction. Front. Bioinform. 2023, 3, 17. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K. The Prospects and Opportunities of Protein Structure Prediction with AI. Nat. Rev. Mol. Cell Biol. 2022, 23, 445–446. [Google Scholar] [CrossRef]
- Abanades, B.; Georges, G.; Bujotzek, A.; Deane, C.M. ABlooper: Fast Accurate Antibody CDR Loop Structure Prediction with Accuracy Estimation. Bioinformatics 2022, 38, 1877–1880. [Google Scholar] [CrossRef] [PubMed]
- Ruffolo, J.A.; Gray, J.J.; Sulam, J. Deciphering Antibody Affinity Maturation with Language Models and Weakly Supervised Learning. arXiv 2021, arXiv:2112.07782. [Google Scholar]
- Wang, S.; Sun, S.; Li, Z.; Zhang, R.; Xu, J. Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model. PLoS Comput. Biol. 2017, 13, e1005324. [Google Scholar] [CrossRef]
- Fernández-Quintero, M.L.; Kokot, J.; Waibl, F.; Fischer, A.L.M.; Quoika, P.K.; Deane, C.M.; Liedl, K.R. Challenges in Antibody Structure Prediction. MAbs 2023, 15, 2175319. [Google Scholar] [CrossRef] [PubMed]
- Ruffolo, J.A.; Sulam, J.; Gray, J.J. Antibody Structure Prediction Using Interpretable Deep Learning. Patterns 2022, 3, 100406. [Google Scholar] [CrossRef]
- Ruffolo, J.A.; Guerra, C.; Mahajan, S.P.; Sulam, J.; Gray, J.J. Geometric Potentials from Deep Learning Improve Prediction of CDR H3 Loop Structures. Bioinformatics 2020, 36, i268. [Google Scholar] [CrossRef]
- AlQuraishi, M. Machine Learning in Protein Structure Prediction. Curr. Opin. Chem. Biol. 2021, 65, 1–8. [Google Scholar] [CrossRef]
- Eisenstein, M. Artificial Intelligence Powers Protein-Folding Predictions. Nature 2021, 599, 706–708. [Google Scholar] [CrossRef]
- AlQuraishi, M. Protein-Structure Prediction Revolutionized. Nature 2021, 596, 487–488. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. What’s next for AlphaFold and the AI Protein-Folding Revolution. Nature 2022, 604, 234–238. [Google Scholar] [CrossRef]
- Wu, R.; Ding, F.; Wang, R.; Shen, R.; Zhang, X.; Luo, S.; Su, C.; Wu, Z.; Xie, Q.; Berger, B.; et al. High-Resolution de Novo Structure Prediction from Primary Sequence. bioRxiv 2022. [Google Scholar] [CrossRef]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; Smetanin, N.; Verkuil, R.; Kabeli, O.; Shmueli, Y.; et al. Evolutionary-Scale Prediction of Atomic Level Protein Structure with a Language Model. bioRxiv 2022. [Google Scholar] [CrossRef]
- Schoof, M.; Faust, B.; Saunders, R.A.; Sangwan, S.; Rezelj, V.; Hoppe, N.; Boone, M.; Billesbølle, C.B.; Puchades, C.; Azumaya, C.M.; et al. An Ultrapotent Synthetic Nanobody Neutralizes SARS-CoV-2 by Stabilizing Inactive Spike. Science 2020, 370, 1473–1479. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, Y. How Significant Is a Protein Structure Similarity with TM-Score = 0.5? Bioinformatics 2010, 26, 889. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Su, H.; Wang, W.; Ye, L.; Wei, H.; Peng, Z.; Anishchenko, I.; Baker, D.; Yang, J. The TrRosetta Server for Fast and Accurate Protein Structure Prediction. Nat. Protoc. 2021, 16, 5634–5651. [Google Scholar] [CrossRef]
- Hong, J.; Kwon, H.J.; Cachau, R.; Chen, C.Z.; Butay, K.J.; Duan, Z.; Li, D.; Ren, H.; Liang, T.; Zhu, J.; et al. Camel Nanobodies Broadly Neutralize SARS-CoV-2 Variants. bioRxiv 2021. [Google Scholar] [CrossRef]
- Frosi, Y.; Lin, Y.C.; Shimin, J.; Ramlan, S.R.; Hew, K.; Engman, A.H.; Pillai, A.; Yeung, K.; Cheng, Y.X.; Cornvik, T.; et al. Engineering an Autonomous VH Domain to Modulate Intracellular Pathways and to Interrogate the EIF4F Complex. Nat. Commun. 2022, 13, 4854. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein Complex Prediction with AlphaFold-Multimer. bioRxiv 2022. [Google Scholar] [CrossRef]
- Zimmermann, I.; Egloff, P.; Hutter, C.A.J.; Arnold, F.M.; Stohler, P.; Bocquet, N.; Hug, M.N.; Hub er, S.; Siegrist, M.; Hetemann, L.; et al. Synthetic Single Domain Antibodies for the Conformational Trapping of Membrane Proteins. Elife 2018, 7, e34317. [Google Scholar] [CrossRef] [PubMed]
- Moreno, E.; Valdés-Tresanco, M.S.; Molina-Zapata, A.; Sánchez-Ramos, O. Structure-Based Design and Construction of a Synthetic Phage Display Nanobody Library. BMC Res. Notes 2022, 15, 124. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Tresanco, M.S.; Molina-Zapata, A.; Pose, A.G.; Moreno, E. Structural Insights into the Design of Synthetic Nanobody Libraries. Molecules 2022, 27, 2198. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, J.; Krawczyk, K.; Leem, J.; Baker, T.; Fuchs, A.; Georges, G.; Shi, J.; Deane, C.M. SAbDab: The Structural Antibody Database. Nucleic Acids Res. 2014, 42, D1140–D1146. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Dunbar, J.; Deane, C.M. ANARCI: Antigen Receptor Numbering and Receptor Classification. Bioinformatics 2016, 32, 298–300. [Google Scholar] [CrossRef]
- Yang, J.; Anishchenko, I.; Park, H.; Peng, Z.; Ovchinnikov, S.; Baker, D. Improved Protein Structure Prediction Using Predicted Interresidue Orientations. Proc. Natl. Acad. Sci. USA 2020, 117, 1496–1503. [Google Scholar] [CrossRef]
- Pereira, J.; Simpkin, A.J.; Hartmann, M.D.; Rigden, D.J.; Keegan, R.M.; Lupas, A.N. High-Accuracy Protein Structure Prediction in CASP14. Proteins 2021, 89, 1687–1699. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Steinegger, M.; Söding, J. MMseqs2 Desktop and Local Web Server App for Fast, Interactive Sequence Searches. Bioinformatics 2019, 35, 2856–2858. [Google Scholar] [CrossRef] [PubMed]
- Steinegger, M.; Söding, J. MMseqs2 Enables Sensitive Protein Sequence Searching for the Analysis of Massive Data Sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef] [PubMed]
- Rives, A.; Meier, J.; Sercu, T.; Goyal, S.; Lin, Z.; Liu, J.; Guo, D.; Ott, M.; Zitnick, C.L.; Ma, J.; et al. Biol.ogical Structure and Function Emerge from Scaling Unsupervised Learning to 250 Million Protein Sequences. Proc. Natl. Acad. Sci. USA 2021, 118, e2016239118. [Google Scholar] [CrossRef] [PubMed]
- Suzek, B.E.; Huang, H.; McGarvey, P.; Mazumder, R.; Wu, C.H. UniRef: Comprehensive and non-redundant UniProt reference clusters. Bioinformatics 2007, 23, 1282–1288. [Google Scholar] [CrossRef]
- Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.F.G.; Qin, C.; Zidek, A.; Nelson, A.; Bridgland, A.; Penedones, H.; et al. De Novo Structure Prediction with Deep-Learning Based Scoring. In Proceedings of the Thirteenth Critical Assessment of Techniques for Protein Structure Prediction (ProteinStructure Prediction Center), Cancun, Mexico, 1–4 December 2018; pp. 11–12. [Google Scholar]
- Dunbar, J.; Krawczyk, K.; Leem, J.; Marks, C.; Nowak, J.; Regep, C.; Georges, G.; Kelm, S.; Popovic, B.; Deane, C.M. SAbPred: A Structure-Based Antibody Prediction Server. Nucleic Acids Res. 2016, 44, W474–W478. [Google Scholar] [CrossRef]
- Chaudhury, S.; Lyskov, S.; Gray, J.J. PyRosetta: A Script-Based Interface for Implementing Molecular Modeling Algorithms Using Rosetta. Bioinformatics 2010, 26, 689–691. [Google Scholar] [CrossRef]
- Šali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. Scoring Function for Automated Assessment of Protein Structure Template Quality. Proteins 2004, 57, 702–710. [Google Scholar] [CrossRef]
- Zemla, A. LGA: A Method for Finding 3D Similarities in Protein Structures. Nucleic Acids Res. 2003, 31, 3370. [Google Scholar] [CrossRef]
- Shirts, M.R.; Klein, C.; Swails, J.M.; Yin, J.; Gilson, M.K.; Mobley, D.L.; Case, D.A.; Zhong, E.D. Lessons Learned from Comparing Molecular Dynamics Engines on the SAMPL5 Dataset. J. Comput. Aided Mol. Des. 2016, 31, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Bedre, R. Bioinfokit: Bioinformatics Data Analysis and Visualization Toolkit, version 1.0.5; Zenodo: Geneva, Switzerland, 2021. [Google Scholar] [CrossRef]
- Eastman, P.; Swails, J.; Chodera, J.D.; McGibbon, R.T.; Zhao, Y.; Beauchamp, K.A.; Wang, L.P.; Simmonett, A.C.; Harrigan, M.P.; Stern, C.D.; et al. OpenMM 7: Rapid Development of High Performance Algorithms for Molecular Dynamics. PLoS Comput. Biol. 2017, 13, e1005659. [Google Scholar] [CrossRef] [PubMed]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins Struct. Funct. Bioinform. 2006, 65, 712–725. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Jiménez-Gutiérrez, D.E.; Moreno, E. Structural Modeling of Nanobodies: A Benchmark of State-of-the-Art Artificial Intelligence Programs. Molecules 2023, 28, 3991. https://doi.org/10.3390/molecules28103991
Valdés-Tresanco MS, Valdés-Tresanco ME, Jiménez-Gutiérrez DE, Moreno E. Structural Modeling of Nanobodies: A Benchmark of State-of-the-Art Artificial Intelligence Programs. Molecules. 2023; 28(10):3991. https://doi.org/10.3390/molecules28103991
Chicago/Turabian StyleValdés-Tresanco, Mario S., Mario E. Valdés-Tresanco, Daiver E. Jiménez-Gutiérrez, and Ernesto Moreno. 2023. "Structural Modeling of Nanobodies: A Benchmark of State-of-the-Art Artificial Intelligence Programs" Molecules 28, no. 10: 3991. https://doi.org/10.3390/molecules28103991
APA StyleValdés-Tresanco, M. S., Valdés-Tresanco, M. E., Jiménez-Gutiérrez, D. E., & Moreno, E. (2023). Structural Modeling of Nanobodies: A Benchmark of State-of-the-Art Artificial Intelligence Programs. Molecules, 28(10), 3991. https://doi.org/10.3390/molecules28103991





