Honey Quality Control: Review of Methodologies for Determining Entomological Origin
Abstract
1. Introduction
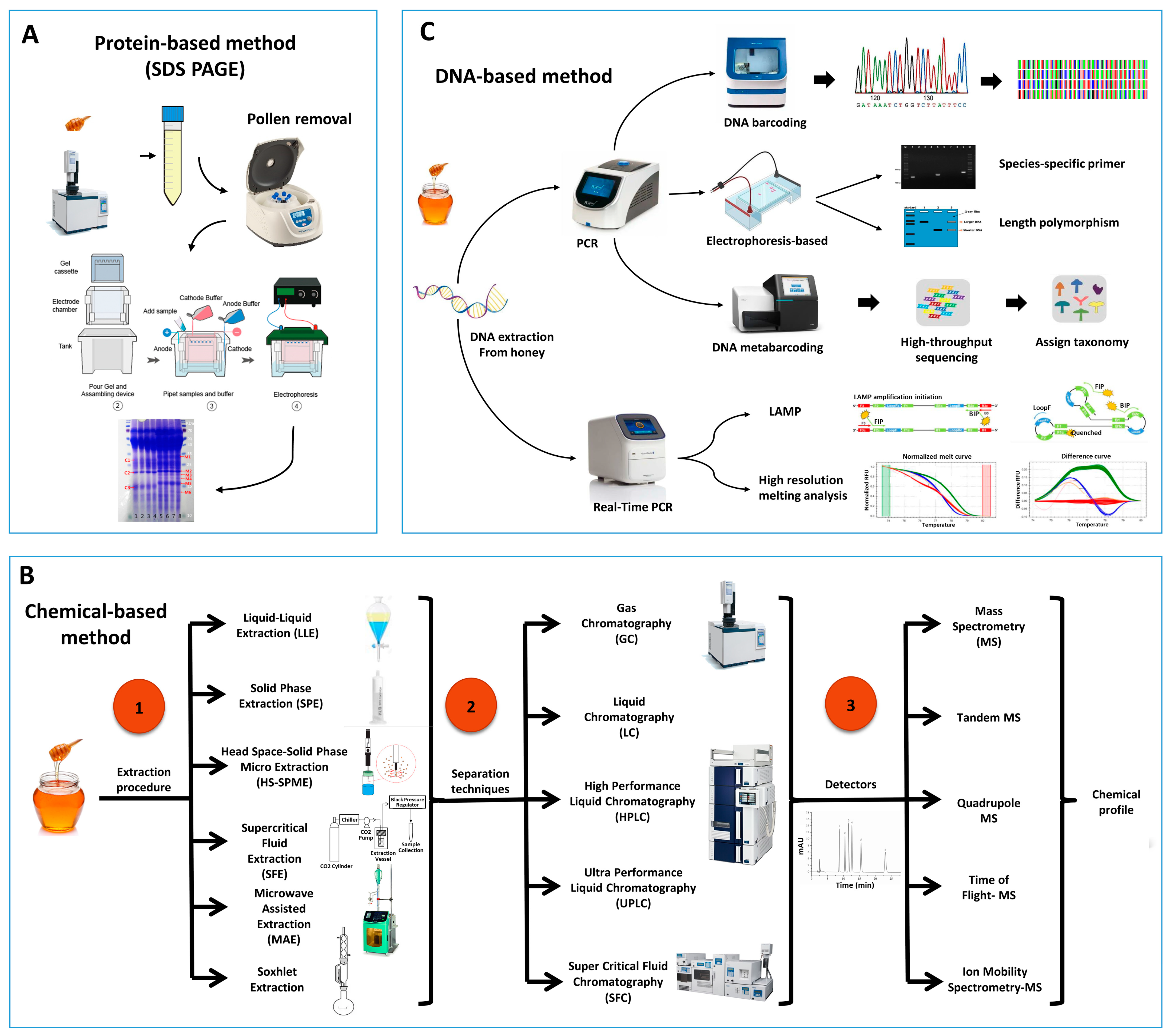
2. Identification Methods of Entomological Origin of Honey
2.1. Protein Based Method
2.2. Physicochemical Properties and Bioactive Characterization of Honey to Determine Entomological Origin
2.3. Chemical Profiling to Authenticate Entomological Origin of Honey
2.4. DNA-Based Method
2.4.1. DNA Barcoding
2.4.2. Usage of Species-Specific Primers
2.4.3. Real-Time PCR-Based Methods
High-Resolution Melting Analysis
Loop-Mediated Isothermal Amplification (LAMP) Technology
2.4.4. Length Polymorphism of PCR Product
2.4.5. DNA Metabarcoding Approach
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Won, S.-R.; Lee, D.-C.; Ko, S.H.; Kim, J.-W.; Rhee, H.-I. Honey major protein characterization and its application to adulteration detection. Food Res. Int. 2008, 41, 952–956. [Google Scholar] [CrossRef]
- Pavlova, T.; Stamatovska, V.; Kalevska, T.; Dimov, I.; Nakov, G. Quality characteristics of honey: A review. Proc. Univ. RUSE 2018, 57, 31–37. [Google Scholar]
- Guler, A.; Kocaokutgen, H.; Garipoglu, A.V.; Onder, H.; Ekinci, D.; Biyik, S. Detection of adulterated honey produced by honeybee (Apis mellifera L.) colonies fed with different levels of commercial industrial sugar (C3 and C4 plants) syrups by the carbon isotope ratio analysis. Food Chem. 2014, 155, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Lee, D.G.; Jung, C. A comparative study of the two different methods IRMS and CRDS for estimation of δ13C (%) of Honey Samples. J. Apic. 2018, 33, 99–105. [Google Scholar] [CrossRef]
- Anklam, E. A review of the analytical methods to determine the geographical and botanical origin of honey. Food Chem. 1998, 63, 549–562. [Google Scholar] [CrossRef]
- Aries, E.; Burton, J.; Carrasco, L.; De Rudder, O.; Maquet, A. Scientific Support to the Implementation of a Coordinated Control Plan with a View to Establishing the Prevalence of Fraudulent Practices in the Marketing of Honey. NSANTE/2015; JRC Technical Report 2016, JRC104749. Available online: https://ec.europa.eu/food/sites/food/files/safety/docs/oc_control-progs_honey_jrc-tech-report_2016.pdf (accessed on 14 December 2018).
- Wu, L.; Du, B.; Vander Heyden, Y.; Chen, L.; Zhao, L.; Wang, M.; Xue, X. Recent advancements in detecting sugar-based adulterants in honey–A challenge. TrAC Trends Anal. Chem. 2017, 86, 25–38. [Google Scholar] [CrossRef]
- Kim, C.-K.; Lee, D.-C.; Choi, S.-H. Detection of Korean Native Honey and European Honey by Using Duplex Polymerase Chain Reaction and Immunochromatographic Assay. Korean J. Food Sci. Anim. Resour. 2017, 37, 599. [Google Scholar] [CrossRef]
- Oroian, M.; Ropciuc, S.; Paduret, S.; Todosi, E. Rheological analysis of honeydew honey adulterated with glucose, fructose, inverted sugar, hydrolysed inulin syrup and malt wort. LWT 2018, 95, 1–8. [Google Scholar] [CrossRef]
- Zhang, Y.-Z.; Wang, S.; Chen, Y.-F.; Wu, Y.-Q.; Tian, J.; Si, J.-J.; Zhang, C.-P.; Zheng, H.-Q.; Hu, F.-L. Authentication of Apis cerana honey and Apis mellifear honey based on major royal jelly protein 2 gene. Molecules 2019, 24, 289. [Google Scholar] [CrossRef]
- Soares, S.; Grazina, L.; Mafra, I.; Costa, J.; Pinto, M.A.; Duc, H.P.; Oliveira, M.B.P.; Amaral, J.S. Novel diagnostic tools for Asian (Apis cerana) and European (Apis mellifera) honey authentication. Food Res. Int. 2018, 105, 686–693. [Google Scholar] [CrossRef]
- Ruttner, F. Biogeography and Taxonomy of Honeybees; Springer: Berlin/Heidelberg, Germany, 1988. [Google Scholar]
- Hepburn, C.; Radloff, S.E. Honeybees of Asia; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Anderson, D.L.; Annand, N.; Lacey, M.; Ete, S. Control of Asian Honey Bees in Solomon Islands; Australian Centre for International Agricultural Research (ACIAR): Canberra, Australia, 2012. [Google Scholar]
- Barry, S.; Cook, D.; Duthie, R.; Clifford, D.; Anderson, D. Future Surveillance Needs for Honeybee Biosecurity; Rural Industries Research and Development Corporation: Canberra, Australia, 2010. [Google Scholar]
- Shield, J. The Asian Honey Bee: Report of an Incursion in Cairns 2007: Technical Aspects of the Response; Department of Primary Industries and Fisheries: Brisbane, Australia, 2007; pp. 1–106. [Google Scholar]
- Sammataro, D.; Avitabile, A. The Beekeeper’s Handbook, 3rd ed.; Comstock Publishing Associates: Ithaca, NY, USA, 1998. [Google Scholar]
- Winston, M.; Dropkin, J.; Taylor, O. Demography and life history characteristics of two honey bee races (Apis mellifera). Oecologia 1981, 48, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Partap, L.; Verma, L. Asian bees and beekeeping: Issues and initiatives. In Proceedings of the 4th Asian Apiculture Association International Conference, Kathmandu, Nepal, 23–28 March 1998; pp. 3–14. [Google Scholar]
- Verma, L. Beekeeping in Integrated Mountain Development; Oxford & IBH Publishing: New Delhi, India, 1991; pp. 1–237. [Google Scholar]
- Garnery, L.; Cornuet, J.M.; Solignac, M. Evolutionary history of the honey bee Apis mellifera inferred from mitochondrial DNA analysis. Mol. Ecol. 1992, 1, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.C.; Lee, S.Y.; Cha, S.H.; Choi, Y.S.; Rhee, H.I. Discrimination of native bee-honey and foreign bee-honey by SDS–PAGE. Korean J. Food Sci. 1998, 30, 1–5. [Google Scholar]
- Ramón-Sierra, J.M.; Ruiz-Ruiz, J.C.; Ortiz-Vázquez, E. Electrophoresis characterization of protein as a method to establish the entomological origin of stingless bee honeys. Food Chem. 2015, 138, 43–48. [Google Scholar] [CrossRef]
- Chua, L.S.; Lee, J.Y.; Chan, G.F. Honey protein extraction and determination by mass spectrometry. Anal. Bioanal. Chem. 2013, 405, 3063–3074. [Google Scholar] [CrossRef]
- Erban, T.; Shcherbachenko, E.; Talacko, P.; Harant, K. A single honey proteome dataset for identifying adulteration by foreign analyses and mining various protein markers natural to honey. J. Proteom. 2021, 239, 104157. [Google Scholar] [CrossRef]
- Schmitzová, J.; Klaudiny, J.; Albert, Š.; Schröder, W.; Schreckengost, W.; Hanes, J.; Júdová, J.; Simúth, J. A family of major royal jelly proteins of the honeybee Apis mellifera L. Cell. Mol. Life Sci. 1998, 54, 1020–1030. [Google Scholar] [CrossRef]
- Albert, Š.; Klaudiny, J. The MRJP/YELLOW protein family of Apis mellifera: Identification of new members in the EST library. J. Insect Physiol. 2004, 50, 51–59. [Google Scholar] [CrossRef]
- Drapeau, M.D.; Albert, Š.; Kucharski, R.; Prusko, C.; Maleszka, R. Evolution of the Yellow/Major Royal Jelly Protein family and the emergence of social behavior in honey bees. Genome Res. 2006, 16, 1385–1394. [Google Scholar] [CrossRef]
- Di Girolamo, F.; D’Amato, A.; Righetti, P.G. Assessment of the floral origin of honey via proteomic tools. J. Proteom. 2012, 75, 3688–3693. [Google Scholar] [CrossRef]
- Zhang, Y.-Z.; Chen, Y.-F.; Wu, Y.-Q.; Si, J.-J.; Zhang, C.-P.; Zheng, H.-Q.; Hu, F.-L. Discrimination of the entomological origin of honey according to the secretions of the bee (Apis cerana or Apis mellifera). Food Res. Int. 2019, 116, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Zannat, R.; Rahman, M.M.; Joaty, J.Y.; Miah, M.R.U.; Al Mamun, M.A.; Hassan, J. Towards authentication of entomological origin of honey in Bangladesh through molecular and biochemical approaches. J. Agric. Food Res. 2023, 12, 100543. [Google Scholar] [CrossRef]
- Buawangpong, N.; Burgett, M. Capped honey moisture content from four honey bee species; Apis dorsata F., Apis florea F., Apis cerana F. and Apis mellifera L. (Hymenoptera: Apidae) in Northern Thailand. J. Apic. 2019, 34, 157–160. [Google Scholar] [CrossRef]
- Rodríguez-Malavera, A.J.; Rasmussenb, C.; Gutiérrezc, M.G.; Gild, F.; Nievesd, B.; Vitc, P. Properties of honey from ten species of Peruvian stingless bees. Nat. Prod. Commun. 2009, 4, 1221–1226. [Google Scholar] [CrossRef]
- Kek, S.P.; Chin, N.L.; Tan, S.W.; Yusof, Y.A.; Chua, L.S. Classification of honey from its bee origin via chemical profiles and mineral content. Food Anal. Methods 2017, 10, 19–30. [Google Scholar] [CrossRef]
- Ávila, S.; Beux, M.R.; Ribani, R.H.; Zambiazi, R.C. Stingless bee honey: Quality parameters, bioactive compounds, health-promotion properties and modification detection strategies. Trends Food Sci. Technol. 2018, 81, 37–50. [Google Scholar] [CrossRef]
- Wu, J.; Han, B.; Zhao, S.; Zhong, Y.; Han, W.; Gao, J. Bioactive characterization of multifloral honeys from Apis cerana, Apis dorsata, and Lepidoptrigona flavibasis. Food Res. Int. 2022, 161, 111808. [Google Scholar] [CrossRef]
- Kek, S.P.; Chin, N.L.; Yusof, Y.A.; Tan, S.W.; Chua, L.S. Classification of entomological origin of honey based on its physicochemical and antioxidant properties. Int. J. Food Prop. 2017, 20, S2723–S2738. [Google Scholar] [CrossRef]
- Beitlich, N.; Koelling-Speer, I.; Oelschlaegel, S.; Speer, K. Differentiation of Manuka Honey from Kanuka Honey and from Jelly Bush Honey using HS-SPME-GC/MS and UHPLC-PDA-MS/MS. J. Agric. Food Chem. 2014, 62, 6435–6444. [Google Scholar] [CrossRef]
- Biluca, F.C.; Braghini, F.; Gonzaga, L.V.; Costa, A.C.O.; Fett, R. Physico-chemical profiles, minerals and bioactive compounds of stingless bee honey (Meliponinae). J. Food Compos. Anal. 2016, 50, 61–69. [Google Scholar] [CrossRef]
- Alissandrakis, E.; Tarantilis, P.A.; Harizanis, P.C.; Polissiou, M. Comparison of the volatile composition in thyme honeys from several origins in Greece. J. Agric. Food Chem. 2007, 55, 8152–8157. [Google Scholar] [CrossRef] [PubMed]
- da Costa, A.C.V.; Sousa, J.M.B.; Bezerra, T.K.A.; da Silva, F.L.H.; Pastore, G.M.; da Silva, M.A.A.P.; Madruga, M.S. Volatile profile of monofloral honeys produced in Brazilian semiarid region by stingless bees and key volatile compounds. LWT-Food Sci. Technol. 2018, 94, 198–207. [Google Scholar] [CrossRef]
- Wang, X.; Rogers, K.M.; Li, Y.; Yang, S.; Chen, L.; Zhou, J. Untargeted and targeted discrimination of honey collected by Apis cerana and Apis mellifera based on volatiles using HS-GC-IMS and HS-SPME-GC-MS. J. Agric. Food Chem. 2019, 67, 12144–12152. [Google Scholar] [CrossRef] [PubMed]
- Sharin, S.N.; Sani, M.S.A.; Jaafar, M.A.; Yuswan, M.Y.; Kassim, N.K.; Manaf, Y.N.; Wasoh, H.; Zaki, N.N.M.; Hashim, A.M. Discrimination of Malaysian stingless bee honey from different entomological origins based on physicochemical properties and volatile compound profiles using chemometrics and machine learning. Food Chem. 2021, 346, 128654. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Liang, Y.; Yi, S.; Qiu, S.; Liu, M.; Ning, F.; Luo, L. Honeycomb, a new food resource with health care function: The difference of volatile compounds found in Apis cerana and Apis mellifera honeycombs. Foods 2022, 11, 3204. [Google Scholar] [CrossRef]
- Xu, J.; Zhou, Q.; Lin, S.; Luo, L. Study on the composition of beeswax of Apis cerana cerana and Apis mellifera ligustica. Chromatography 1989, 7, 175–176. [Google Scholar]
- Wang, X.; Li, Y.; Chen, L.; Zhou, J. Analytical strategies for LC-MS based untargeted and targeted metabolomics approaches reveal the entomological origins of honey. J. Agric. Food Chem. 2022, 70, 1358–1366. [Google Scholar] [CrossRef]
- Zuccato, V.; Finotello, C.; Menegazzo, I.; Peccolo, G.; Schievano, E. Entomological authentication of stingless bee honey by 1H NMR-based metabolomics approach. Food Control 2017, 82, 145–153. [Google Scholar] [CrossRef]
- Schievano, E.; Stocchero, M.; Zuccato, V.; Conti, I.; Piana, L. NMR assessment of European acacia honey origin and composition of EU-blend based on geographical floral markers. Food Chem. 2019, 288, 96–101. [Google Scholar] [CrossRef]
- Razali, M.T.A.; Zainal, Z.A.; Maulidiani, M.; Shaari, K.; Zamri, Z.; Idrus, M.Z.M.; Khatib, A.; Abas, F.; Ling, Y.S.; Rui, L.L.; et al. Classification of raw stingless bee honeys by bee species origins using the NMR- and LC-MS-based metabolomics approach. Molecules 2018, 23, 216. [Google Scholar] [CrossRef]
- Grabato, J.R.; Pilario, K.E.; Micor, J.R.L.; Mojica, E.-R.E. Geographical and entomological differentiation pf Phillipine honey by multivariate analysis of FTIR spectra. J. Food Compos. Anal. 2022, 114, 104853. [Google Scholar] [CrossRef]
- Sahlan, M.; Karwita, S.; Gozan, M.; Hermansyah, H.; Yohda, M.; Yoo, Y.J.; Pratami, D.K. Identification and classification of honey’s authenticity by attenuated total reflectance Fourier-transform infrared spectroscopy and chemometric method. Vet. World 2019, 12, 1304–1310. [Google Scholar] [CrossRef] [PubMed]
- Bottero, M.T.; Dalmasso, A. Animal species identification in food products: Evolution of biomolecular methods. Vet. J. 2011, 190, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kumar, R.R.; Sharma, B.D.; Gokulakrishnan, P.; Mendiratta, S.K.; Sharma, D. Identification of species origin of meat and meat products on the DNA basis: A review. Crit. Rev. Food Sci. Nutr. 2015, 55, 1340–1351. [Google Scholar] [CrossRef]
- Amaral, J.; Meira, L.; Oliveira, M.; Mafra, I. Advances in authenticity testing for meat speciation. In Advances in Food Authenticity Testing; Elsevier: Amsterdam, The Netherlands, 2016; pp. 369–414. [Google Scholar]
- Willette, D.A.; Simmonds, S.E.; Cheng, S.H.; Esteves, S.; Kane, T.L.; Nuetzel, H.; Pilaud, N.; Rachmawati, R.; Barber, P.H. Using DNA barcoding to track seafood mislabeling in Los Angeles restaurants. Conserv. Biol. 2017, 31, 1076–1085. [Google Scholar] [CrossRef] [PubMed]
- Bruni, I.; Galimberti, A.; Caridi, L.; Scaccabarozzi, D.; De Mattia, F.; Casiraghi, M.; Labra, M. A DNA barcoding approach to identify plant species in multiflower honey. Food Chem. 2015, 170, 308–315. [Google Scholar] [CrossRef]
- Barcaccia, G.; Lucchin, M.; Cassandro, M. DNA Barcoding as a molecular tool to track down Mislabeling and food piracy. Diversity 2015, 8, 2. [Google Scholar] [CrossRef]
- Kek, S.P.; Chin, N.L.; Tan, S.W.; Yusof, Y.A.; Chua, L.S. Molecular identification of honey entomological origin based on bee mitochondrial 16S rRNA and COI gene sequences. Food Control 2017, 78, 150–159. [Google Scholar] [CrossRef]
- Schnell, I.B.; Fraser, M.; Willerslev, E.; Gilbert, M.T. Characterisation of insect and plant origins using DNA extracted from small volumes of bee honey. Arthropod-Plant Interact. 2010, 4, 107–116. [Google Scholar] [CrossRef]
- Utzeri, V.J.; Ribani, A.; Taurisano, V.; Fontanesi, L. Entomological authentication of honey based on a DNA method that distinguishes Apis mellifera mitochondrial C mitotypes: Application to honey produced by A. m. ligustica and A. m. carnica. Food Control. 2022, 134, 108713. [Google Scholar] [CrossRef]
- Prosser, S.W.; Hebert, P.D. Rapid identification of the botanical and entomological sources of honey using DNA metabarcoding. Food Chem. 2017, 214, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Raffiudin, R.; Shullia, N.I.; Febiriani, T.V.; Nisa, N.R.; Rahmadini, J.; Purwanto, H. Entomological origin detection of honey from Apis mellifera and A. cerana javana in Indonesia based on the Major Royal Jelly Protein 2 (mrjp2) gene. J. Apic. Res. 2023, 62, 330–333. [Google Scholar] [CrossRef]
- Mohamadzade Namin, S.; Yeasmin, F.; Choi, H.W.; Jung, C. DNA-Based Method for Traceability and Authentication of Apis cerana and A. dorsata Honey (Hymenoptera: Apidae), Using the NADH dehydrogenase 2 Gene. Foods 2022, 11, 928. [Google Scholar] [CrossRef] [PubMed]
- Moritz, C.; Dowling, T.E.; Brown, W.M. Evolution of animal mitochondrial DNA: Relevance for population biology and systematics. Ann. Rev. Ecol. Syst. 1987, 18, 269–292. [Google Scholar] [CrossRef]
- Song, S.; Pursell, Z.F.; Copeland, W.C.; Longley, M.J.; Kunkel, T.A.; Mathews, C.K. DNA precursor asymmetries in mammalian tissue mitochondria and possible contribution to mutagenesis through reduced replication fidelity. Proc. Natl. Acad. Sci. USA 2005, 102, 14. [Google Scholar] [CrossRef]
- Zink, R.M.; Barrowclough, G.E. Mitochondrial DNA under siege in avian phylogeography. Mol. Ecol. 2008, 17, 2107–2121. [Google Scholar] [CrossRef]
- Yi, S.; Ellsworth, D.L.; Li, W.H. Slow molecular clocks in old world monkeys, apes and humans. Mol. Biol. Evol. 2002, 19, 2191–2198. [Google Scholar] [CrossRef]
- McClellan, D.A. The codon-degeneracy model of molecular evolution. J. Mol. Evol. 2000, 50, 131–140. [Google Scholar] [CrossRef]
- Garritano, S.; Gemignani, F.; Voegele, C.; Nguyen-Dumont, T.; Calvez-Kelm, F.L.; De Silva, D.; Lesueur, F.; Landi, S.; Tavtigian, S.V. Determining the effectiveness of high resolution melting analysis for SNP genotyping and mutation scanning at the TP53 locus. BMC Genet. 2009, 10, 5. [Google Scholar] [CrossRef]
- Soares, S.; Grazina, L.; Mafra, I.; Costa, J.; Pinto, M.A.; Oliveira, M.B.P.O.; Amaral, J.S. Towards honey authentication: Differentiation of Apis mellifera subspecies in European honey based on mitochondrial DNA marker. Food Chem. 2019, 283, 294–301. [Google Scholar] [CrossRef]
- Honrado, M.; Lopes, A.R.; Pinto, M.A.; Amaral, J.S. A novel real-time PCR coupled with high resolution melting analysis as a simple and fast tool for the entomological authentication of honey by targeting Apis mellifera mitochondrial DNA. Food Res. Int. 2022, 161, 111761. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplifcation using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Foo, P.C.; Najian, A.B.N.; Muhamad, N.A.; Ahamad, M.; Mohamed, M.; Yean, C.Y.; Lim, B.H. Loop-mediated isothermal amplification (LAMP) reaction as viable PCR substitute for diagnostic applications: A comparative analysis study of LAMP, comventional PCR, nested PCR (nPCR0 and real-time PCR (qPCR) based on Entamoeba histolytica DNA derived from faecal sample. BMC Biotechnol. 2020, 20, 34. [Google Scholar]
- Gao, J.; Jin, X.; Gong, B.; Li, J.; Chen, A.; Tan, J.; Wang, J. Identification of insect sources of honey in China based on real-time fluorescent LAMP technology. J. Food Compos. Anal. 2023, 115, 104875. [Google Scholar] [CrossRef]
- Utzeri, V.J.; Ribani, A.; Fontanesi, L. Authentication of honey based on a DNA method to differentiate Apis mellifera subspecies: Application to Sicilian honey bee (A. m. siciliana) and Iberian honey bee (A. m. iberiensis) honeys. Food Control 2018, 91, 294–301. [Google Scholar] [CrossRef]
- Hawkins, J.; de Vere, N.; Griffith, A.; Ford, C.R. Using DNA metabarcoding to Identify the floral composition of honey: A new tool for investigating honey bee foraging preferences. PLoS ONE 2015, 10, e0134735. [Google Scholar] [CrossRef]
- de Vere, N.; Jones, L.E.; Gilmore, T.; Moscrop, J.; Lowe, A.; Smith, D.; Hegarty, M.J.; Creer, S.; Ford, C.R. Using DNA metabarcoding to investigate honey bee foraging reveals limited flower use despite high floral availability. Sci. Rep. 2017, 7, 42838. [Google Scholar] [CrossRef]
- Mohamadzade Namin, S.; Kim, M.-J.; Son, M.; Jung, C. Honey DNA metabarcoding revealed foraging resource partitioning between Korean native and introduced honey bees (Hymentoptera: Apidae). Sci. Rep. 2022, 12, 14394. [Google Scholar] [CrossRef]
- Bovo, S.; Utzeri, V.J.; Ribani, A.; Cabbri, R.; Fontanesi, L. Shortgun sequencing of honey DNA can describe honey bee derived environmental signatures and the honey bee hologenome complexity. Sci. Rep. 2020, 10, 9279. [Google Scholar] [CrossRef]
- Brudzynski, K.; Abubaker, K.; Miotto, D. Unraveling a mechanism of honey antibacterial action: Polyphenol/H2O2-induced oxidative effect on bacterial cell growth and on DNA degradation. Food Chem. 2012, 133, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Gryson, N. Effect of food processing on plant DNA degradation and PCR-based GMO analysis: A review. Anal. Bioanal. Chem. 2010, 396, 2003–2022. [Google Scholar] [CrossRef]
- Kek, S.P.; Chin, N.L.; Tan, S.W.; Yusof, Y.A.; Chua, L.S. Comparison of DNA extraction methods for entomological origin identification of honey using simple additive weighting method. Int. J. Food Sci. Technol. 2018, 53, 2490–2499. [Google Scholar] [CrossRef]
- Turci, M.; Sardaro, M.L.S.; Visioli, G.; Maestri, E.; Marmiroli, M.; Marmiroli, N. Valuation of DNA extraction procedures for traceability of various tomato products. Food Control 2010, 21, 143–149. [Google Scholar] [CrossRef]
- Hyatt, S. Asian Honey Bee (Apis cerana Javana) in Cairns, Far North Queensland; Report of Field Observations April 2007–September 2011; State of Queensland, Department of Employment, Economic Development and Innovation: Brisbane, Australia, 2012; pp. 1–24. [Google Scholar]
- Zheng, H.; Cao, L.; Huang, S.; Neumann, P.; Hu, F. Current Status of the Beekeeping Industry in China. In Asian Beekeeping in the 21st Century; Chantawannakul, P., Williams, G., Neumann, P., Eds.; Springer: Singapore, 2018. [Google Scholar]
- Bell, K.L.; Petit, R.A.; Cutler, A.; Dobbs, E.K.; Macpherson, J.M.; Read, T.D.; Burgess, K.S.; Brosi, B.J. Comparing whole-genome shotgun sequencing and DNA metabarcoding approaches for species identification and quantification of pollen species mixtures. Ecol. Evol. 2021, 11, 16082–16098. [Google Scholar] [CrossRef]
| Primer | Primr Sequence | Target Gene | Size of Amplicon (bp) | Method of Discrimination | Reference |
|---|---|---|---|---|---|
| COI-300F | GGATTTATTGTCTGAGCACATC | COI | 316 | DNA barcoding | Kek et al., [58] |
| COI-300R | TTGCGAATACTGCTCCTATT | ||||
| 16S-300F | GGACGATAAGACCCTATAGAA | 16S rRNA | 296–307 | ||
| 16S-300R | TTGTTAAAAGTCGAACAGAC | ||||
| AncientLepF2 | ATTRRWRATGATCAARTWTATAAT | COI | 120 | DNA metabarcoding | Prosser and Hebert, [61]. |
| Apis238_R1 | TAATCAAAATCTAATATTATTTATTCG |
| Primer | Primr Sequence | Target Species | Target Gene | Size of Amplicon (bp) | Method of Discrimination | Reference |
|---|---|---|---|---|---|---|
| M2-F | CATCACTTGAATGATTAAATTTTTTACCA | A. mellifera | COI | 206 | Duplex PCR | Kim et al., [8] |
| M2-R | TTATTTTTAAATTAATATGAATTAAGTGGGG | |||||
| M5-F | CACTTGAATGATTAAATTTTTTACCACCT | A. mellifera | COI | 133 | ||
| M5-R | CTTAAGTTCAATGCACTTATTCTGCC | |||||
| C6-F | GGAGGGGGAGATCCAATTTA | A. cerana | COI | 178 | ||
| C5-R | ACCTAATATTGCGTAAATTATACCTAGATTT | |||||
| AC1-F | TCTGAATTCAAACTCAAAGTAAAA | A. cerana | tRNAleu-cox2 | 111 | PCR | Suares et al., [11] |
| AC1-R | ATAATATGAGTTTGATTCTTGAAA | |||||
| AM1-F | AGCTAATTAAAACAACAATACA | Both species | 16S rRNA | 140 | Melting curve analysis by Real-Time PCR | |
| AM1-R | AAGGTAGTAAATGTTGAATCATT | |||||
| C-F | TTTAACAATAAAAATAATCAGAAGA | A. cerana | MRJP2 | 212 | Duplex PCR | Zhang et al., [10] |
| C-R | TTACATCCTAATTGATTTTAATGCG | |||||
| M-F | GCCATCCCTTGAAATTGTCACTCGT | A. mellifera | MRJP2 | 560 | Melting curve analysis by Real-Time PCR | |
| M-R | TCTGCAAACGACCAATCAGGATAT | |||||
| AC-F | TCATTAGRTTTTACAAAATCWGATCA | A. cerana | NADH2 | 223 | Duplex PCR | Mohamadzade Namin et al., [63] |
| AC-R | CTTATAACTAAATATGTTAATGATCATA | |||||
| AM-F | CYATTAGATTTACTAAAACAGATACT | A. mellifera | NADH2 | 376 | Duplex PCR | |
| AM-R | ATAATTAAATGAATATAAAATAATTATAGCA | |||||
| AD-F | TATATTAATTGTTATAACTTACATAAATAA | A. dorsata | NADH2 | 301 | Duplex PCR | |
| AD-R | GGATTAAGAATATATAATATTCATATTTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamadzade Namin, S.; Ghosh, S.; Jung, C. Honey Quality Control: Review of Methodologies for Determining Entomological Origin. Molecules 2023, 28, 4232. https://doi.org/10.3390/molecules28104232
Mohamadzade Namin S, Ghosh S, Jung C. Honey Quality Control: Review of Methodologies for Determining Entomological Origin. Molecules. 2023; 28(10):4232. https://doi.org/10.3390/molecules28104232
Chicago/Turabian StyleMohamadzade Namin, Saeed, Sampat Ghosh, and Chuleui Jung. 2023. "Honey Quality Control: Review of Methodologies for Determining Entomological Origin" Molecules 28, no. 10: 4232. https://doi.org/10.3390/molecules28104232
APA StyleMohamadzade Namin, S., Ghosh, S., & Jung, C. (2023). Honey Quality Control: Review of Methodologies for Determining Entomological Origin. Molecules, 28(10), 4232. https://doi.org/10.3390/molecules28104232







