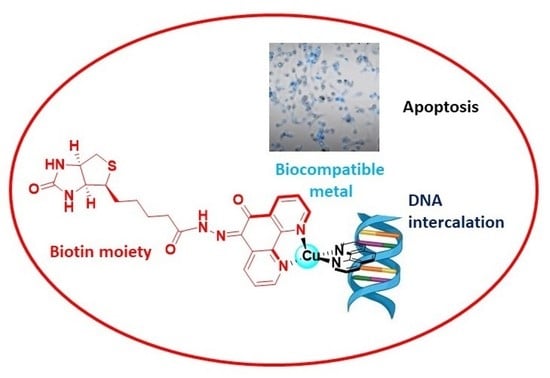
Novel Biotinylated Cu(II)-Phenanthroline Complexes: 2D and 3D Cytotoxic Activity and Mechanistic Insight
Abstract
:1. Introduction
2. Results and Discussion
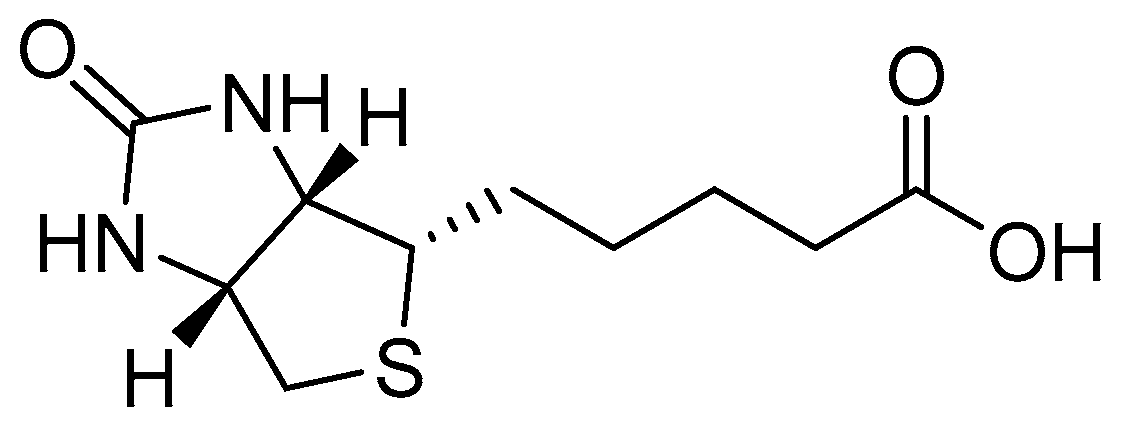
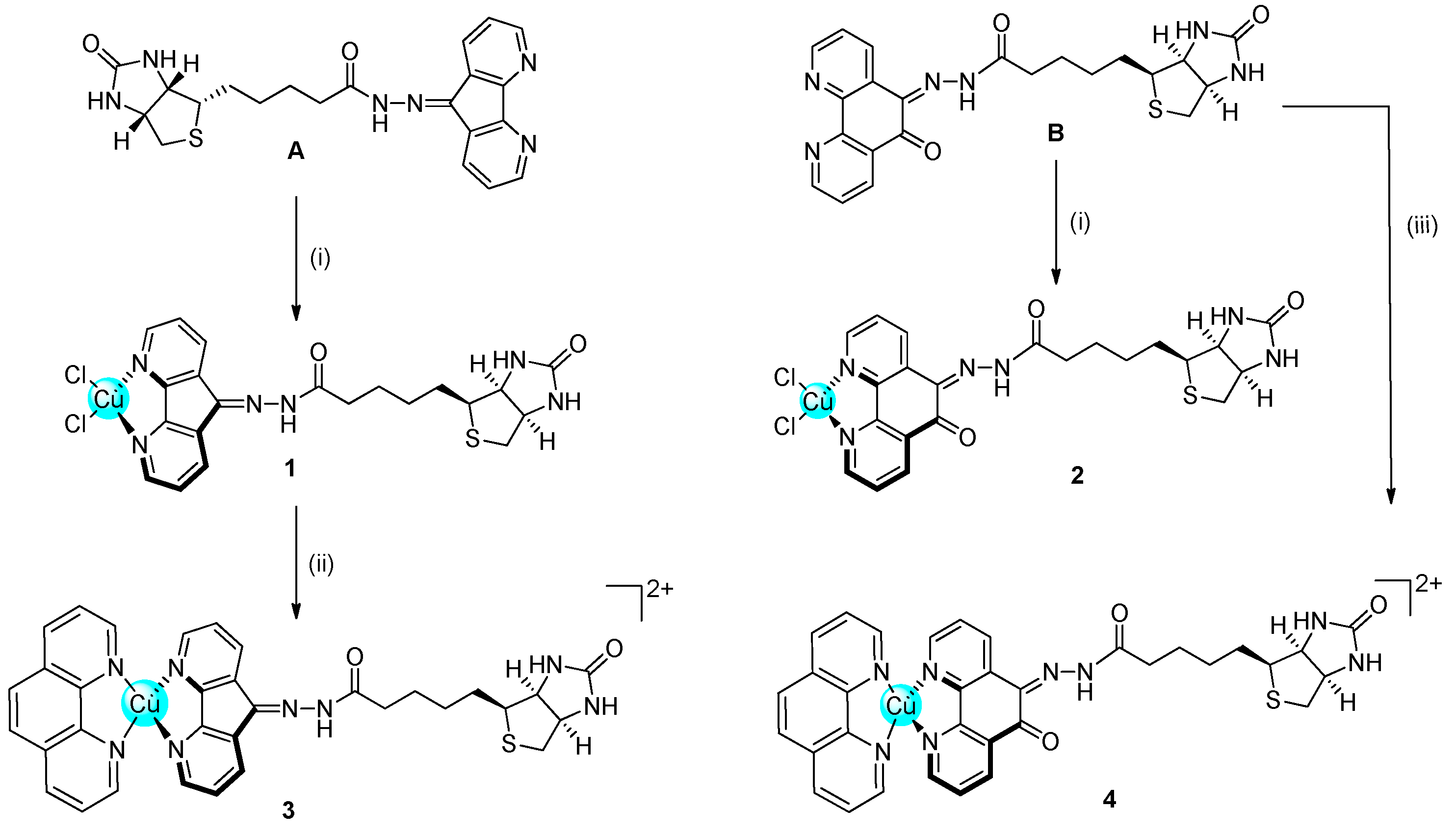
2.1. Synthesis and Characterisation
2.2. Cytotoxicity
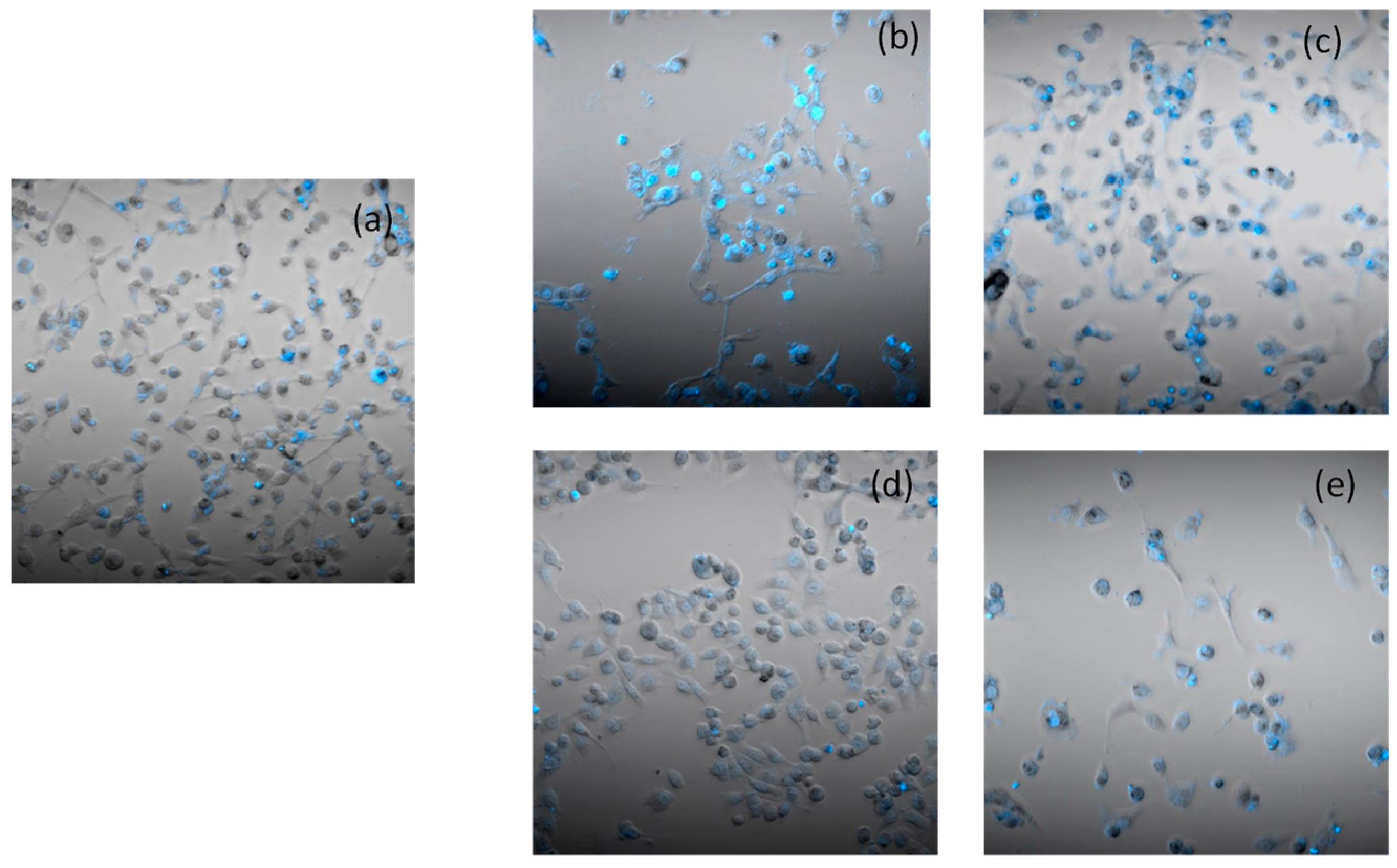
2.3. Cellular Uptake
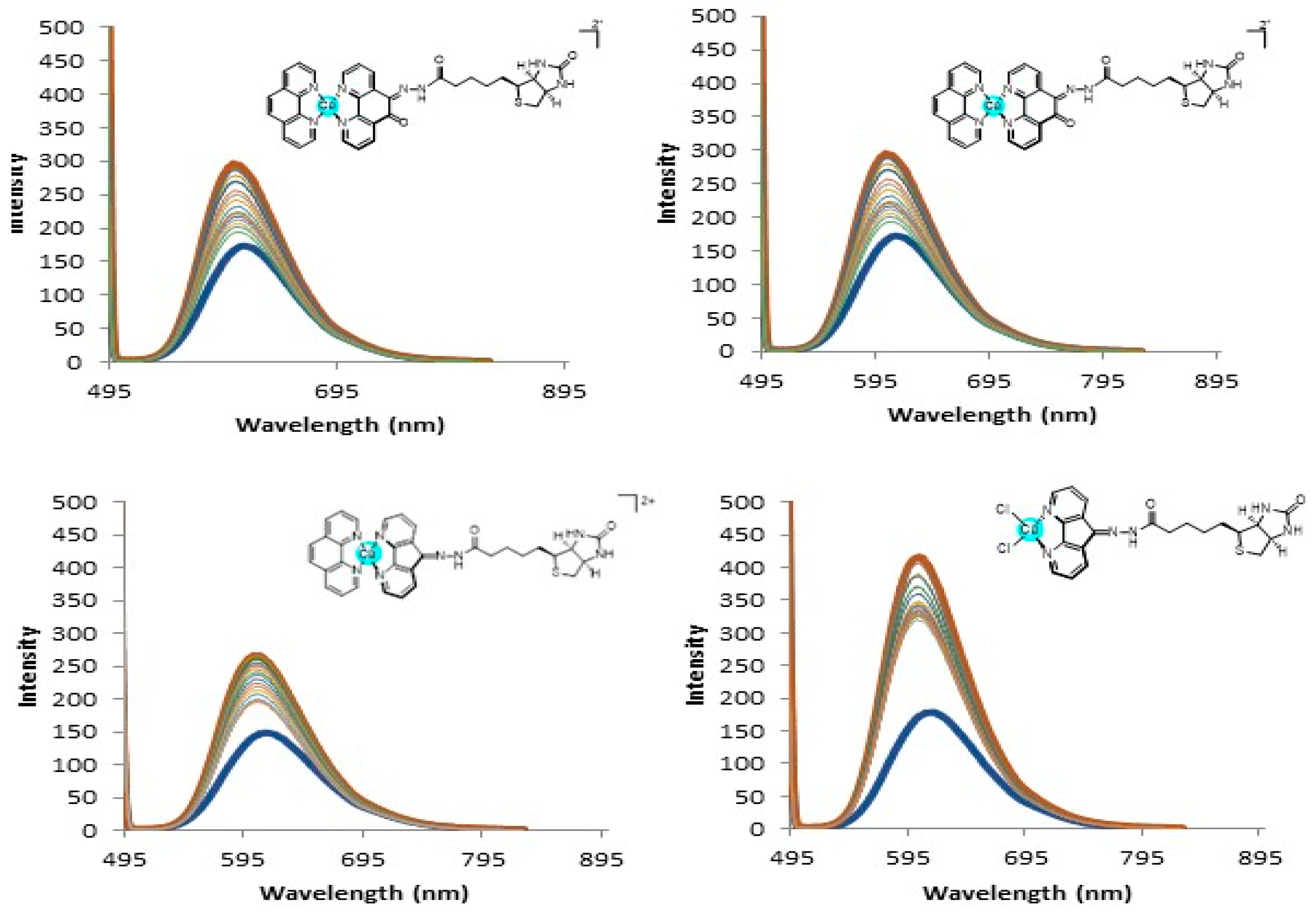
2.4. DNA Interaction and PDI Inhibition
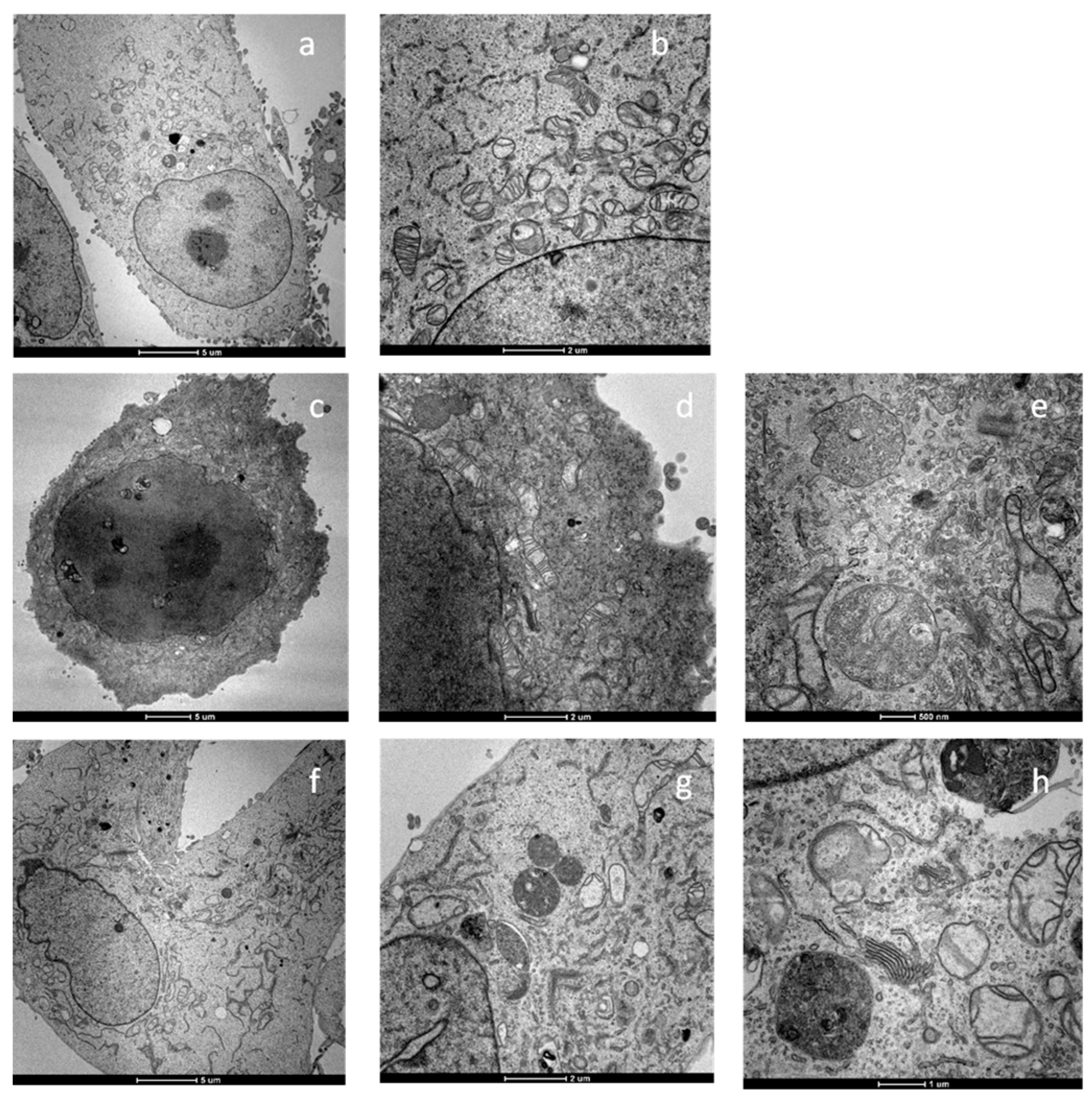
2.5. Apoptosis and Morphological Studies
3. Experimental Part
3.1. Material and Methods
3.1.1. Stability
3.1.2. Ethidium Bromide Displacement Assay
3.1.3. Experiments with Cultured Human Cancer Cells
3.1.4. Cell Cultures
3.1.5. MTT Test
3.1.6. Spheroid Cultures
3.1.7. APH Assay
3.1.8. Copper Cellular Content
3.1.9. Comet Assay
3.1.10. Protein Disulfide Isomerase (PDI) Activity
3.1.11. Transmission Electron Microscopy (TEM) Analyses
3.1.12. Cell Death Induction
3.1.13. Statistical Analysis
3.2. Synthesis and Characterisation
3.2.1. 1,10-Phenanthroline-5,6-dione (phendione)
3.2.2. Dafone (4,5-Diazafluoren-9-one)
3.2.3. Biotin Methyl Ester (1)
3.2.4. Biotin Hydrazide (2)
3.2.5. Biotin-Dafone (A)
3.2.6. Biotin-Phen (B)
3.2.7. [Cu(phen)(ONO2)2]
3.2.8. [Cu(dafone-biotin)Cl2] (1)
3.2.9. [Cu(phendione-biotin)Cl2] (2)
3.2.10. [Cu(phen)(dafone-biotin)](NO3)2 (3)
3.2.11. [Cu(phen)(phendione-biotin)](NO3)2 (4)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Yu, C.; Wang, Z.; Sun, Z.; Zhang, L.; Zhang, W.; Xu, Y.; Zhang, J.-J. Platinum-Based Combination Therapy: Molecular Rationale, Current Clinical Uses, and Future Perspectives. J. Med. Chem. 2020, 63, 13397–13412. [Google Scholar] [CrossRef] [PubMed]
- Wheate, N.J.; Walker, S.S.; Craig, E.G.; Oun, R. The status of platinum anticancer drugs in the clinic and in clinical trials. Dalton Trans. 2010, 39, 8113–8127. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, T.C.; Suntharalingam, K.; Lippard, S.J. The Next Generation of Platinum Drugs: Targeted Pt(II) Agents, Nanoparticle Delivery, and Pt(IV) Prodrugs. Chem. Rev. 2016, 116, 3436–3486. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.V.; Hambley, T.W. Platinum Drug Distribution in Cancer Cells and Tumors. Chem. Rev. 2009, 109, 4911–4920. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhu, C.; He, Y.; Zhang, Z.; Zhou, W.; Muhammad, N.; Guo, Y.; Wang, X.; Guo, Z. Restraining Cancer Cells by Dual Metabolic Inhibition with a Mitochondrion-Targeted Platinum(II) Complex. Angew. Chem. Int. Ed. 2019, 58, 4638–4643. [Google Scholar] [CrossRef]
- Huang, K.-B.; Wang, F.-Y.; Feng, H.-W.; Luo, H.; Long, Y.; Zou, T.; Chan, A.S.C.; Liu, R.; Zou, H.; Chen, Z.F.; et al. An aminophosphonate ester ligand-containing platinum(ii) complex induces potent immunogenic cell death in vitro and elicits effective anti-tumour immune responses in vivo. Chem. Commun. 2019, 55, 13066–13069. [Google Scholar] [CrossRef]
- Harper, B.W.; Krause-Heuer, A.M.; Grant, M.P.; Manohar, M.; Garbutcheon-Singh, K.B.; Aldrich-Wright, J.R. Advances in Platinum Chemotherapeutics. Chem. Eur. J. 2010, 16, 7064–7077. [Google Scholar] [CrossRef] [PubMed]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in copper complexes as anticancer agents. Chem. Rev. 2014, 114, 815–862. [Google Scholar] [CrossRef] [PubMed]
- Maiti, S.; Paira, P. Biotin conjugated organic molecules and proteins for cancer therapy: A review. Eur. J. Med. Chem. 2018, 45, 206–223. [Google Scholar] [CrossRef]
- Russell-Jones, G.; McTavish, K.; McEwan, J.; Rice, J.; Nowotnik, D. Vitamin-mediated targeting as a potential mechanism to increase drug uptake by tumours. J. Inorg. Biochem. 2004, 98, 1625–1633. [Google Scholar] [CrossRef]
- Lo, K.K.-W.; Hiu, W.-K.; Chung, C.-K.; Tsang, K.H.-K.; Lee, T.K.-M.; Ng, D.C.-M. Luminescent Transition Metal Polypyridine Biotin Complexes. J. Chin. Chem. Soc. 2006, 53, 53–65. [Google Scholar] [CrossRef]
- Zhao, J.; Hua, W.; Xu, G.; Gou, S. Biotinylated platinum(IV) complexes designed to target cancer cells. J. Inorg. Biochem. 2017, 176, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Fang, L.; Hua, W.; Gou, S. Biotin-Pt (IV)-indomethacin hybrid: A targeting anticancer prodrug providing enhanced cancer cellular uptake and reversing cisplatin resistance. J. Inorg. Biochem. 2017, 175, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, N.; Sadia, N.; Zhu, C.; Luo, C.; Guo, Z.; Wang, X. Biotin-tagged platinum(iv) complexes as targeted cytostatic agents against breast cancer cells. Chem. Commun. 2017, 53, 9971–9974. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; He, W.; Song, Y.; Zhao, J.; Song, Z.; Shan, Y.; Hua, W.; Sun, Y. Multifunctional platinum(iv) complex bearing HDAC inhibitor and biotin moiety exhibits prominent cytotoxicity and tumor-targeting ability. Dalton Trans. 2022, 51, 7343–7351. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.; Gautam, S.; Ramu, V.; Kondaiah, P.; Chakravarty, A.R. Photocytotoxic cancer cell-targeting platinum(II) complexes of glucose-appended curcumin and biotinylated 1,10-phenanthroline. Dalton Trans. 2019, 48, 17556–17565. [Google Scholar] [CrossRef] [PubMed]
- Marloye, M.; Inam, H.; Moore, C.J.; Debaille, V.; Pritchard, J.R.; Gelbcke, M.; Meyer, F.; Dufrasne, F.; Berger, G. Synthesis, structure and anticancer properties of new biotin- and morpholine-functionalized ruthenium and osmium half-sandwich complexes. J. Biolog. Inorg. Chem. 2021, 26, 535–549. [Google Scholar] [CrossRef]
- Zhang, K.Y.; Lo, K.K.-W. Synthesis, Properties, and Live-Cell Imaging Studies of Luminescent Cyclometalated Iridium(III) Polypyridine Complexes Containing Two or Three Biotin Pendants. Inorg. Chem. 2009, 48, 6011–6025. [Google Scholar] [CrossRef]
- Leung, C.-H.; Zhong, H.-J.; Chan, D.S.-H.; Ma, D.-L. Bioactive iridium and rhodium complexes as therapeutic agents. Coord. Chem. Rev. 2013, 257, 1764–1776. [Google Scholar] [CrossRef]
- Kallus, S.; Uhlik, L.; Van Schoonhoven, S.; Pelivan, K.; Berger, W.; Enyedy, E.A.; Hofman, T.; Heffeter, P.; Kowol, C.R.; Keppler, B.K. Synthesis and biological evaluation of biotin-conjugated anticancer thiosemicarbazones and their iron(III) and copper(II) complexes. J. Inorg. Biochem. 2019, 190, 85–97. [Google Scholar] [CrossRef]
- Graham, D.R.; Marshall, L.E.; Reich, K.A.; Sigman, D.S. Cleavage of DNA by coordination complexes. Superoxide formation in the oxidation of 1,10-phenanthroline-cuprous complexes by oxygen—Relevance to DNA-cleavage reaction. J. Am. Chem. Soc. 1980, 102, 5419–5542. Available online: https://pubs.acs.org/doi/pdf/10.1021/ja00536a063 (accessed on 1 July 1980). [CrossRef]
- Musib, D.; Upadhyay, A.; Pal, M.; Kausar Raza, M.D.; Saha, I.; Kunwar, A.; Roy, M. Red light-activable biotinylated copper(II) complex-functionalized gold nanocomposite (Biotin-Cu@AuNP) towards targeted photodynamic therapy. J. Inorg. Biochem. 2023, 243, 112183. [Google Scholar] [CrossRef] [PubMed]
- Ng, N.S.; Laverett, P.; Hibbs, D.E.; Yang, Q.; Bulanadi, J.C.; Wu, M.J.; Aldrich-Wright, J.R. The antimicrobial properties of some copper(ii) and platinum(ii) 1,10-phenanthroline complexes. Dalton Trans. 2013, 42, 3196–3209. [Google Scholar] [CrossRef]
- Vadlapudi, A.D.; Vadlapatla, R.K.; Mitra, A.K. Sodium Dependent Multivitamin Transporter (SMVT): A Potential Target for Drug Delivery. Curr. Drug Targets 2012, 13, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Barrett, S.; De Franco, M.; Kellett, A.; Dempsey, E.; Marzano, C.; Erxleben, A.; Gandin, V.; Montagner, D. Anticancer activity, DNA binding and cell mechanistic studies of estrogen-functionalised Cu(II) complexes. J. Biol. Inorg. Chem. 2020, 25, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Molphy, Z.; McKee, V.; Kellett, A. Copper bis-Dipyridoquinoxaline Is a Potent DNA Intercalator that Induces Superoxide-Mediated Cleavage via the Minor Groove. Molecules 2019, 24, 4301. [Google Scholar] [CrossRef]
- Pellei, M.; Santini, C.; Bagnarelli, L.; Battocchio, C.; Lucci, G.; Venditti, I.; Meneghini, C.; Amatori, S.; Sgarbossa, P.; Marzano, C.; et al. Exploring the Antitumor Potential of Copper Complexes Based on Ester Derivatives of Bis(pyrazol-1-yl)acetate Ligands. Int. J. Mol. Sci. 2022, 23, 9397. [Google Scholar] [CrossRef]
- Menon, S.; Rajasekharan, M.V. Bis chelate Cu(II) complexes of dafone—Synthesis, structural, EPR and optical spectral studies. Polyhedron 1998, 17, 2463–2476. [Google Scholar] [CrossRef]
- Molphy, Z.; Prisecaru, A.; Slator, C.; Barron, N.; McCann, M.; Colleran, J.; Chandran, D.; Gathergood, N.; Kellett, A. Copper Phenanthrene Oxidative Chemical Nucleases. Inorg. Chem. 2014, 53, 5392–5404. [Google Scholar] [CrossRef]
- Barrett, S.; Delaney, S.; Kavanagh, K.; Montagner, D. Evaluation of in vitro and in vivo antibacterial activity of novel Cu(II)-steroid complexes. Inorg. Chim. Acta 2018, 479, 261–265. [Google Scholar] [CrossRef]
- Moynihan, E.; Bassi, G.; Ruffini, A.; Panseri, S.; Montesi, M.; Velasco-Torrijos, T.; Montagner, D. Click Pt(IV)-Carbohydrates Pro-Drugs for Treatment of Osteosarcoma. Front. Chem. 2021, 9, 795997. [Google Scholar] [CrossRef] [PubMed]
- Moynihan, E.; Panseri, S.; Bassi, G.; Rossi, A.; Campodoni, E.; Dempsey, E.; Montesi, M.; Velasco-Torrijos, T.; Montagner, D. Development of Novel Pt(IV)-Carbohydrate Derivatives as Targeted Anticancer Agents against Osteosarcoma. Int. J. Mol. Sci. 2023, 24, 6028. [Google Scholar] [CrossRef] [PubMed]
- Boger, D.L.; Fink, B.E.; Brunette, S.R.; Tse, W.C.; Hedrick, M.P. A simple, high-resolution method for establishing DNA binding affinity and sequence selectivity. J. Am. Chem. Soc. 2001, 123, 5878–5891. [Google Scholar] [CrossRef] [PubMed]
- Dall’Acqua, S.; Peron, G.; Ferrari, S.; Gandin, V.; Bramucci, M.; Quassinti, L.; Mártonfi, P.; Maggi, F. Phytochemical investigations and antiproliferative secondary metabolites from Thymus alternans growing in Slovakia. Pharm. Biol. 2017, 55, 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Harper, B.W.J.; Petruzzella, E.; Sirota, R.; Faccioli, F.F.; Aldrich-Wright, J.R.; Gandin, V.; Gibson, D. Synthesis, characterization and in vitro and in vivo anticancer activity of Pt(iv) derivatives of [Pt(1S,2S-DACH)(5,6-dimethyl-1,10-phenanthroline)]. Dalton Trans. 2017, 30, 7005–7019. [Google Scholar] [CrossRef]
- Doboszewski, B.; Groaz, E.; Herdewijn, P. Synthesis of phosphonoglycine backbone units for the development of phosphono peptide nucleic acids. Eur. J. Org. Chem. 2013, 22, 4804–4815. [Google Scholar] [CrossRef]
- Sittiwong, W.; Cordonier, E.L.; Zampleni, J.; Dussault, P.H. β-Keto and β-hydroxyphosphonate analogs of biotin-5’-AMP are inhibitors of holocarboxylase synthetase. Bioorg. Med. Chem. Lett. 2014, 24, 5568–5571. [Google Scholar] [CrossRef]
- Murar, C.E.; Thuaud, F.; Bode, J.W. KAHA Ligations That Form Aspartyl Aldehyde Residues as Synthetic Handles for Protein Modification and Purification. J. Am. Chem. Soc. 2014, 136, 18140–18148. [Google Scholar] [CrossRef] [PubMed]


| Complex | PSN-1 | HCT-15 | 2008 | A431 | MDA-MB-231 | U-1285 |
|---|---|---|---|---|---|---|
| 1 | 0.6 ± 0.2 | 0.4 ± 0.1 | 4.5 ± 0.6 | 7.2 ± 0.6 | 2.1 ± 0.4 | 5.9 ± 0.5 |
| 2 | 0.4 ± 0.1 | 0.4 ± 0.1 | 1.1 ± 0.3 | 1.2 ± 0.5 | 1.4 ± 0.1 | 3.8 ± 0.5 |
| 3 | 0.7 ± 0.2 | 1.8 ± 0.2 | 3.3 ± 0.1 | 0.9 ± 0.2 | 1.4 ± 0.4 | 3.0 ± 0.3 |
| 4 | 0.6 ± 0.2 | 0.4 ± 0.02 | 0.6 ± 0.2 | 0.3 ± 0.03 | 0.6 ± 0.1 | 2.0 ± 0.6 |
| A | >50 | >50 | >50 | >50 | >50 | >50 |
| B | N/A | N/A | 21.48 ± 0.72 | N/A | N/A | 28.13 ± 2.77 |
| Cisplatin | 12.1 ± 2.9 | 13.9 ± 1.7 | 2.2 ± 1.4 | 2.1 ± 0.9 | 30.5 ± 2.6 | 2.1 ± 0.9 |
| Complex | 2008 | C13* | R.F. |
|---|---|---|---|
| 1 | 4.5 ± 0.6 | 2.0 ± 0.8 | 0.4 |
| 2 | 1.1 ± 0.3 | 1.1 ± 0.4 | 1.0 |
| 3 | 3.3 ± 0.1 | 2.5 ± 0.2 | 0.7 |
| 4 | 0.6 ± 0.2 | 1.1 ± 0.3 | 1.9 |
| Cisplatin | 2.2 ± 1.4 | 24.1 ± 3.0 | 11.1 |
| Complex | CHO | S.I. |
|---|---|---|
| 1 | 13.1 ± 0.9 | 3.9 |
| 2 | 5.2 ± 0.3 | 3.7 |
| 3 | 2.1 ± 0.8 | 1.1 |
| 4 | 1.8 ± 0.7 | 2.3 |
| Cisplatin | 24.1 ± 3.0 | 11.1 |
| Complex | PSN-1 |
|---|---|
| 2 | 1.3 ± 0.5 |
| 4 | 6.9 ± 1.3 |
| Cisplatin | 52.6 ± 3.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrett, S.; De Franco, M.; Donati, C.; Marzano, C.; Gandin, V.; Montagner, D. Novel Biotinylated Cu(II)-Phenanthroline Complexes: 2D and 3D Cytotoxic Activity and Mechanistic Insight. Molecules 2023, 28, 4112. https://doi.org/10.3390/molecules28104112
Barrett S, De Franco M, Donati C, Marzano C, Gandin V, Montagner D. Novel Biotinylated Cu(II)-Phenanthroline Complexes: 2D and 3D Cytotoxic Activity and Mechanistic Insight. Molecules. 2023; 28(10):4112. https://doi.org/10.3390/molecules28104112
Chicago/Turabian StyleBarrett, Stephen, Michele De Franco, Chiara Donati, Cristina Marzano, Valentina Gandin, and Diego Montagner. 2023. "Novel Biotinylated Cu(II)-Phenanthroline Complexes: 2D and 3D Cytotoxic Activity and Mechanistic Insight" Molecules 28, no. 10: 4112. https://doi.org/10.3390/molecules28104112
APA StyleBarrett, S., De Franco, M., Donati, C., Marzano, C., Gandin, V., & Montagner, D. (2023). Novel Biotinylated Cu(II)-Phenanthroline Complexes: 2D and 3D Cytotoxic Activity and Mechanistic Insight. Molecules, 28(10), 4112. https://doi.org/10.3390/molecules28104112



.jpg)