Enhancement Effects and Mechanism Studies of Two Bismuth-Based Materials Assisted by DMSO and Glycerol in GC-Rich PCR
Abstract
1. Introduction
2. Results and Discussion
2.1. Optimization of Conditions for PCR Systems
- (1)
- Optimization for amplifying GNAS1 promoters
- (2)
- Optimization of the System for Amplifying the APOE Gene
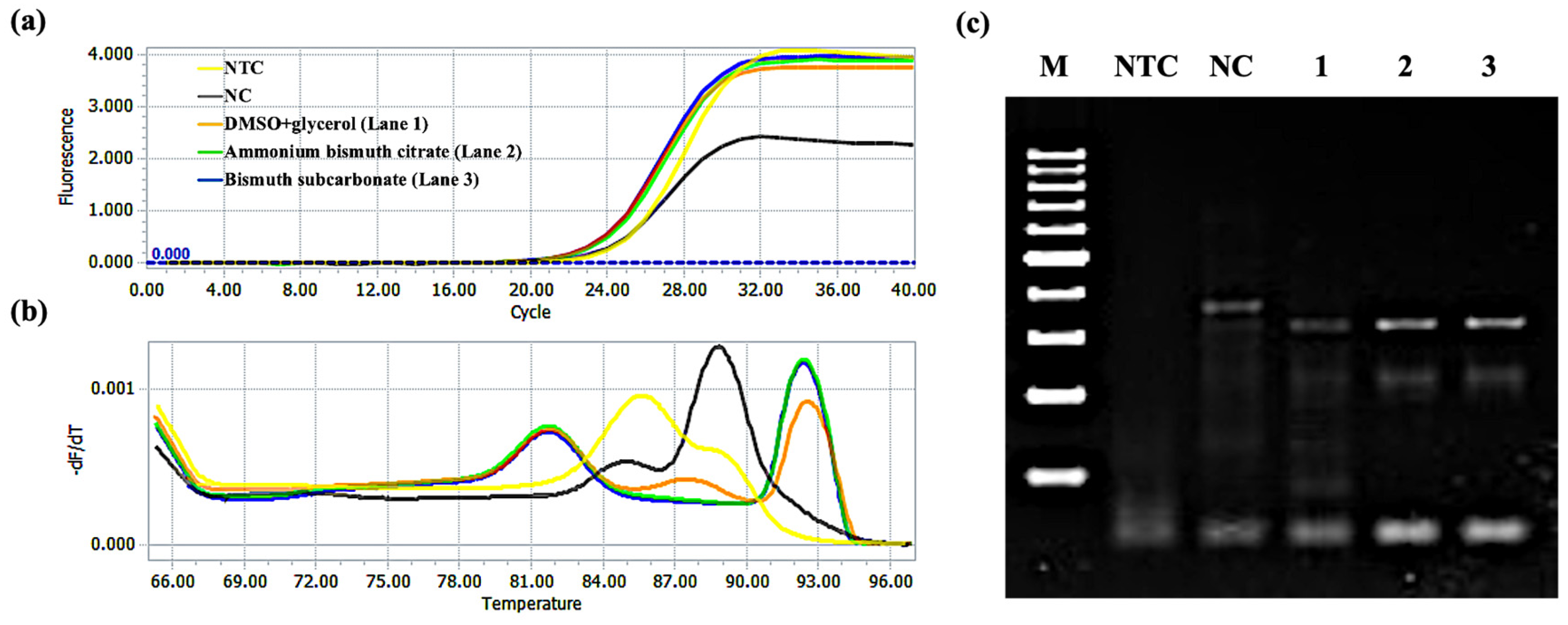
2.2. Validation of Amplification Products
2.3. Bismuth-Based Materials Enhanced PCR Amplification
2.3.1. Amplification of the GNAS1 Gene
2.3.2. Amplification of the APOE Gene
2.3.3. Applicability with Different Enzymes
2.4. Mechanism of Materials with PCR Components
2.4.1. Bismuth-Based Materials Decrease the Melting Temperature (Tm) of Primers
2.4.2. Bismuth-Based Materials Adsorb Polymerases and Regulate the Number of Active Polymerases in PCR
2.4.3. Bismuth-Based Materials Promote the Dissociation of the Product
3. Experiment
3.1. Materials and Apparatus
3.2. PCR Systems
3.2.1. Preparation of Working Solution
3.2.2. Conventional PCR Amplification
- (1)
- Materials used in amplification reactions
- (2)
- Different Taq DNA polymerases in amplification reactions
- (3)
- The effect of the materials on polymerase
3.3. Gel Electrophoresis
3.4. Fluorescence Measurement of Melting Temperature (Tm) of Primers
3.5. Fluorescence Measurement of Dissociation Percentage of PCR Products
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Assal, N.; Lin, M. PCR procedures to amplify GC-rich DNA sequences of Mycobacterium bovis. J. Microbiol. Methods 2021, 181, 106121. [Google Scholar] [CrossRef]
- Bai, S.; Xu, B.; Zhang, Y.; Zhang, Y.; Dang, H.; Yang, S.; Zuo, C.; Zhang, L.; Li, J.; Xie, G. Tuning the specificity of DNA probes using bulge-loops for low-abundance SNV detection. Biosens. Bioelectron. 2020, 154, 112092. [Google Scholar] [CrossRef] [PubMed]
- Kinimi, E.; Hakizimana, J.N.; Misinzo, G. Nucleotide amplification and sequencing of the GC-rich region between matrix and fusion protein genes of peste des petits ruminants virus. J. Virol. Methods 2022, 300, 114390. [Google Scholar] [CrossRef] [PubMed]
- Sahdev, S.; Saini, S.; Tiwari, P.; Saxena, S.; Saini, K.S. Amplification of GC-rich genes by following a combination strategy of primer design, enhancers and modified PCR cycle conditions. Mol. Cell. Probes 2007, 21, 303–307. [Google Scholar] [CrossRef]
- Baskaran, N.; Kandpal, R.P.; Bhargava, A.K.; Glynn, M.W.; Bale, A.; Weissman, S.M. Uniform amplification of a mixture of deoxyribonucleic acids with varying GC content. Genome Res. 1996, 6, 633–638. [Google Scholar] [CrossRef]
- Henke, W.; Herdel, K.; Jung, K.; Schnorr, D.; Loening, S.A. Betaine Improves the PCR Amplification of GC-Rich DNA Sequences. Nucleic Acids Res. 1997, 25, 3957–3958. [Google Scholar] [CrossRef]
- Musso, M.; Bocciardi, R.; Parodi, S.; Ravazzolo, R.; Ceccherini, I. Betaine, Dimethyl Sulfoxide, and 7-Deaza-dGTP, a Powerful Mixture for Amplification of GC-Rich DNA Sequences. J. Mol. Diagn. 2006, 8, 544–550. [Google Scholar] [CrossRef]
- Mutter, G.L.; Boynton, K.A. PCR bias in amplification of androgen receptor alleles, a trinucleotide repeat marker used in clonality studies. Nucleic Acids Res. 1995, 23, 1411–1418. [Google Scholar]
- Karunanathie, H.; Kee, P.S.; Ng, S.F.; Kennedy, M.A.; Chua, E.W. PCR enhancers: Types, mechanisms, and applications in long-range PCR. Biochimie 2022, 197, 130–143. [Google Scholar] [CrossRef]
- Korbie, D.J.; Mattick, J.S. Touchdown PCR for Increased Specificity and Sensitivity in PCR Amplification. Nat. Protoc. 2008, 3, 1452–1456. [Google Scholar] [CrossRef]
- Yang, Z.; Shen, B.; Yue, L.; Miao, Y.; Hu, Y.; Ouyang, R. Application of Nanomaterials to Enhance Polymerase Chain Reaction. Molecules 2022, 27, 8854. [Google Scholar] [CrossRef]
- Duan, M.; Zhu, X.; Fan, L.; He, Y.; Yang, C.; Guo, R.; Chen, S.; Sun, X.; Liu, J. Phase-Transitional Bismuth-Based Metals enable Rapid Embolotherapy, Hyperthermia, and Biomedical Imaging. Adv. Mater. 2022, 34, 2205002. [Google Scholar] [CrossRef] [PubMed]
- Orellana-Tavra, C.; Köppen, M.; Li, A.; Stock, N.; Fairen-Jimenez, D. Biocompatible, Crystalline, and Amorphous Bismuth-Based Metal–Organic Frameworks for Drug Delivery. ACS Appl. Mater. Interfaces 2020, 12, 5633–5641. [Google Scholar] [CrossRef] [PubMed]
- Shahbazi, M.-A.; Faghfouri, L.; Ferreira, M.P.A.; Figueiredo, P.; Maleki, H.; Sefat, F.; Hirvonen, J.; Santos, H.A. The versatile biomedical applications of bismuth-based nanoparticles and composites: Therapeutic, diagnostic, biosensing, and regenerative properties. Chem. Soc. Rev. 2020, 49, 1253–1321. [Google Scholar] [CrossRef]
- Song, K.; Du, J.; Wang, X.; Zheng, L.; Ouyang, R.; Li, Y.; Miao, Y.; Zhang, D. Biodegradable Bismuth-Based Nano-Heterojunction for Enhanced Sonodynamic Oncotherapy through Charge Separation Engineering. Adv. Healthc. Mater. 2022, 11, 2102503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Liu, J.; Yu, X.; Deng, Y.; Sun, Y.; Liu, T.; Dong, L.; Zhu, C.; Shen, X.; Zhu, J.; et al. Bismuth-Based Mesoporous Nanoball Carrying Sorafenib for Computed Tomography Imaging and Synergetic Chemoradiotherapy of Hepatocellular Carcinoma. Adv. Healthc. Mater. 2020, 9, e2000650. [Google Scholar] [CrossRef]
- Gan, H.-Y.; Peng, T.-L.; Huang, Y.-M.; Su, K.-H.; Zhao, L.-L.; Yao, L.-Y.; Yang, R.-J. Efficacy of two different dosages of levofloxacin in curing Helicobacter pylori infection: A Prospective, Single-Center, randomized clinical trial. Sci. Rep. 2018, 8, 9045. [Google Scholar] [CrossRef]
- Mcnicholl, A.G.; Gisbert, J.P. Addition of Bismuth to the Standard Triple Therapy for Helicobacter Pylori Infection Reply. Clin. Gastroenterol. Hepatol. 2019, 17, 2822–2823. [Google Scholar] [CrossRef]
- Park, J.Y.; Back, S.H.; Chang, S.-J.; Lee, S.J.; Lee, K.G.; Park, T.J. Dopamine-Assisted Synthesis of Carbon-Coated Silica for PCR Enhancement. ACS Appl. Mater. Interfaces 2015, 7, 15633–15640. [Google Scholar] [CrossRef]
- Sun, C.; Cheng, Y.; Pan, Y.; Yang, J.; Wang, X.; Xia, F. Efficient polymerase chain reaction assisted by metal–organic frameworks. Chem. Sci. 2020, 11, 797–802. [Google Scholar] [CrossRef]
- Vu, B.V.; Litvinov, D.; Willson, R.C. Gold Nanoparticle Effects in Polymerase Chain Reaction: Favoring of Smaller Products by Polymerase Adsorption. Anal. Chem. 2008, 80, 5462–5467. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhu, Y.; Jiang, Y.; Qiao, R.; Zhu, S.; Chen, W.; Xu, C. Effects of Quantum Dots in Polymerase Chain Reaction. J. Phys. Chem. B 2009, 113, 7637–7641. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, F.; Wang, H.; Song, M. Graphene oxide enhances the specificity of the polymerase chain reaction by modifying primer-template matching. Sci. Rep. 2017, 7, 16510. [Google Scholar] [CrossRef]
- Kambli, P.; Kelkar-Mane, V. Nanosized Fe3O4 an efficient PCR yield enhancer—Comparative study with Au, Ag nanoparticles. Colloids Surf. B Biointerfaces 2016, 141, 546–552. [Google Scholar] [CrossRef]
- Lin, Y.; Li, J.; Yao, J.; Liang, Y.; Zhang, J.; Zhou, Q.; Jiang, G. Mechanism of gold nanoparticle induced simultaneously increased PCR efficiency and specificity. Chin. Sci. Bull. 2013, 58, 4593–4601. [Google Scholar] [CrossRef]
- Li, A.; Zhou, B.; Alves, C.S.; Xu, B.; Guo, R.; Shi, X.; Cao, X. Mechanistic Studies of Enhanced PCR Using PEGylated PEI-Entrapped Gold Nanoparticles. ACS Appl. Mater. Interfaces 2016, 8, 25808–25817. [Google Scholar] [CrossRef]
- Nie, L.; Gao, L.; Yan, X.; Wang, T. Functionalized tetrapod-like ZnO nanostructures for plasmid DNA purification, polymerase chain reaction and delivery. Nanotechnology 2007, 18, 015101. [Google Scholar] [CrossRef]
- Yüce, M.; Uysal, E.; Budak, H. Amplification yield enhancement of short DNA templates using bulk and surface-attached amine-functionalized single-wall carbon nanotubes. Appl. Surf. Sci. 2015, 349, 147–155. [Google Scholar] [CrossRef]
- Lou, X.; Zhang, Y. Mechanism Studies on NanoPCR and Applications of Gold Nanoparticles in Genetic Analysis. ACS Appl. Mater. Interfaces 2013, 5, 6276–6284. [Google Scholar] [CrossRef]
- Lestarini, I.A.; Suryani, D.; Sabrina, Y. An Efficient Polymerase Chain Reaction (PCR) Enhancer for Highly Guanine-Cytosine (GC)-Rich DNA Sequences. Bali Med. J. 2019, 8, 415. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, X.; Meng, L.; Liu, F.; Shen, C.; Yang, W. Enhanced amplification of GC-rich DNA with two organic reagents. Biotechniques 2009, 47, 775–779. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, H.S.; Siffert, W.; Frey, U.H. Successful amplification of extremely GC-rich promoter regions using a novel ‘slowdown PCR’ technique. Pharmacogenetics 2003, 13, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Frey, U.H.; Bachmann, H.S.; Peters, J.; Siffert, W. PCR-amplification of GC-rich regions: ‘slowdown PCR’. Nat. Protoc. 2008, 3, 1312–1317. [Google Scholar] [CrossRef]
- Yang, W.; Li, X.; Sun, J.; Shao, Z. Enhanced PCR Amplification of GC-Rich DNA Templates by Gold Nanoparticles. ACS Appl. Mater. Interfaces 2013, 5, 11520–11524. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-Y.; Li, Q.; Yu, Y.-H.; Zhong, M.; Yang, L.; Wu, Q.-H.; Qiu, Y.-R.; Luo, S.-Q. A primer design strategy for PCR amplification of GC-rich DNA sequences. Clin. Biochem. 2011, 44, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Pyle, J.R.; Piecco, K.W.E.S.; Vicente, J.R.; Chen, J. In Situ Sensing of Reactive Oxygen Species on Dye-Stained Single DNA Molecules under Illumination. Langmuir 2019, 35, 11308–11314. [Google Scholar] [CrossRef]
- Hinkle, K.R. Molecular dynamics simulations reveal single-stranded DNA (ssDNA) forms ordered structures upon adsorbing onto single-walled carbon nanotubes (SWCNT). Colloids Surf. B Biointerfaces 2022, 212, 112343. [Google Scholar] [CrossRef]
- Ma, H.; Xu, Z.; Fang, H.; Lei, X. Unexpected sequence adsorption features of polynucleotide ssDNA on graphene oxide. Phys. Chem. Chem. Phys. 2020, 22, 11740–11746. [Google Scholar] [CrossRef]
- Qiu, Q.; Chen, H.; Ying, S.; Sharif, S.; You, Z.; Wang, Y.; Ying, Y. Simultaneous fluorometric determination of the DNAs of Salmonella enterica, Listeria monocytogenes and Vibrio parahemolyticus by using an ultrathin metal-organic framework (type Cu-TCPP). Microchim. Acta 2019, 186, 93. [Google Scholar] [CrossRef]
- Pan, D.; Mi, L.; Huang, Q.; Hu, J.; Fan, C. Genetic analysis with nanoPCR. Integr. Biol. 2012, 4, 1155–1163. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Jung, J.Y.; Kim, D.-H.; Moon, S.; Lee, W.-H.; Chun, B.-W.; Choi, D.-H. DMSO Improves the Ski-Slope Effect in Direct PCR. Appl. Sci. 2021, 11, 1943. [Google Scholar] [CrossRef]
- Varadharajan, B.; Parani, M. DMSO and betaine significantly enhance the PCR amplification of ITS2 DNA barcodes from plants. Genome 2021, 64, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Jurišić, V.; Obradović, J.; Tošić, N.; Pavlović, S.; Kulić, M.; Djordjević, N. Effects of DMSO, glycerol, betaine and their combinations in detecting single nucleotide polymorphisms of epidermal growth factor receptor (EGFR) gene promoter sequence in non-small-cell lung cancer (NSCLC) patients. J. Pharm. Biomed. Anal. 2016, 128, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Hong, Y.; Fan, G. Bismuth Reduces Cisplatin-Induced Nephrotoxicity Via Enhancing Glutathione Conjugation and Vesicular Transport. Front. Pharmacol. 2022, 13, 887876. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Schmitt, C.; Gorgette, O.; Marbouty, M.; Duchateau, M.; Gianetto, Q.G.; Matondo, M.; Guigner, J.-M.; De Reuse, H. Bacterial Membrane Vesicles as a Novel Strategy for Extrusion of Antimicrobial Bismuth Drug in Helicobacter pylori. mBio 2022, 13, e0163322. [Google Scholar] [CrossRef]
- Korkola, N.C.; Hudson, E.; Stillman, M.J. Structurally restricted Bi(III) metallation of apo-βMT1a: Metal-induced tangling. Metallomics 2021, 13, mfab020. [Google Scholar] [CrossRef]
- He, X.; Liao, X.; Li, H.; Xia, W.; Sun, H. Bismuth-Induced Inactivation of Ferric Uptake Regulator from Helicobacter pylori. Inorg. Chem. 2017, 56, 15041–15048. [Google Scholar] [CrossRef]
- Cheek, G.T.; Peña, D.V. Electrochemical Investigations of L-Cysteine Interactions with Bismuth Ions. J. Electrochem. Soc. 2020, 167, 155522. [Google Scholar] [CrossRef]
- Jiang, X.; Li, K.; Xie, B.; Zhou, J. An amplification velocity-controlled PCR device for accurately detecting the initial content of target DNA templates. Chem. Eng. J. 2023, 456, 141123. [Google Scholar] [CrossRef]
- Blaizot, R.; Simon, S.; Ginouves, M.; Prévot, G.; Blanchet, D.; Ravel, C.; Couppie, P.; Demar, M.; Nabet, C. Validation of Swab Sampling and SYBR Green-Based Real-Time PCR for the Diagnosis of Cutaneous Leishmaniasis in French Guiana. J. Clin. Microbiol. 2021, 59, e02218-20. [Google Scholar] [CrossRef]










| Test | Enzyme | DMSO | Glycerol |
|---|---|---|---|
| Test 1 | 1.25U | 3% | 5% |
| Test 2 | 1.25U | 3% | 10% |
| Test 3 | 1.25U | 6% | 5% |
| Test 4 | 1.25U | 6% | 10% |
| Test 5 | 2.50U | 3% | 5% |
| Test 6 | 2.50U | 3% | 10% |
| Test 7 | 2.50U | 6% | 5% |
| Test 8 | 2.50U | 6% | 10% |
| Name | Sequence (5′-3′) | Description |
|---|---|---|
| ASPro4se-FP | GAGCGTTGGCGTCGTGC (17 bp) | Forward primer for GNAS1 promoter |
| GAGCGTTGGCGTCGTGC-Cy5-3’ | ||
| ASPro4as-RP | GAGGAGGAGGGCCGAGGA (18 bp) | Reverse primer for GNAS1 promoter |
| 5’-Cy5-GAGGAGGAGGGCCGAGGA | ||
| ASPro4se-FP-C | GCACGACGCCAACGCTC-BHQ3-3’ | Complementary to ASPro4se-FP |
| ASPro4as-RP-C | 5’-BHQ3-TCCTCGGCCCTCCTCCTC | Complementary to ASPro4as-RP |
| APOE-FP | CCCGGTGGCGGAGGAGACG (19 bp) | Forward primer for APOE gene |
| CCCGGTGGCGGAGGAGACG-Cy5-3’ | ||
| APOE-RP | GTCGCGGCCCTGTTCCACCAG (21 bp) | Reverse primer for APOE gene |
| GTCGCGGCCCTGTTCCACCAG-BHQ3-3’ | ||
| APOE-FP-C | CGTCTCCTCCGCCACCGGG-BHQ3-3’ | Complementary to APOE-FP |
| APOE-RP-C | CTGGTGGAACAGGGCCGCGAC-Cy5-3’ | Complementary to APOE-RP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; Yang, J.; Yue, L.; Shen, B.; Wang, J.; Miao, Y.; Ouyang, R.; Hu, Y. Enhancement Effects and Mechanism Studies of Two Bismuth-Based Materials Assisted by DMSO and Glycerol in GC-Rich PCR. Molecules 2023, 28, 4515. https://doi.org/10.3390/molecules28114515
Yang Z, Yang J, Yue L, Shen B, Wang J, Miao Y, Ouyang R, Hu Y. Enhancement Effects and Mechanism Studies of Two Bismuth-Based Materials Assisted by DMSO and Glycerol in GC-Rich PCR. Molecules. 2023; 28(11):4515. https://doi.org/10.3390/molecules28114515
Chicago/Turabian StyleYang, Zhu, Junlei Yang, Lihuan Yue, Bei Shen, Jing Wang, Yuqing Miao, Ruizhuo Ouyang, and Yihong Hu. 2023. "Enhancement Effects and Mechanism Studies of Two Bismuth-Based Materials Assisted by DMSO and Glycerol in GC-Rich PCR" Molecules 28, no. 11: 4515. https://doi.org/10.3390/molecules28114515
APA StyleYang, Z., Yang, J., Yue, L., Shen, B., Wang, J., Miao, Y., Ouyang, R., & Hu, Y. (2023). Enhancement Effects and Mechanism Studies of Two Bismuth-Based Materials Assisted by DMSO and Glycerol in GC-Rich PCR. Molecules, 28(11), 4515. https://doi.org/10.3390/molecules28114515






