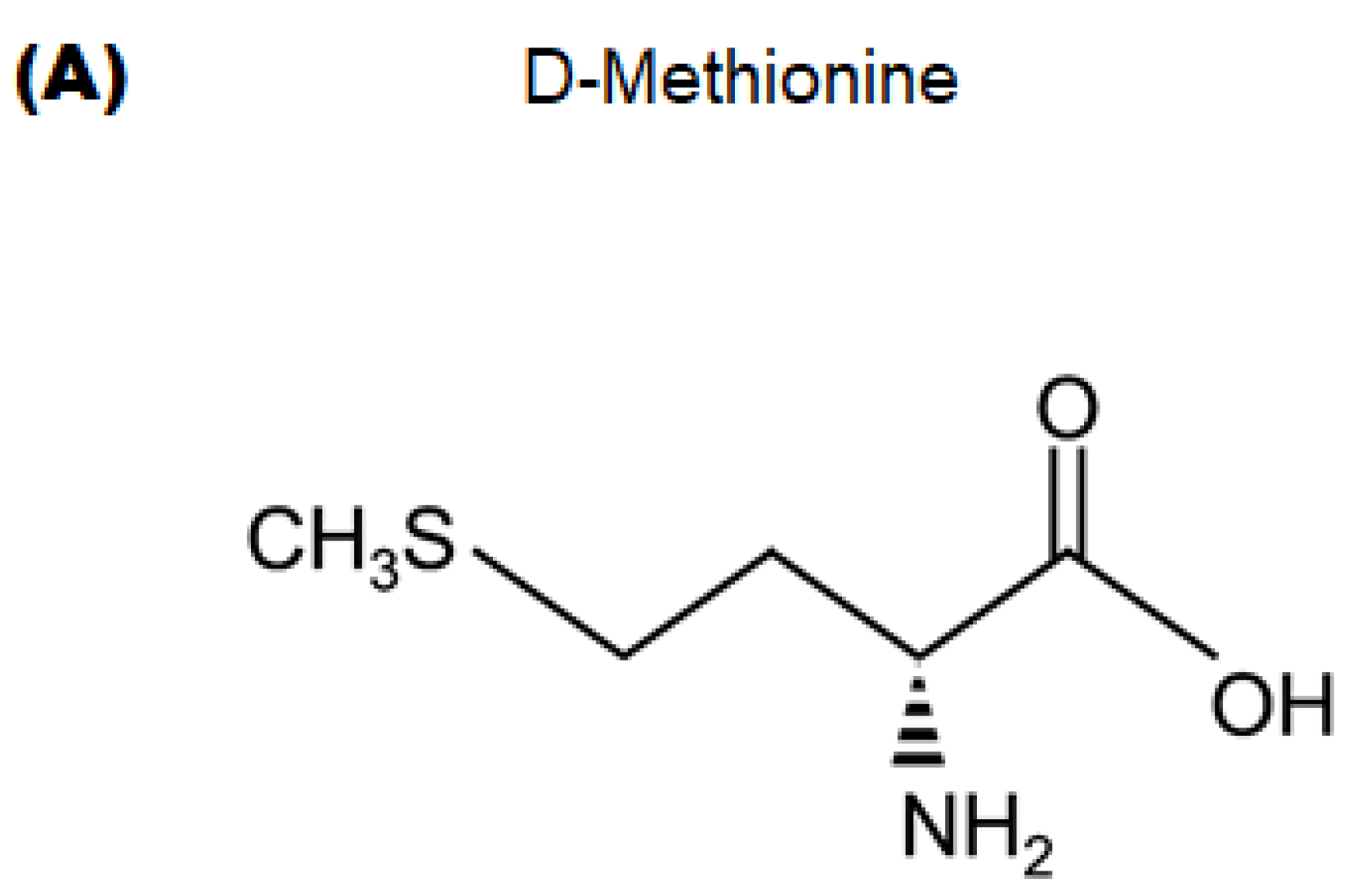
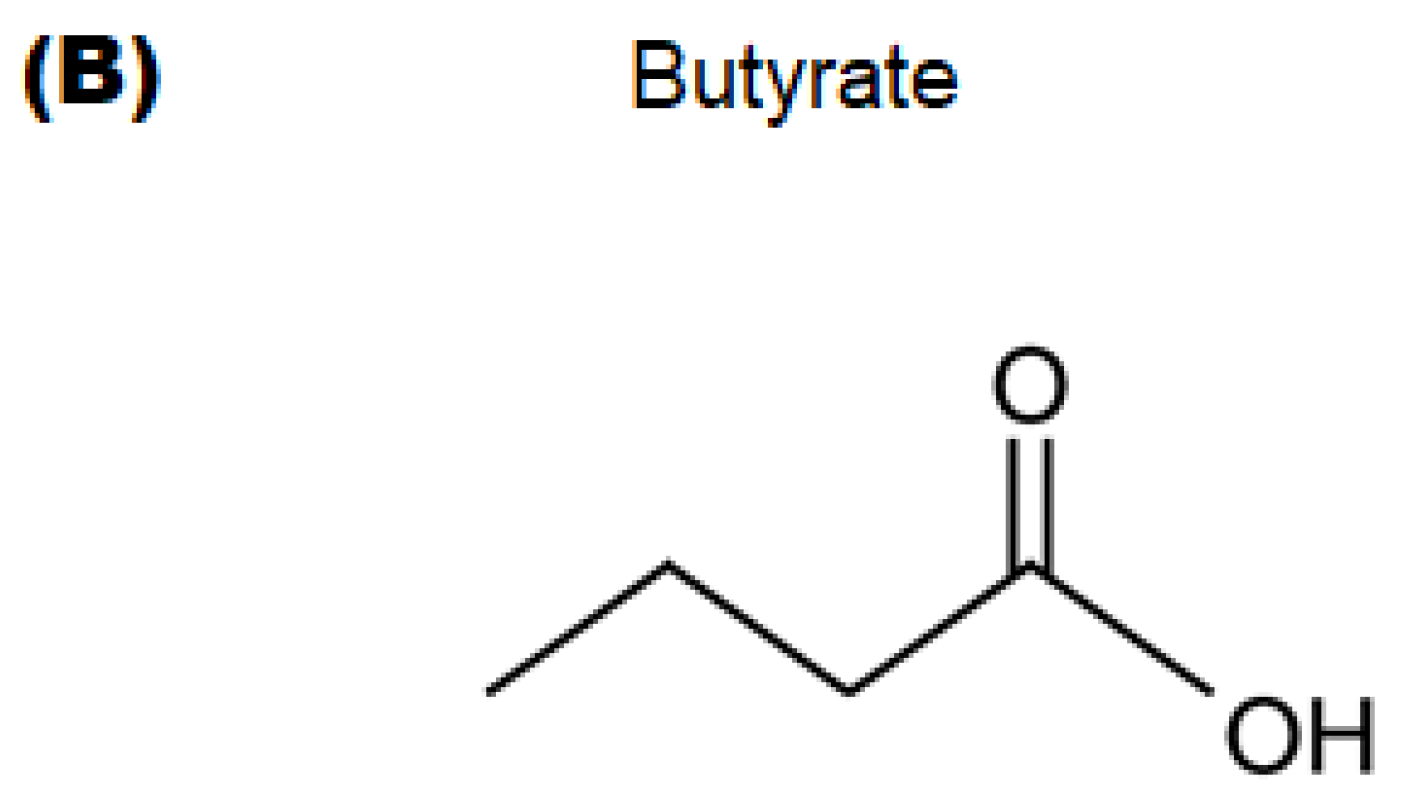
Gut Protective Effect from D-Methionine or Butyric Acid against DSS and Carrageenan-Induced Ulcerative Colitis
Abstract
:1. Introduction
2. Results
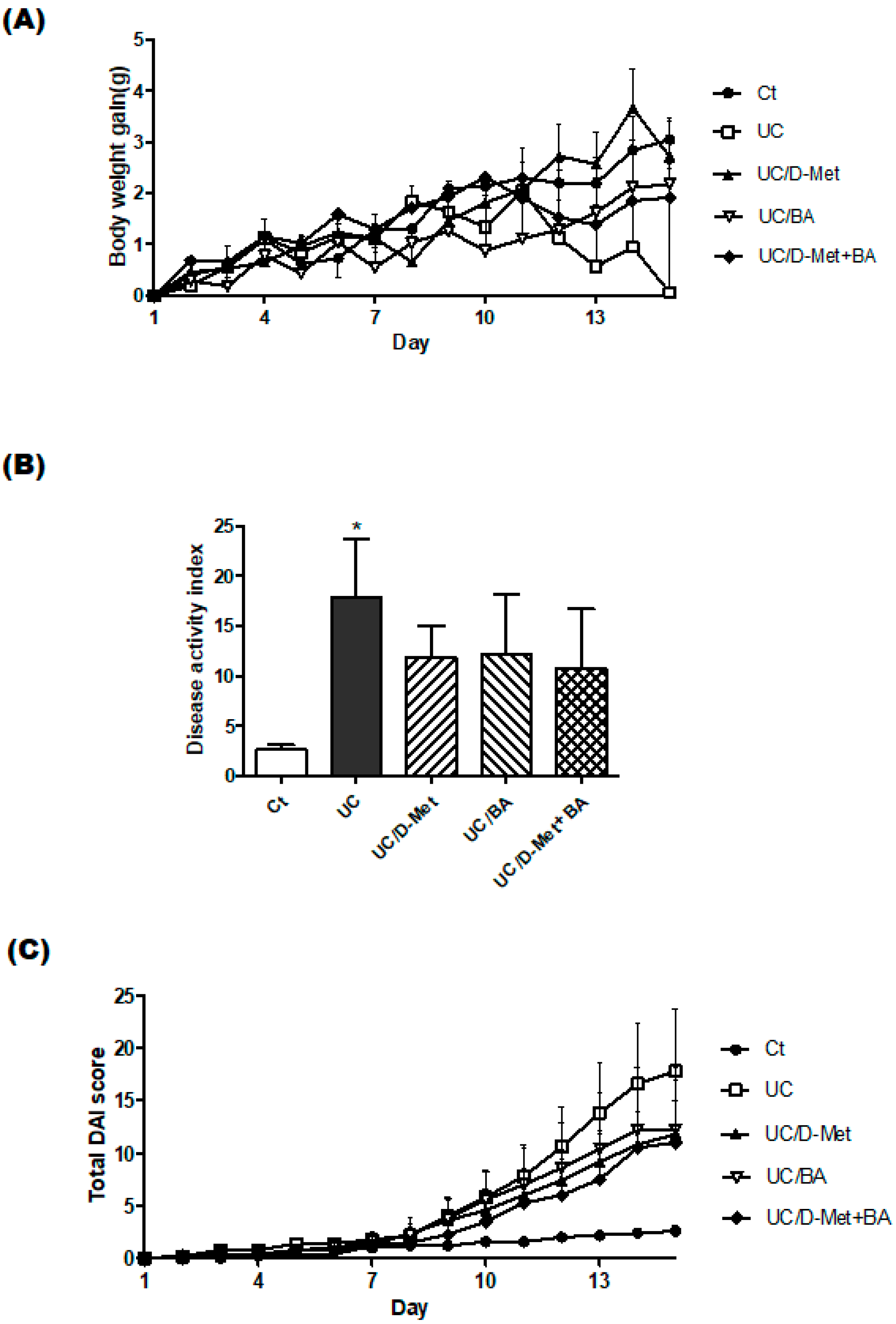
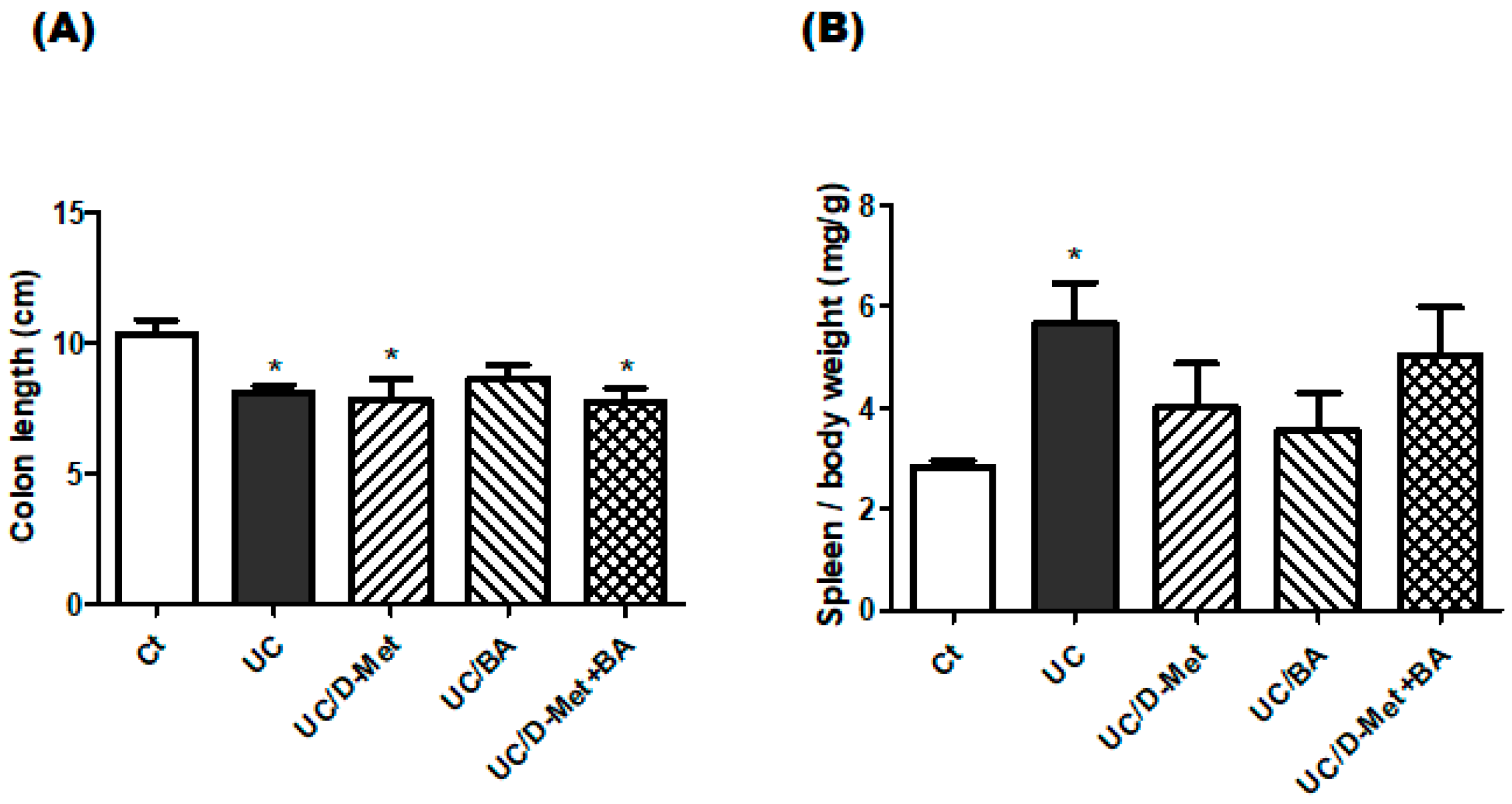
2.1. Characteristics and Disease Activity Index of UC Model Mice
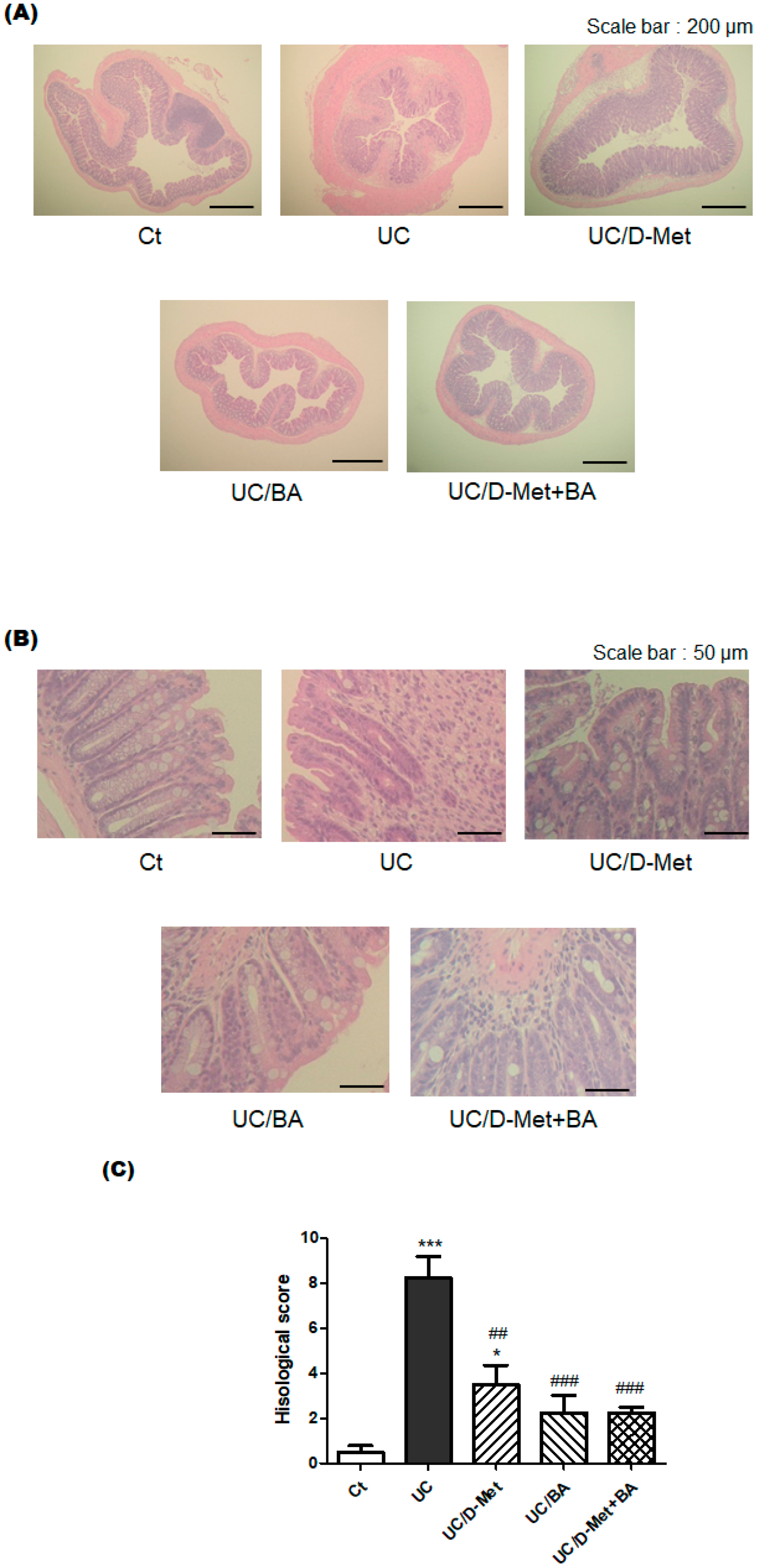
2.2. Histological Finding in the Colon
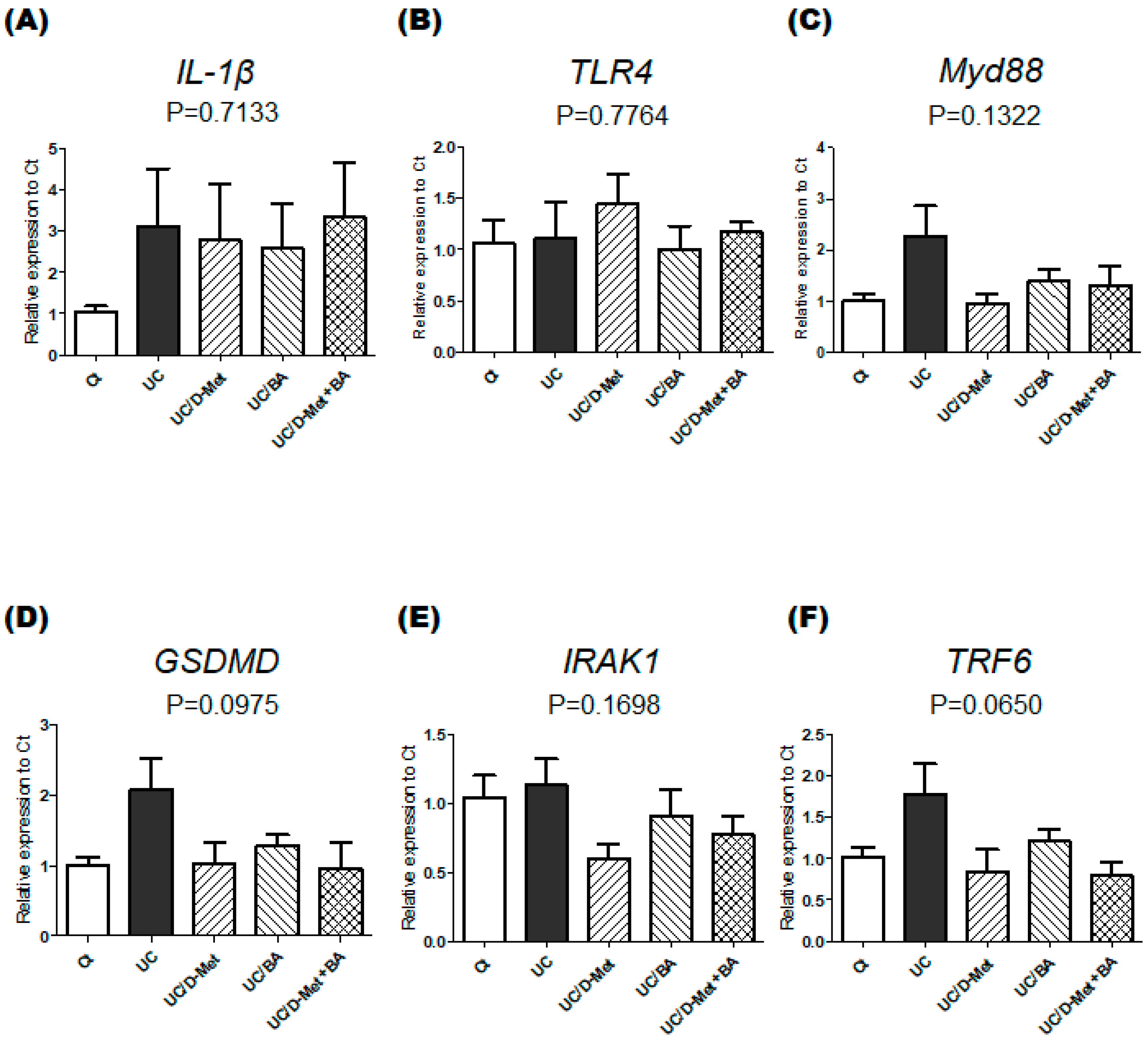
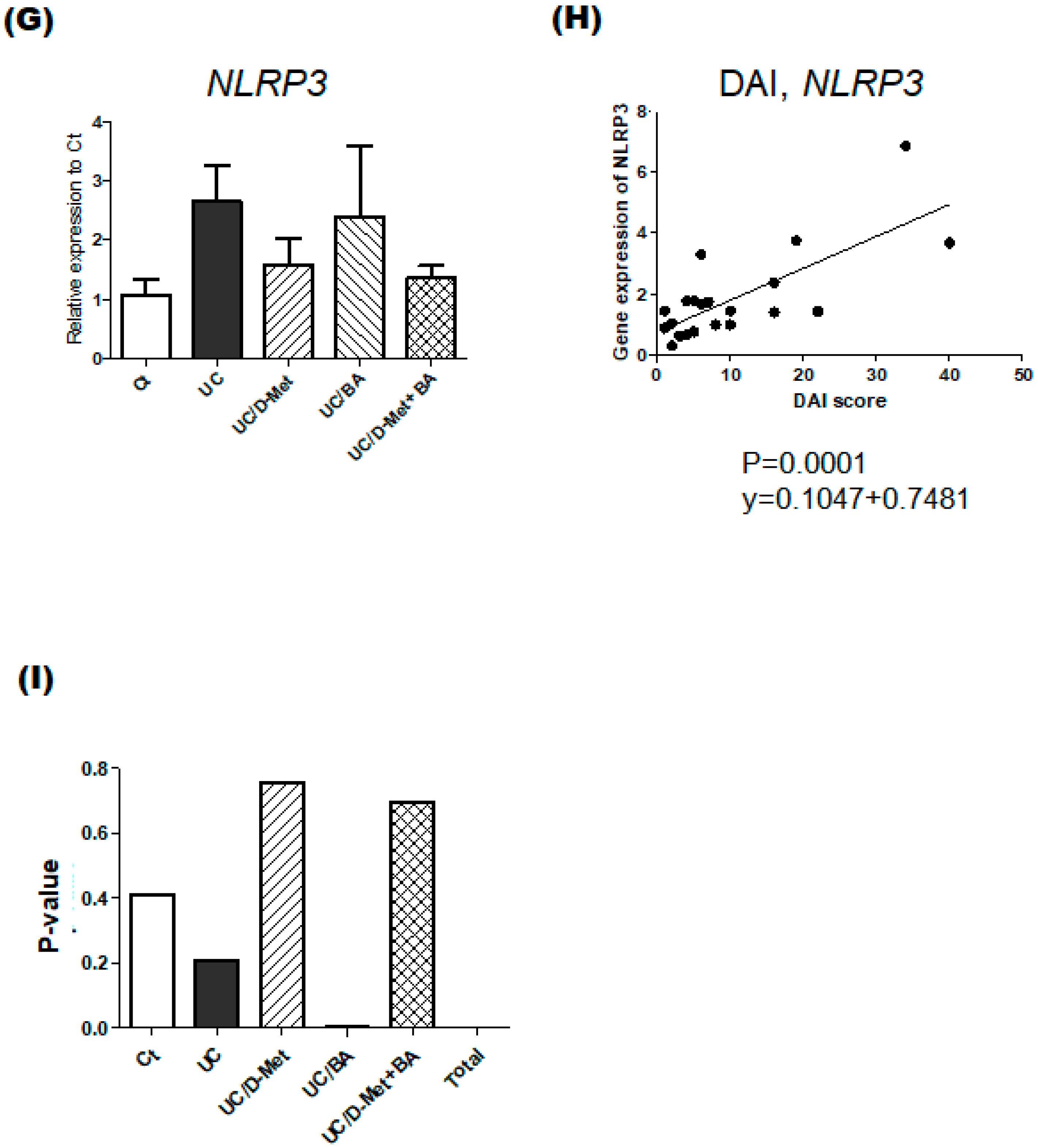
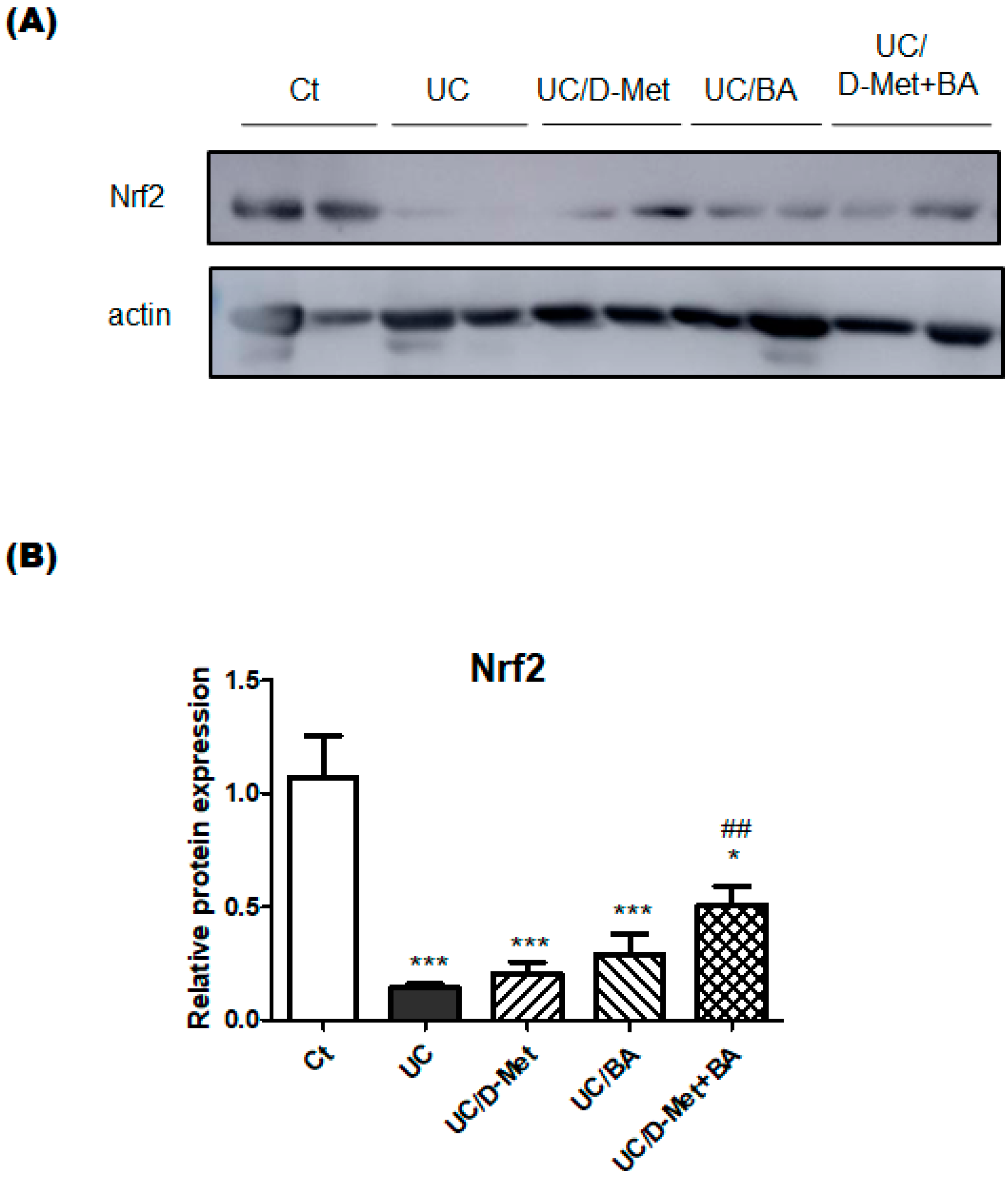
2.3. Analyses for Genes and Protein Expression in the Colon of UC Mice
3. Discussion
4. Materials and Methods
4.1. Mice and Water
4.2. Materials
4.3. Disease Activity Index
4.4. Histopathological Analysis
4.5. Real-Time PCR Gene Expression Analysis
4.6. Western Blot Analysis
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| BA | butyric acid, butyrate |
| D-Met | d-methionine |
| DSS | dextran sodium sulfate |
| GSDMD | gasdermin D |
| IBD | inflammatory bowel diseases |
| IL-1 | βinterleukin 1β |
| IRAK1 | IL-1 receptor-associated kinase |
| LPS | lipopolysaccharide |
| Myd88 | myeloid differentiation factor 88 |
| NLRP3 | NLR family pyrin domain containing 3 |
| Nrf2 | nuclear factor-erythroid 2-related factor-2 |
| QOL | quality of life |
| ROS | reactive oxygen species |
| SCFAs | short-chain fatty acids |
| TLR4 | toll-like receptor 4 |
| TRAF6 | TNF receptor-associated factor 6 |
| UC | ulcerative colitis |
References
- Podolsky, D.K. Inflammatory bowel disease. N. Engl. J. Med. 2002, 347, 417–429. [Google Scholar] [CrossRef]
- Xavier, R.J.; Podolsky, D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature 2007, 448, 427–434. [Google Scholar] [CrossRef]
- Bouma, G.; Strober, W. The immunological and genetic basis of inflammatory bowel disease. Nat. Rev. Immunol. 2003, 3, 521–533. [Google Scholar] [CrossRef]
- Atreya, R.; Neurath, M.F. New therapeutic strategies for treatment of inflammatory bowel disease. Mucosal. Immunol. 2008, 1, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Reinisch, W.; Sandborn, W.J.; Hommes, D.W.; D’Haens, G.; Hanauer, S.; Schreiber, S.; Panaccione, R.; Fedorak, R.N.; Tighe, M.B.; Huang, B.; et al. Adalimumab for induction of clinical remission in moderately to severely active ulcerative colitis: Results of a randomised controlled trial. Gut 2011, 60, 780–787. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N.; McGinley, E.L.; Binion, D.G. Inflammatory bowel disease in the elderly is associated with worse outcomes: A national study of hospitalizations. Inflamm. Bowel. Dis. 2009, 15, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Britzen-Laurent, N.; Weidinger, C.; Stürzl, M. Contribution of Blood Vessel Activation, Remodeling and Barrier Function to Inflammatory Bowel Diseases. Int. J. Mol. Sci. 2023, 24, 5517. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Li, Y.Y. Inflammatory bowel disease: Pathogenesis. World J. Gastroenterol. 2014, 20, 91–99. [Google Scholar] [CrossRef]
- Thoo, L.; Noti, M.; Krebs, P. Keep calm: The intestinal barrier at the interface of peace and war. Cell Death. Dis. 2019, 10, 849. [Google Scholar] [CrossRef]
- Niu, W.; Yang, F.; Fu, Z.; Dong, Y.; Zhang, Z.; Ju, J. The role of enteric dysbacteriosis and modulation of gut microbiota in the treatment of inflammatory bowel disease. Microb. Pathog. 2022, 165, 105381. [Google Scholar] [CrossRef]
- Sheng, K.; Xu, Y.; Kong, X.; Wang, J.; Zha, X.; Wang, Y. Probiotic Bacillus cereus Alleviates Dextran Sulfate Sodium-Induced Colitis in Mice through Improvement of the Intestinal Barrier Function, Anti-Inflammation, and Gut Microbiota Modulation. J. Agric. Food Chem. 2021, 69, 14810–14823. [Google Scholar] [CrossRef]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef]
- Rubinstein, M.M.; Makker, V. Optimizing immunotherapy for gynecologic cancers. Curr. Opin. Obs. Gynecol. 2020, 32, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, Y.; Taniguchi, K.; Sawamura, H.; Tsuji, A.; Matsuda, S. Promising role of D-amino acids in irritable bowel syndrome. World J. Gastroenterol. 2022, 28, 4471–4474. [Google Scholar] [CrossRef] [PubMed]
- Mi, Y.; Chin, Y.X.; Cao, W.X.; Chang, Y.G.; Lim, P.E.; Xue, C.H.; Tang, Q.J. Native κ-carrageenan induced-colitis is related to host intestinal microecology. Int. J. Biol. Macromol. 2020, 147, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Morohoshi, Y.; Matsuoka, K.; Chinen, H.; Kamada, N.; Sato, T.; Hisamatsu, T.; Okamoto, S.; Inoue, N.; Takaishi, H.; Ogata, H.; et al. Inhibition of neutrophil elastase prevents the development of murine dextran sulfate sodium-induced colitis. J. Gastroenterol. 2006, 41, 318–324. [Google Scholar] [CrossRef]
- Li, J.; Sung, C.Y.; Lee, N.; Ni, Y.; Pihlajamäki, J.; Panagiotou, G.; El-Nezami, H. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc. Natl. Acad. Sci. USA 2016, 113, E1306–E1315. [Google Scholar] [CrossRef]
- Dai, Y.C.; Zheng, L.; Zhang, Y.L.; Chen, X.; Chen, D.L.; Wang, L.J.; Tang, Z.P. Jianpi Qingchang decoction regulates intestinal motility of dextran sulfate sodium-induced colitis through reducing autophagy of interstitial cells of Cajal. World J. Gastroenterol. 2017, 23, 4724–4734. [Google Scholar] [CrossRef]
- Mitani, T.; Yoshioka, Y.; Furuyashiki, T.; Yamashita, Y.; Shirai, Y.; Ashida, H. Enzymatically synthesized glycogen inhibits colitis through decreasing oxidative stress. Free Radic. Biol. Med. 2017, 106, 355–367. [Google Scholar] [CrossRef]
- Stokkers, P.C.; van Aken, B.E.; Basoski, N.; Reitsma, P.H.; Tytgat, G.N.; van Deventer, S.J. Five genetic markers in the interleukin 1 family in relation to inflammatory bowel disease. Gut 1998, 43, 33–39. [Google Scholar] [CrossRef]
- Yang, Q.Y.; Ma, L.L.; Zhang, C.; Lin, J.Z.; Han, L.; He, Y.N.; Xie, C.G. Exploring the Mechanism of Indigo Naturalis in the Treatment of Ulcerative Colitis Based on TLR4/MyD88/NF-κB Signaling Pathway and Gut Microbiota. Front. Pharmacol. 2021, 12, 674416. [Google Scholar] [CrossRef]
- Li, J.; Ma, C.M.; Di, D.L. A narrative review of pyrolysis and its role in ulcerative colitis. Eur. Rev. Med. Pharm. Sci. 2022, 26, 1156–1163. [Google Scholar]
- Chen, Y.L.; Zhang, Y.L.; Dai, Y.C.; Tang, Z.P. Systems pharmacology approach reveals the antiinflammatory effects of Ampelopsis grossedentata on dextran sodium sulfate-induced colitis. World J. Gastroenterol. 2018, 24, 1398–1409. [Google Scholar] [CrossRef] [PubMed]
- Cosin-Roger, J.; Simmen, S.; Melhem, H.; Atrott, K.; Frey-Wagner, I.; Hausmann, M.; de Vallière, C.; Spalinger, M.R.; Spielmann, P.; Wenger, R.H.; et al. Hypoxia ameliorates intestinal inflammation through NLRP3/mTOR downregulation and autophagy activation. Nat. Commun. 2017, 8, 98. [Google Scholar] [CrossRef]
- Miao, F.; Shan, C.; Ma, T.; Geng, S.; Ning, D. Walnut oil alleviates DSS-induced colitis in mice by inhibiting NLRP3 inflammasome activation and regulating gut microbiota. Microb. Pathog. 2021, 154, 104866. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.C.; Ji, J.A.; Jiang, Y.L.; Chen, Z.Y.; Yuan, Z.W.; You, Q.D.; Jiang, Z.Y. An inhibitor of the Keap1-Nrf2 protein-protein interaction protects NCM460 colonic cells and alleviates experimental colitis. Sci. Rep. 2016, 6, 26585. [Google Scholar] [CrossRef]
- Dodson, M.; Redmann, M.; Rajasekaran, N.S.; Darley-Usmar, V.; Zhang, J. KEAP1-NRF2 signalling and autophagy in protection against oxidative and reductive proteotoxicity. Biochem. J. 2015, 469, 347–355. [Google Scholar] [CrossRef] [PubMed]
- De Souza, H.S.; Fiocchi, C. Immunopathogenesis of IBD: Current state of the art. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 13–27. [Google Scholar] [CrossRef]
- Calder, P.C.; Albers, R.; Antoine, J.M.; Blum, S.; Bourdet-Sicard, R.; Ferns, G.A.; Folkerts, G.; Friedmann, P.S.; Frost, G.S.; Guarner, F.; et al. Inflammatory disease processes and interactions with nutrition. Br. J. Nutr. 2009, 101, S1–S45. [Google Scholar] [CrossRef]
- Ahmad, T.; Ishaq, M.; Karpiniec, S.; Park, A.; Stringer, D.; Singh, N.; Ratanpaul, V.; Wolfswinkel, K.; Fitton, H.; Caruso, V.; et al. Oral Macrocystis pyrifera Fucoidan Administration Exhibits Anti-Inflammatory and Antioxidant Properties and Improves DSS-Induced Colitis in C57BL/6J Mice. Pharmaceutics 2022, 14, 2383. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Feferman, L.; Borthakur, S.; Tobacman, J.K. Common food additive carrageenan stimulates Wnt/ β-catenin signaling in colonic epithelium by inhibition of nucleoredoxin reduction. Nutr. Cancer. 2014, 66, 117–127. [Google Scholar] [CrossRef]
- Li, L.; Fang, Z.; Liu, X.; Hu, W.; Lu, W.; Lee, Y.K.; Zhao, J.; Zhang, H.; Chen, W. Lactobacillus reuteri attenuated allergic inflammation induced by HDM in the mouse and modulated gut microbes. PLoS ONE 2020, 15, e0231865. [Google Scholar] [CrossRef] [PubMed]
- Brain, J.D.; Hsu, Y.H.; Vasanthakumar, A.; Kim, J.; Mitchell, R.; Chang-Sheng, M.; Iinomi, M.; Akatsuka, K.; Molina, R.M. Effects of a vinegar-based multi-micronutrient supplement in rats: A multi-pronged assessment of dietary impact. J. Funct. Foods. 2018, 42, 371–378. [Google Scholar] [CrossRef]
- Cheng, X.; Yang, Y.L.; Yang, H.; Wang, Y.H.; Du, G.H. Kaempferol alleviates LPS-induced neuroinflammation and BBB dysfunction in mice via inhibiting HMGB1 release and down-regulating TLR4/MyD88 pathway. Int. Immunopharmacol. 2018, 56, 29–35. [Google Scholar] [CrossRef]
- Kobayashi, M.; Yamamoto, M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv. Enzym. Regul. 2006, 46, 113–140. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, D.; Wang, H.; Wang, T.; Weng, Y.; Zhang, Y.; Luo, Y.; Lu, Y.; Wang, Y. Therapeutic Targeting of Nrf2 Signaling by Maggot Extracts Ameliorates Inflammation-Associated Intestinal Fibrosis in Chronic DSS-Induced Colitis. Front. Immunol. 2021, 12, 670159. [Google Scholar] [CrossRef]
- Scapagnini, G.; Vasto, S.; Abraham, N.G.; Caruso, C.; Zella, D.; Fabio, G. Modulation of Nrf2/ARE pathway by food polyphenols: A nutritional neuroprotective strategy for cognitive and neurodegenerative disorders. Mol. Neurobiol. 2011, 44, 192–201. [Google Scholar] [CrossRef]
- Shopit, A.; Niu, M.; Wang, H.; Tang, Z.; Li, X.; Tesfaldet, T.; Ai, J.; Ahmad, N.; Al-Azab, M.; Tang, Z. Protection of diabetes-induced kidney injury by phosphocreatine via the regulation of ERK/Nrf2/HO-1 signaling pathway. Life Sci. 2020, 242, 117248. [Google Scholar] [CrossRef]
- Hejazian, S.M.; Hosseiniyan Khatibi, S.M.; Barzegari, A.; Pavon-Djavid, G.; Razi Soofiyani, S.; Hassannejhad, S.; Ahmadian, E.; Ardalan, M.; Zununi Vahed, S. Nrf-2 as a therapeutic target in acute kidney injury. Life Sci. 2021, 264, 118581. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhou, B.; Pang, X.; Song, X.; Gu, Y.; Xie, R.; Liu, T.; Xu, X.; Wang, B.; Cao, H. Clostridium butyricum, a butyrate-producing potential probiotic, alleviates experimental colitis through epidermal growth factor receptor activation. Food Funct. 2022, 13, 7046–7061. [Google Scholar] [CrossRef]
- Jiang, P.; Yang, W.; Jin, Y.; Huang, H.; Shi, C.; Jiang, Y.; Wang, J.; Kang, Y.; Wang, C.; Yang, G. Lactobacillus reuteri protects mice against Salmonella typhimurium challenge by activating macrophages to produce nitric oxide. Microb. Pathog. 2019, 137, 103754. [Google Scholar] [CrossRef]
- Schroeder, B.O.; Birchenough, G.M.H.; Ståhlman, M.; Arike, L.; Johansson, M.E.V.; Hansson, G.C.; Bäckhed, F. Bifidobacteria or Fiber Protects against Diet-Induced Microbiota-Mediated Colonic Mucus Deterioration. Cell Host Microbe 2018, 23, 27–40.e7. [Google Scholar] [CrossRef]
- Seki, H.; Shiohara, M.; Matsumura, T.; Miyagawa, N.; Tanaka, M.; Komiyama, A.; Kurata, S. Prevention of antibiotic-associated diarrhea in children by Clostridium butyricum MIYAIRI. Pediatr. Int. 2003, 45, 86–90. [Google Scholar] [CrossRef]
- Shashkova, T.; Popenko, A.; Tyakht, A.; Peskov, K.; Kosinsky, Y.; Bogolubsky, L.; Raigorodskii, A.; Ischenko, D.; Alexeev, D.; Govorun, V. Agent Based Modeling of Human Gut Microbiome Interactions and Perturbations. PLoS ONE 2016, 11, e0148386. [Google Scholar] [CrossRef] [PubMed]
- Federica, R.; Edda, R.; Daniela, R.; Simone, B.; Giulia, N.; Gabriele, L.; Marta, M.; Marco, P.; Gianluca, B.; Elena, N.; et al. Characterization of the “gut microbiota-immunity axis” and microbial lipid metabolites in atrophic and potential celiac disease. Front Microbiol. 2022, 13, 886008. [Google Scholar] [PubMed]
- Seo, M.; Inoue, I.; Tanaka, M.; Matsuda, N.; Nakano, T.; Awata, T.; Katayama, S.; Alpers, D.H.; Komoda, T. Clostridium butyricum MIYAIRI 588 improves high-fat diet-induced non-alcoholic fatty liver disease in rats. Dig. Dis. Sci. 2013, 58, 3534–3544. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Li, Z.; Xu, D.; Wang, Y.; Bai, Q.; Feng, Y.; Su, G.; Chen, P.; Wang, Y.; Liu, H.; et al. Cepharanthine Hydrochloride Improves Cisplatin Chemotherapy and Enhances Immunity by Regulating Intestinal Microbes in Mice. Front Cell Infect. Microbiol. 2019, 9, 225. [Google Scholar] [CrossRef]
- Zhai, S.; Qin, S.; Li, L.; Zhu, L.; Zou, Z.; Wang, L. Dietary butyrate suppresses inflammation through modulating gut microbiota in high-fat diet-fed mice. FEMS Microbiol. Lett. 2019, 366, fnz153. [Google Scholar] [CrossRef]
- Yang, B.; El Nahas, A.M.; Thomas, G.L.; Haylor, J.L.; Watson, P.F.; Wagner, B.; Johnson, T.S. Caspase-3 and apoptosis in experimental chronic renal scarring. Kidney Int. 2001, 60, 1765–1776. [Google Scholar] [CrossRef]
- Asakawa, T.; Onizawa, M.; Saito, C.; Hikichi, R.; Yamada, D.; Minamidate, A.; Mochimaru, T.; Asahara, S.I.; Kido, Y.; Oshima, S.; et al. Oral administration of D-serine prevents the onset and progression of colitis in mice. J. Gastroenterol. 2021, 56, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.; Fonseca, J.R.; Rock, T.M.; Krauss-Etschmann, S.; Schmitt-Kopplin, P. Enantioseparation and selective detection of D-amino acids by ultra-high-performance liquid chromatography/mass spectrometry in analysis of complex biological samples. J. Chromatogr. A 2014, 1324, 109–114. [Google Scholar] [CrossRef]
- Wu, C.H.; Ko, J.L.; Liao, J.M.; Huang, S.S.; Lin, M.Y.; Lee, L.H.; Chang, L.Y.; Ou, C.C. D-methionine alleviates cisplatin-induced mucositis by restoring the gut microbiota structure and improving intestinal inflammation. Ther. Adv. Med. Oncol. 2019, 11, 1758835918821021. [Google Scholar] [CrossRef]
- Nakade, Y.; Iwata, Y.; Furuichi, K.; Mita, M.; Hamase, K.; Konno, R.; Miyake, T.; Sakai, N.; Kitajima, S.; Toyama, T.; et al. Gut microbiota-derived D-serine protects against acute kidney injury. JCI Insight 2018, 3, e97957. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H. The physiological role of hydrogen sulfide and beyond. Nitric Oxide 2014, 41, 4–10. [Google Scholar] [CrossRef]
- Souza, L.K.; Araújo, T.S.; Sousa, N.A.; Sousa, F.B.; Nogueira, K.M.; Nicolau, L.A.; Medeiros, J.V. Evidence that d-cysteine protects mice from gastric damage via hydrogen sulfide produced by d-amino acid oxidase. Nitric Oxide 2017, 64, 1–6. [Google Scholar] [CrossRef] [PubMed]
- De Rosa, V.; Secondo, A.; Pannaccione, A.; Ciccone, R.; Formisano, L.; Guida, N.; Crispino, R.; Fico, A.; Polishchuk, R.; D’Aniello, A.; et al. D-Aspartate treatment attenuates myelin damage and stimulates myelin repair. EMBO Mol. Med. 2019, 11, e9278. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Macfarlane, S. Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 2012, 95, 50–60. [Google Scholar] [CrossRef]
- Cava, F.; Lam, H.; de Pedro, M.A.; Waldor, M.K. Emerging knowledge of regulatory roles of D-amino acids in bacteria. Cell Mol. Life Sci. 2011, 68, 817–831. [Google Scholar] [CrossRef]
- Sasabe, J.; Miyoshi, Y.; Rakoff-Nahoum, S.; Zhang, T.; Mita, M.; Davis, B.M.; Hamase, K.; Waldor, M.K. Interplay between microbial d-amino acids and host d-amino acid oxidase modifies murine mucosal defence and gut microbiota. Nat. Microbiol. 2016, 1, 16125. [Google Scholar] [CrossRef]
- Kawase, T.; Nagasawa, M.; Ikeda, H.; Yasuo, S.; Koga, Y.; Furuse, M. Gut microbiota of mice putatively modifies amino acid metabolism in the host brain. Br. J. Nutr. 2017, 117, 775–783. [Google Scholar] [CrossRef]
- Nagano, T.; Yamao, S.; Terachi, A.; Yarimizu, H.; Itoh, H.; Katasho, R.; Kawai, K.; Nakashima, A.; Iwasaki, T.; Kikkawa, U.; et al. d-amino acid oxidase promotes cellular senescence via the production of reactive oxygen species. Life Sci. Alliance 2019, 2, e201800045. [Google Scholar] [CrossRef] [PubMed]
- Canteros, M.G. D-Arginine as a neuroprotective amino acid: Promising outcomes for neurological diseases. Drug. Discov. Today 2014, 19, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Shao, B.Z.; Wei, W.; Ke, P.; Xu, Z.Q.; Zhou, J.X.; Liu, C. Activating cannabinoid receptor 2 alleviates pathogenesis of experimental autoimmune encephalomyelitis via activation of autophagy and inhibiting NLRP3 inflammasome. CNS Neurosci. Ther. 2014, 20, 1021–1028. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Sun, Y.; Liu, W.; Wu, X.; Guo, L.; Cai, P.; Wu, X.; Wu, X.; Shen, Y.; Shu, Y.; et al. Small molecule-driven mitophagy-mediated NLRP3 inflammasome inhibition is responsible for the prevention of colitis-associated cancer. Autophagy 2014, 10, 972–985. [Google Scholar] [CrossRef]

| Score | Weight Loss (%) | Occult Blood | Stool Consistency |
|---|---|---|---|
| 0 | <1 | Negative | Normal |
| 1 | 1–5 | Occult blood stool | Soft stool |
| 2 | 5–10 | Loose stool | |
| 3 | 10–20 | Bloody stool | Muddy stool |
| 4 | >20 | Hematochezia | Diarrhea |
| Score | Epithelium Loss (%) | Crypt Damage (%) | Depletion of Goblet Cells | Infiltration of Inflammatory Cells |
|---|---|---|---|---|
| 0 | None | None | None | None |
| 1 | 0–5 | 0–5 | Mild | Mild |
| 2 | 5–10 | 5–10 | Moderate | Moderate |
| 3 | >10 | >10 | Severe | Severe |
| Gene | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| β-actin | TTCTACAATGAGCTGCGTGTG | CTTTTCACGGTTGGCCTTAG |
| IL-1β | CCCTGCAGCTGGAGAGTGTGGA | CTGAGCGACCTGTCTTGGCCG |
| TLR4 | GGCAGCAGGTGGAATTGTAT | AGGCCCCAGAGTTTTGTTCT |
| MyD88 | TTGCCAGCGAGCTAATTGAGA | TTCTGTTGGACACCTGGAGA |
| GSDMD | ATCTCATTCCGGTGGACAGC | AAACACTCCGGTTCTGGTTCT |
| IRAK1 | GAGACCCTTGCTGGTCAGAG | GCTACACCCACCCACAGAGT |
| TRF6 | ATTTCATTGTCAACTGGGCA | TGAGTGTCCCATCTGCTTGA |
| NLRP3 | ATGCTGCTTCGACATCTCCT | AACCAATGCGAGATCCTGAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikeda, Y.; Matsuda, S. Gut Protective Effect from D-Methionine or Butyric Acid against DSS and Carrageenan-Induced Ulcerative Colitis. Molecules 2023, 28, 4392. https://doi.org/10.3390/molecules28114392
Ikeda Y, Matsuda S. Gut Protective Effect from D-Methionine or Butyric Acid against DSS and Carrageenan-Induced Ulcerative Colitis. Molecules. 2023; 28(11):4392. https://doi.org/10.3390/molecules28114392
Chicago/Turabian StyleIkeda, Yuka, and Satoru Matsuda. 2023. "Gut Protective Effect from D-Methionine or Butyric Acid against DSS and Carrageenan-Induced Ulcerative Colitis" Molecules 28, no. 11: 4392. https://doi.org/10.3390/molecules28114392
APA StyleIkeda, Y., & Matsuda, S. (2023). Gut Protective Effect from D-Methionine or Butyric Acid against DSS and Carrageenan-Induced Ulcerative Colitis. Molecules, 28(11), 4392. https://doi.org/10.3390/molecules28114392








